Abstract
African trypanosomes are protected by a densely packed surface monolayer of variant surface glycoprotein (VSG). A haptoglobin–hemoglobin receptor (HpHbR) within this VSG coat mediates heme acquisition. HpHbR is also exploited by the human host to mediate endocytosis of trypanolytic factor (TLF)1 from serum, contributing to innate immunity. Here, the crystal structure of HpHbR from Trypanosoma congolense has been solved, revealing an elongated three α-helical bundle with a small membrane distal head. To understand the receptor in the context of the VSG layer, the dimensions of Trypanosoma brucei HpHbR and VSG have been determined by small-angle X-ray scattering, revealing the receptor to be more elongated than VSG. It is, therefore, likely that the receptor protrudes above the VSG layer and unlikely that the VSG coat can prevent immunoglobulin binding to the receptor. The HpHb-binding site has been mapped by single-residue mutagenesis and surface plasmon resonance. This site is located where it is readily accessible above the VSG layer. A single HbHpR polymorphism unique to human infective T. brucei gambiense has been shown to be sufficient to reduce binding of both HpHb and TLF1, modulating ligand affinity in a delicate balancing act that allows nutrient acquisition but avoids TLF1 uptake.
African trypanosomes infect humans and domestic and game animals, causing disease and placing a large constraint on the agricultural productivity of rural sub-Saharan Africa (1). Infection is transmitted by tsetse flies, and, once established in the mammalian host, the trypanosomes multiply in the bloodstream and tissue spaces. Infection can persist for years because of a population-survival strategy based on autoregulation of parasitaemia and a sophisticated system of antigenic variation that produces novel variants at a frequency sufficient to avoid complete clearance by the immune response (2, 3). This antigenic variation is based on a single protein, the variant surface glycoprotein (VSG). Only one VSG is expressed at any one time and an antigenic switch follows either a gene conversion from the genomic reservoir of VSG genes or an epigenetic switch that activates a VSG gene in a different expression site (4). In addition to its role in antigenic variation, VSG also protects the underlying plasma membrane as it forms a coat that covers the entire external surface with a packing density approaching the maximum possible (5) and sufficient to shield epitopes adjacent to the plasma membrane (6).
Receptors within the VSG coat mediate uptake of large ligands from the host, the two best-characterized being the transferrin receptor for iron (7, 8) and the haptoglobin–hemoglobin receptor (HpHbR) for heme (9). VSG is an elongated homodimer attached to the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor (10, 11), and any receptor must be able to bind ligand in the context of the VSG coat. The structure of the transferrin receptor has not been determined, but there is evidence that it has a GPI-anchor, is structurally related to VSGs (13, 14), and that the ligand-binding site is distal to the plasma membrane (14). Modeling has suggested that this location and the number, size, and position of N-linked oligosaccharides facilitate ligand access (15). The HpHbR shows little apparent sequence similarity to VSGs and is less well characterized but is also linked to the plasma membrane through a C-terminal GPI anchor.
HpHbR also plays a central role in determining whether humans can be infected by trypanosomes. Most African trypanosomes, such as Trypanosoma brucei brucei and Trypanosoma congolense, cannot infect humans because of innate immunity (16). For T. brucei brucei, this is mediated by trypanolytic factors 1 and 2 (TLF1 and TLF2) (17–19). TLFs are characterized by the presence of apolipoprotein (Apo)L1 and haptoglobin-related protein (Hpr) (9, 20). In TLF1, Hpr is complexed with hemoglobin (HprHb) (21), and TLF1 uptake occurs via binding of the HprHb component to the receptor (22–24). After endocytosis of TLF1, ApoL1 is trafficked to the lysosome, where it causes swelling and rupture, resulting in cell death (25–27). Human infective trypanosomes have overcome this innate immunity. In East Africa, T. brucei rhodesiense expresses the serum resistance-associated (SRA) protein (28–30), which binds to and inactivates ApoL1 (23). T. brucei gambiense, the human infective form present in West Africa, does not contain the SRA gene, and it has been proposed that resistance to TLF1 results from reduced uptake caused by reduced expression and sequence polymorphisms in HpHbR (31). Indeed, HpHbR deletion in T. brucei brucei disrupts TLF1 uptake, and expression of the receptor from T. brucei gambiense cannot restore this (31).
Here, the structure of a trypanosome receptor, HpHbR, from T. congolense is reported, and the HpHb-binding site is identified. HpHbR is an elongated three α-helical bundle with a small head structure that is distal to the C-terminal GPI-anchor attachment site. This head structure contains the ligand-binding site. The relative dimensions of HpHbR and VSG suggest that the receptor protrudes above the VSG layer, rendering the binding site accessible to ligand but also making it unlikely that the VSG coat can prevent immunoglobulin binding to the receptor. A single HbHpR polymorphism unique to human infective T. brucei gambiense is sufficient to reduce binding of both HpHb and TLF1, altering ligand affinity in a delicate balancing act that retains nutrient acquisition but avoids uptake of TLF1.
Results
Identification of T. congolense HpHbR.
To investigate the molecular basis for HpHb uptake and resistance to innate immunity, we screened receptors from different African trypanosome species for HpHb binding and the ability to crystallize. Mature T. congolense HpHbR is significantly shorter than T. brucei brucei HpHbR, with the mature protein containing 252 residues compared with 340 (9). The two proteins align with 28% sequence identity from the N terminus, with the additional residues in T. brucei brucei HpHbR found predominantly at the C terminus (Figs. S1 and S2). Both were expressed as recombinant proteins in Escherichia coli and purified. The dissociation constants (KD) for binding of the receptors to immobilized HpHb were measured using surface plasmon resonance (SPR). The values obtained were 1 μM for T. brucei brucei HpHbR and 8 μM for T. congolense HpHbR (Fig. S3), confirming the identification of T. congolense HpHbR as a haptoglobin–hemoglobin receptor. Measurement of the affinity in the reverse orientation, with HpHb binding to immobilized T. brucei brucei HpHbR, gave an apparent KD of 4.5 nM (Fig. S3), in line with the 12–13 nM reported previously (9, 21, 22). The most likely cause of differences in KD values measured in the two orientations is due to the binding of a dimeric HpHb to either one or two monomeric receptors immobilized on the surface. This will result in a complex mixture of monovalent and bivalent binding and cause a reduction in the apparent KD. In contrast, the use of immobilized HpHb produced values for the monovalent interaction because the receptor is monomeric. Indeed, measurement of the binding of T. congolense HpHbR to HpHb using isothermal titration calorimetry gave an affinity in the region of 3 μM and a stoichiometry of two receptors binding to each HpHb complex (Fig. S4), and equilibrium analytical ultracentrifugation confirmed the dimeric nature of HpHb found in a recent crystal structure (32, 33) (Fig. S5).
Structure of T. congolense HpHbR.
Crystals of T. congolense HpHbR grew in the form of hexagonal rods that, after dehydration, diffracted to 1.6-Å resolution. They were of space group P6522, with a single molecule in the asymmetric unit. Phases were obtained using single isomorphous replacement with anomalous signal (SIRAS) with a gold derivative and a 3.0-Å resolution isomorphous native dataset (Table S1), An initial model containing three long α-helices was used as a search model in a 1.6 Å resolution native dataset, allowing a final model containing residues Gly3 to Val247 to be built and refined to this resolution (Table S2). This structure is complete except for four residues at the C terminus (including the GPI-anchor attachment site) and two at the N terminus.
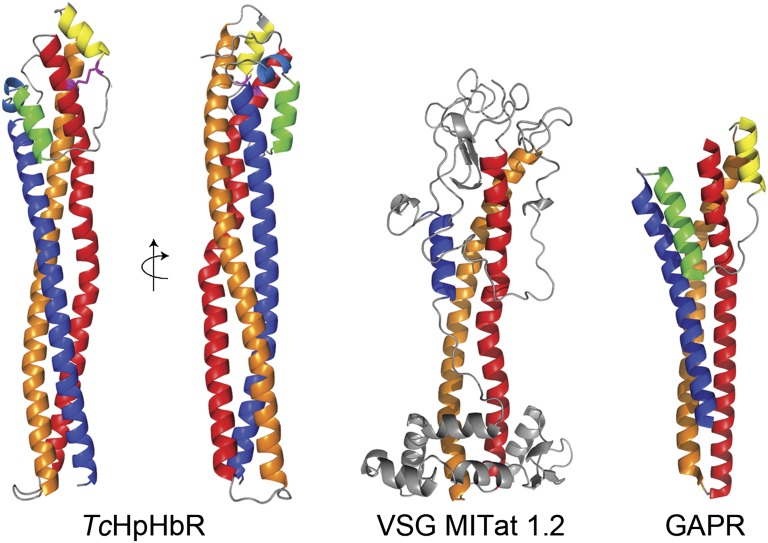
The receptor is monomeric and has an elongated structure, with a total length of 112 Å (Fig. 1). It consists principally of a three-helix bundle: helix I (red; residues 12–74), helix II (orange; residues 82–149), and helix VI (dark blue; residues 192–246). The C-terminal end of helix VI is membrane-proximal, with five residues linking it to the GPI anchor. At the membrane distal end, the receptor widens to form a compact head structure. In addition to the ends of the three major helices, this head includes the N terminus and a 42-residue-long loop that links helices II and VI and contains three small helices: helix III (yellow; residues 153–161), helix IV (green; residues 172–181), and helix V (light blue; resides 183–187). Helices III and IV lie at either side of helix I, making the broadest face of the head. The head structure is stabilized by a core of conserved hydrophobic residues and a single conserved disulphide bond between Cys19 and Cys162, which links helices I and III.
Fig. 1.
Structure of T. congolense HpHbR and the conserved three-helical bundle architecture of trypanosome surface proteins. The structure of T. congolense HpHbR reveals an elongated three-helical bundle with a small head structure at the membrane-distal end. The head contains a single disulphide bond (shown in pale blue). HpHbR shares a three-helical architecture with the N-terminal domain of VSG MITat1.2 (PDB ID code 1VSG) and GARP (PDB ID code 2Y44). Helices I (red), II (orange), and VI (blue) form the core architecture of all three proteins. The single disulphide bond is shown in magenta.
Conserved Three-Helix Motif.
The T. congolense HpHbR structure shows striking similarity to two other trypanosome GPI-anchored surface proteins that have been structurally characterized to date. The N-terminal domains of VSGs have two long helices and a third, partially helical strand that runs back along the helical bundle (10) (Fig. 1). In HpHbR, the first two helices adopt a similar path to the two long helices of the VSGs, and the third helix shares the path of the helical part of the third VSG strand. Similarly, the T. congolense glutamic acid/alanine rich protein (GARP) expressed in the insect procyclic form of the parasite and of unknown function shares the same basic architecture (34) (Fig. 1). This suggests that it is likely that a range of GPI-anchored surface proteins adopt this common, and possibly ancestral, structural theme of a three-helical bundle. The conserved basic architecture remains simple in the monomeric HpHbR and GARP, with the presence of a small head structure the only modification. In contrast, it is highly decorated in dimeric VSGs, with loops and extensions, and even the breakdown of the third helical strand, allowing greater structural diversity to support the antigenic variation of VSG. It seems likely that other trypanosome cell surface proteins will also be built on this basic fold, decorated in different ways to allow the evolution of unique functions.
Elongation of T. brucei brucei HpHbR Relative to T. congolense HpHbR.
The only structurally characterized VSGs come from T. brucei, with separate structures available for both N- and C-terminal domains. The VSG N-terminal domain is ∼100 Å long (10), whereas the C-terminal domains are closer to ∼30 Å (35, 36) in length, consistent with estimates of 120- to 150-Å thickness for the VSG coat from electron micrographs (37). In T. congolense, VSGs do not contain the C-terminal domain(s) present in T. brucei and contain ∼70 fewer residues. A similar difference is observed in HpHbR, where a structure-based sequence alignment shows that an additional 90 residues are located entirely at the C terminus of T. brucei brucei HpHbR (Fig. S6).
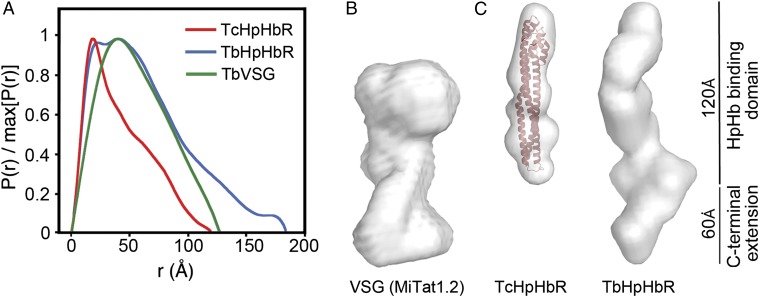
Attempts to obtain crystals of T. brucei brucei HpHbR were unsuccessful, so small-angle X-ray scattering (SAXS) was used to compare T. congolense HpHbR and T. brucei brucei HpHbR with an intact T. brucei VSG (MITat1.2). The radius of gyration, Rg, of VSG was 4.56 nm, and a distance-distribution function indicated a maximum length (Dmax) of 135 Å (Fig. 2 A and B), consistent with predictions from crystal structures and electron microscopy. For the receptors, Rg, was 3.25 nm for T. congolense HpHbR and 5.02 nm for T. brucei brucei HpHbR, indicative of larger dimensions for T. brucei brucei HpHbR (Table S3) while distance distribution functions indicate both to be elongated with a Dmax of 120 Å for T. congolense HpHbR and a longer maximum dimension of 180 Å for T. brucei brucei HpHbR (Fig. 2A). Molecular envelopes were derived from ab initio modeling. The envelope for T. congolense HbHpR (Fig. 2B) has a long axis of 120 Å, comparing well with the 112-Å length of the crystal structure. T. brucei brucei HpHbR is elongated and is significantly longer at ∼180 Å, as a result of the C-terminal extension.
Fig. 2.
T. brucei brucei HpHbR is elongated compared with TcHpHbR. (A) Distance distribution functions derived from SAXS data for VSG MITat1.2 (green), T. congolense HpHbR (red), and T. brucei brucei HpHbR (blue). (B) Molecular envelope derived from SAXS for T. brucei VSG MITat1.2. (C) Molecular envelopes derived from SAXS for T. congolense HpHbR (with the crystal structure docked into the envelope) and T. brucei brucei HpHbR.
These SAXS reconstructions demonstrate that, in T. brucei, the HpHbR is around 60 Å longer than a representative T. brucei VSG, thereby allowing it to reach above the VSG coat. In T. congolense, where VSGs do not contain the C-terminal domain(s) present in T. brucei VSGs, it is also likely that the shorter T. congolense HpHbR protrudes above the VSG layer.
Conserved Residues in the HpHbR Head Are Essential for Ligand Binding.
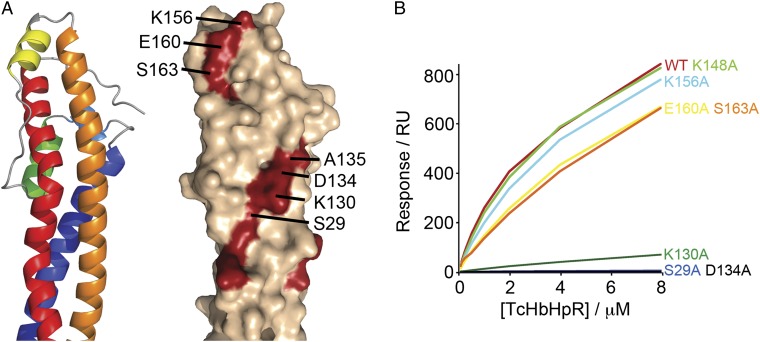
In T. brucei, the likely position of the head structure of HpHbR, protruding above the VSG layer, suggested that it may be the location of a binding site for HpHb, making it readily accessible to this large ligand. Comparison of HpHbRs from T. brucei, T. congolense, and Trypanosoma vivax revealed 34 conserved residues (Fig. S1). Many of these contribute to small hydrophobic cores stabilizing the head structure, whereas the majority of the remainder form two surface-exposed patches (Fig. 3A). At the top, close to the conserved disulphide bond, lies patch A, containing K156 and E160 from helix III and S163 from the subsequent linker. Patch B is ∼25 Å from the top of the head and takes the form of a stripe of residues contributed by helices I and II and containing S29, G33, R36, K130, D134, and A135. A series of alanine-substitution mutants were made for residues in the two patches and binding to HpHb tested by SPR. All of the mutant proteins were indistinguishable from wild type during purification and in circular dichroism spectra, suggesting that the mutations had not affected overall structure. Mutations in patch A (K156A, E160A, or S163A) had little or no effect on HpHb binding. In contrast, mutations in patch B (S29A, K130A, or D134A) reduced, or abolished, binding (Fig. 3B and Table S4).
Fig. 3.
Identification of the HpHb-binding site of T. congolense HpHbR. (A) Location of conserved residues on the surface of T. congolense HpHbR. (B) Binding profiles of T. congolense HpHbR mutants to a HpHb-coated surface. Alanine substitutions in conserved residues localized S29, K130, and D134 to the core of the binding site.
In addition, a mutation, D133A, was made in the T. brucei brucei HpHbR, equivalent to D134A in T. congolense HpHbR, and this also abolished binding, indicating conservation of the binding site (Fig. S7). These data suggest a model in which patch B is central to the HpHb-binding site and the location of this patch toward the top of the receptor permits access to the ligand in the context of the VSG coat.
L175S Polymorphism in Human Infective T. brucei gambiense HpHbR Disrupts TLF-1 Binding.
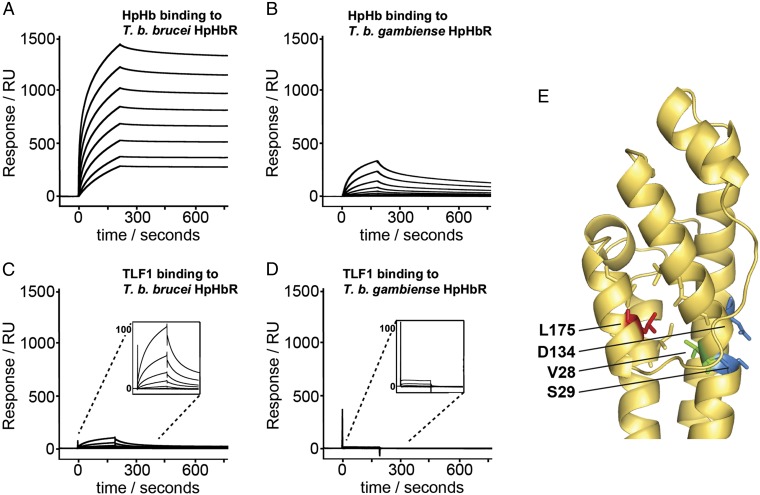
It has been reported that sequence polymorphism in the HpHbR contributes to TLF1 resistance in T. brucei gambiense (31). To identify the residues responsible, the HpHbR gene was sequenced from a range of T. brucei isolates, including both human infective and noninfective genotypes, revealing amino acid sequence polymorphisms at 10 positions (Fig. S8) (31). Only one, L175S, was unique to human infective type 1 T. brucei gambiense. This observation has also been made recently in a study using a much larger sample size (38). To test the effect of this polymorphism, both a T. brucei gambiense HpHbR and a L175S mutant of a T. brucei brucei HpHbR were expressed as a recombinant proteins. The KD values for HpHb were measured as 27 and 30 μM, respectively, compared with 1 μM for wild-type T. brucei brucei HpHbR, revealing a 25- to 30-fold reduction in binding compared with a wild-type T. brucei brucei HpHbR (Fig. S9 and Table S5). These data show that the L175S polymorphism is sufficient to reduce the affinity of a T. brucei brucei HpHbR to that of a T. brucei gambiense HpHbR.
L175 lies on the inner surface of helix IV within the head structure of the receptor, where it interacts with the side chains of L21, L25, V28, L179, I198, and L199, forming part of a small hydrophobic core. This core lies behind the binding site, with V28 at the opposite side of helix I to the conserved binding-site residue S29, mutation of which abolished binding (Fig. 4E). The insertion of a hydrophilic serine side chain will disrupt the hydrophobic core of the head structure, altering the conformation of the binding site, explaining the observed reductions in affinity.
Fig. 4.
Disruption of HpHb and TLF binding in T. brucei gambiense HpHbR by the L175S polymorphism. (A–D) SPR signals after the injection of twofold dilutions of ligand over receptor-coated chip surfaces, from a highest concentration of 1 μM for: HpHb binding to a T. brucei brucei HpHbR-coated surface (A); HpHb binding to a T. brucei gambiense HpHbR coated surface (B); TLF1 binding to a T. brucei brucei HpHbR coated surface (C); and TLF1 binding to a T. brucei gambiense HpHbR coated surface (D). (E) Location of L175 (red) on the structure of T. congolense HpHbR. This residue forms part of a hydrophobic core (including V28 in green) adjacent to the HpHb-binding site (with S29 and D134 in blue).
To determine the effect of the L175S polymorphism on the affinity of HpHbR for TLF1, T. brucei brucei and T. brucei gambiense receptors were coupled to SPR chip surfaces at high densities (7,500 RU). This orientation for the binding assay was chosen because TLF1 is a high-density lipoprotein particle and was unstable when coupled to a chip surface under flow conditions. The binding of HpHb and TLF1 to the immobilized HpHbRs was measured with a maximum ligand concentration of 1 μM.
As described above (Fig. S3), the binding of HpHb to T. brucei brucei HpHbR showed an apparent affinity of around 4.5 nM, because of a complex mixture of monovalent and divalent binding. The binding of HpHb to T. brucei gambiense HpHbR in the same orientation was, again, lower than the binding to T. brucei brucei HpHbR (Fig. 4 A and B), consistent with the lower monovalent KD (Fig. S9).
The binding of TLF1 to T. brucei brucei HpHbR was significantly lower than the binding of HpHb (Fig. 4C). The heterogeneous nature of TLF1, an HDL particle, makes it impossible to obtain a precise KD measurement; however, an approximate fit of the data suggested monovalent binding with an affinity in the region of 5–10 μM. At the concentrations used in these studies, similar to serum concentrations of TLF1 of 0.5 μM (20), no binding was observed to T. brucei gambiense HpHbR (Fig. 4D).
These assays indicate that a single HpHbR binds to both HpHb and TLF1 with affinities in the micromolar range. However, the bivalent binding of HpHb observed on the chip surface is not available to TLF1. The L175S polymorphism, therefore, has greater consequences for reduction of TLF1 binding than for reducing HpHb binding.
Discussion
The VSG coat on the external face of the plasma membrane of African trypanosomes is the best-characterized molecular interface between a protozoan pathogen and its mammalian host. Individual VSGs are selected for diversity in sequence to allow antigenic variation and yet are conserved in structure, presumably to facilitate their function in shielding epitopes close to the plasma membrane and allowing rapid endocytosis and recycling, resulting in antibody clearance. Receptors for host macromolecules, such as HpHbR, lie within the VSG layer, where they allow the trypanosome to take nutrients from the host blood. This paper shows how the structure of HpHbR positions an accessible binding site for a macromolecular ligand in the context of a tightly packed VSG layer and how a polymorphism in the receptor contributes to human infectivity of T. brucei gambiense by reducing ligand affinity to allow HpHb binding while disrupting TLF1 binding.
T. congolense HpHbR is an elongated monomeric receptor consisting of a three-helical bundle with a long axis of 110–120 Å and a GPI-anchor attachment site at the C terminus. The free lateral mobility conferred by the GPI anchor provides a mechanism for bivalent binding of the receptor to a dimeric ligand such as HpHb. Although there is no clear sequence similarity between the receptor and VSGs, this architecture is structurally related, showing that it is an adaptable scaffold for the evolution of diverse functions on a trypanosome cell surface dominated by VSG. The core of the HpHb-binding site is conserved between species and is found in a small head structure some 25 Å from the membrane distal end. An additional C-terminal extension found in T. brucei HpHbR increases the long axis to 180 Å, ensuring that this ligand-binding site is accessible above the VSG layer. This extension of the long axis may be an adaptation to the presence of VSG C-terminal domains that increase the thickness of the VSG layer in T. brucei. The protrusion of HpHbR above the top of the VSG layer contrasts with recent models of the transferrin receptor (15) that suggest that the ligand-binding site is below the top of the VSG layer and that multiple N-linked oligosaccharides keep surrounding VSGs sufficiently distant to permit transferrin binding. The N-linked sites in the HpHbR from both species are located such that they will not occlude access to the ligand-binding site (Fig. S10), but it is possible that they distance the receptor from the VSG.
The small head structure of the HpHbR protrudes beyond the VSG layer. Despite its clear advantages for nutrient uptake, this arrangement breaks the integrity of the cell surface for protection against host immunoglobulins. Why, then, is HpHbR not the basis for immunoglobulin-based attack by the host? The copy number of HpHbR is low (22), and it has been reported to be concentrated in the flagella pocket (9), where it may be protected against immune effectors by an unknown mechanism. Alternatively, HpHbR on the cell surface will have a greater access to ligand and a combination of low copy number and rapidity of movement of any bound immunoglobulin from the cell surface to the endosome may be sufficient to avoid damage by the host immune system. Such rapid endocytosis is characteristic of VSG with bound immunoglobulin, which is subjected to hydrodynamic flow forces and rapidly concentrated in the flagellar pocket (39, 40). Once in the flagellar pocket, it is rapidly endocytosed and the immunoglobulin is degraded (40, 41). It seems likely that ligand- or immunoglobulin-bound HpHbRs are swept up in the same flow. These two models remain to be tested.
Finally, the HpHbR structure rationalizes the reduced uptake of TLF1 in T. brucei gambiense, which is likely to contribute to human infectivity. A single polymorphism within the head structure inserts a serine residue into a small hydrophobic core adjacent to the HpHb-binding site, reducing both HpHb and TLF1 binding. Because the affinity for TLF1 is lower than that of HpHb, this mutation leads to complete disruption of TLF1 binding at concentrations that exceed those found in serum, whereas HpHb binding is still observed. Therefore, the L175S mutation tilts the complex interplay between nutrient and immune effector uptake in favor of the pathogen, allowing it to maintain HbHp uptake while resisting uptake of TLF1, contributing toward the human infectivity of T. brucei gambiense.
Materials and Methods
The T. congolense, T. brucei brucei, and T. brucei gambiense HpHbR genes were amplified from genomic DNA and cloned into a modified pEt15b vector to generate His6-tagged constructs. Mutagenesis was conducted using the QuikChange mutagenesis protocol (Stratagene). Proteins were expressed in E. coli and were purified using metal-affinity and size-exclusion chromatography.
T. congolense HpHbR was crystallized, and data were collected from native crystals, as well as from xenon and gold derivatives, allowing experimental phasing and building of a model.
Binding to HpHb was studied using SPR experiments in which either HpHb or receptors were covalently coupled to a CM5 chip (Biacore), and data were analyzed using BIAevaluation software.
SAXS data were collected from receptors in solution and processed using Primus. Detailed methods are found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank David Staunton for help with biophysical analysis, Wendy Gibson for sharing data on the distribution of the L175S mutation and for genomic DNAs, and Andy Tait for genomic DNAs. This work was supported by Wellcome Trust Project Grant 085256/Z/08/Z (to M.C.) and Wellcome Project Grant 087692/Z/08/Z (to M.K.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. G.C. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4E40).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214943110/-/DCSupplemental.
References
- 1.Shaw A. The economics of African trypanosomiasis. In: Maudlin I, Holmes P, Miles M, editors. The Trypanosomiases. Oxford, UK: CABI Publishing; 2004. pp. 369–402. [Google Scholar]
- 2.Lythgoe KA, Morrison LJ, Read AF, Barry JD. Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc Natl Acad Sci USA. 2007;104(19):8095–8100. doi: 10.1073/pnas.0606206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacGregor P, Savill NJ, Hall D, Matthews KR. Transmission stages dominate trypanosome within-host dynamics during chronic infections. Cell Host Microbe. 2011;9(4):310–318. doi: 10.1016/j.chom.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockdale C, Swiderski MR, Barry JD, McCulloch R. Antigenic variation in Trypanosoma brucei: Joining the DOTs. PLoS Biol. 2008;6(7):e185. doi: 10.1371/journal.pbio.0060185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwede A, Carrington M. Bloodstream form Trypanosome plasma membrane proteins: Antigenic variation and invariant antigens. Parasitology. 2010;137(14):2029–2039. doi: 10.1017/S0031182009992034. [DOI] [PubMed] [Google Scholar]
- 6.Schwede A, Jones N, Engstler M, Carrington M. The VSG C-terminal domain is inaccessible to antibodies on live trypanosomes. Mol Biochem Parasitol. 2011;175(2):201–204. doi: 10.1016/j.molbiopara.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schell D, et al. A transferrin-binding protein of Trypanosoma brucei is encoded by one of the genes in the variant surface glycoprotein gene expression site. EMBO J. 1991;10(5):1061–1066. doi: 10.1002/j.1460-2075.1991.tb08045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steverding D, et al. ESAG 6 and 7 products of Trypanosoma brucei form a transferrin binding protein complex. Eur J Cell Biol. 1994;64(1):78–87. [PubMed] [Google Scholar]
- 9.Vanhollebeke B, et al. A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science. 2008;320(5876):677–681. doi: 10.1126/science.1156296. [DOI] [PubMed] [Google Scholar]
- 10.Blum ML, et al. A structural motif in the variant surface glycoproteins of Trypanosoma brucei. Nature. 1993;362(6421):603–609. doi: 10.1038/362603a0. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson MA, Homans SW, Dwek RA, Rademacher TW. Glycosyl-phosphatidylinositol moiety that anchors Trypanosoma brucei variant surface glycoprotein to the membrane. Science. 1988;239(4841 Pt 1):753–759. doi: 10.1126/science.3340856. [DOI] [PubMed] [Google Scholar]
- 12.Mehlert A, Ferguson MA. Structure of the glycosylphosphatidylinositol anchor of the Trypanosoma brucei transferrin receptor. Mol Biochem Parasitol. 2007;151(2):220–223. doi: 10.1016/j.molbiopara.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Carrington M, Boothroyd J. Implications of conserved structural motifs in disparate trypanosome surface proteins. Mol Biochem Parasitol. 1996;81(2):119–126. doi: 10.1016/0166-6851(96)02706-5. [DOI] [PubMed] [Google Scholar]
- 14.Salmon D, et al. Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. EMBO J. 1997;16(24):7272–7278. doi: 10.1093/emboj/16.24.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehlert A, Wormald MR, Ferguson MA. Modeling of the N-glycosylated transferrin receptor suggests how transferrin binding can occur within the surface coat of Trypanosoma brucei. PLoS Pathog. 2012;8(4):e1002618. doi: 10.1371/journal.ppat.1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laveran A. De l’action du sérum humain sur le trypanosome de Nagana (Tr. brucei) C R Acad Sci. 1902;134:735–739. [Google Scholar]
- 17.Raper J, Nussenzweig V, Tomlinson S. The main lytic factor of Trypanosoma brucei brucei in normal human serum is not high density lipoprotein. J Exp Med. 1996;183(3):1023–1029. doi: 10.1084/jem.183.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajduk SL, et al. Hemoglobin binding plasma protein. J Biol Chem. 1989;264:5210–5219. [Google Scholar]
- 19.Rifkin MR. Identification of the trypanocidal factor in normal human serum: High density lipoprotein. Proc Natl Acad Sci USA. 1978;75(7):3450–3454. doi: 10.1073/pnas.75.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raper J, Fung R, Ghiso J, Nussenzweig V, Tomlinson S. Characterization of a novel trypanosome lytic factor from human serum. Infect Immun. 1999;67(4):1910–1916. doi: 10.1128/iai.67.4.1910-1916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nielsen MJ, et al. Haptoglobin-related protein is a high-affinity hemoglobin-binding plasma protein. Blood. 2006;108(8):2846–2849. doi: 10.1182/blood-2006-05-022327. [DOI] [PubMed] [Google Scholar]
- 22.Drain J, Bishop JR, Hajduk SL. Haptoglobin-related protein mediates trypanosome lytic factor binding to trypanosomes. J Biol Chem. 2001;276(32):30254–30260. doi: 10.1074/jbc.M010198200. [DOI] [PubMed] [Google Scholar]
- 23.Vanhamme L, et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature. 2003;422(6927):83–87. doi: 10.1038/nature01461. [DOI] [PubMed] [Google Scholar]
- 24.Widener J, Nielsen MJ, Shiflett A, Moestrup SK, Hajduk SL. Hemoglobin is a co-factor of human trypanosome lytic factor. PLoS Pathog. 2007;3(9):1250–1261. doi: 10.1371/journal.ppat.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hager KM, et al. Endocytosis of a cytotoxic human high density lipoprotein results in disruption of acidic intracellular vesicles and subsequent killing of African trypanosomes. J Cell Biol. 1994;126(1):155–167. doi: 10.1083/jcb.126.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pérez-Morga D, et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science. 2005;309(5733):469–472. doi: 10.1126/science.1114566. [DOI] [PubMed] [Google Scholar]
- 27.Smith AB, Esko JD, Hajduk SL. Killing of trypanosomes by the human haptoglobin-related protein. Science. 1995;268(5208):284–286. doi: 10.1126/science.7716520. [DOI] [PubMed] [Google Scholar]
- 28.De Greef C, Chimfwembe E, Kihang’a Wabacha J, Bajyana Songa E, Hamers R. Only the serum-resistant bloodstream forms of Trypanosoma brucei rhodesiense express the serum resistance associated (SRA) protein. Ann Soc Belg Med Trop. 1992;72(Suppl 1):13–21. [PubMed] [Google Scholar]
- 29.Welburn SC, et al. Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet. 2001;358(9298):2017–2019. doi: 10.1016/s0140-6736(01)07096-9. [DOI] [PubMed] [Google Scholar]
- 30.Xong HV, et al. A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell. 1998;95(6):839–846. doi: 10.1016/s0092-8674(00)81706-7. [DOI] [PubMed] [Google Scholar]
- 31.Kieft R, et al. Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proc Natl Acad Sci USA. 2010;107(37):16137–16141. doi: 10.1073/pnas.1007074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wejman JC, Hovsepian D, Wall JS, Hainfeld JF, Greer J. Structure and assembly of haptoglobin polymers by electron microscopy. J Mol Biol. 1984;174(2):343–368. doi: 10.1016/0022-2836(84)90342-5. [DOI] [PubMed] [Google Scholar]
- 33.Andersen CB, et al. Structure of the haptoglobin-haemoglobin complex. Nature. 2012;489(7416):456–459. doi: 10.1038/nature11369. [DOI] [PubMed] [Google Scholar]
- 34.Loveless BC, et al. Structural characterization and epitope mapping of the glutamic acid/alanine-rich protein from Trypanosoma congolense: Defining assembly on the parasite cell surface. J Biol Chem. 2011;286(23):20658–20665. doi: 10.1074/jbc.M111.218941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chattopadhyay A, et al. Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2. J Biol Chem. 2005;280(8):7228–7235. doi: 10.1074/jbc.M410787200. [DOI] [PubMed] [Google Scholar]
- 36.Jones NG, et al. Structure of a glycosylphosphatidylinositol-anchored domain from a trypanosome variant surface glycoprotein. J Biol Chem. 2008;283(6):3584–3593. doi: 10.1074/jbc.M706207200. [DOI] [PubMed] [Google Scholar]
- 37.Vickerman K. On the surface coat and flagellar adhesion in trypanosomes. J Cell Sci. 1969;5(1):163–193. doi: 10.1242/jcs.5.1.163. [DOI] [PubMed] [Google Scholar]
- 38.Symula RE, et al. 2012. Trypanosoma brucei gambiense group 1 is distinguished by a unique amino acid substitution in the HpHb receptor implicated in human serum resistance. PLoS Negl Trop Dis 6(7):e1728.
- 39.Engstler M, et al. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131(3):505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 40.Engstler M, et al. Kinetics of endocytosis and recycling of the GPI-anchored variant surface glycoprotein in Trypanosoma brucei. J Cell Sci. 2004;117(Pt 7):1105–1115. doi: 10.1242/jcs.00938. [DOI] [PubMed] [Google Scholar]
- 41.Pal A, Hall BS, Jeffries TR, Field MC. Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. Biochem J. 2003;374(Pt 2):443–451. doi: 10.1042/BJ20030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.