Abstract
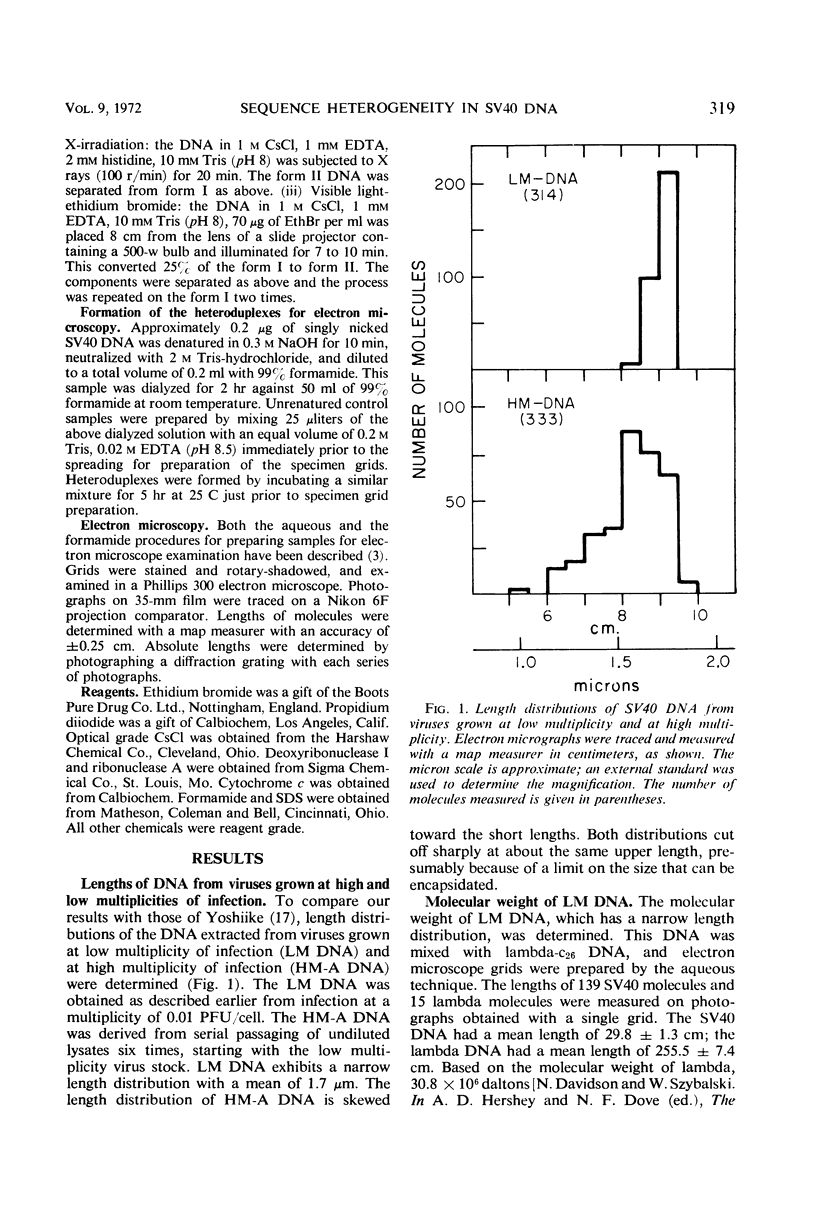
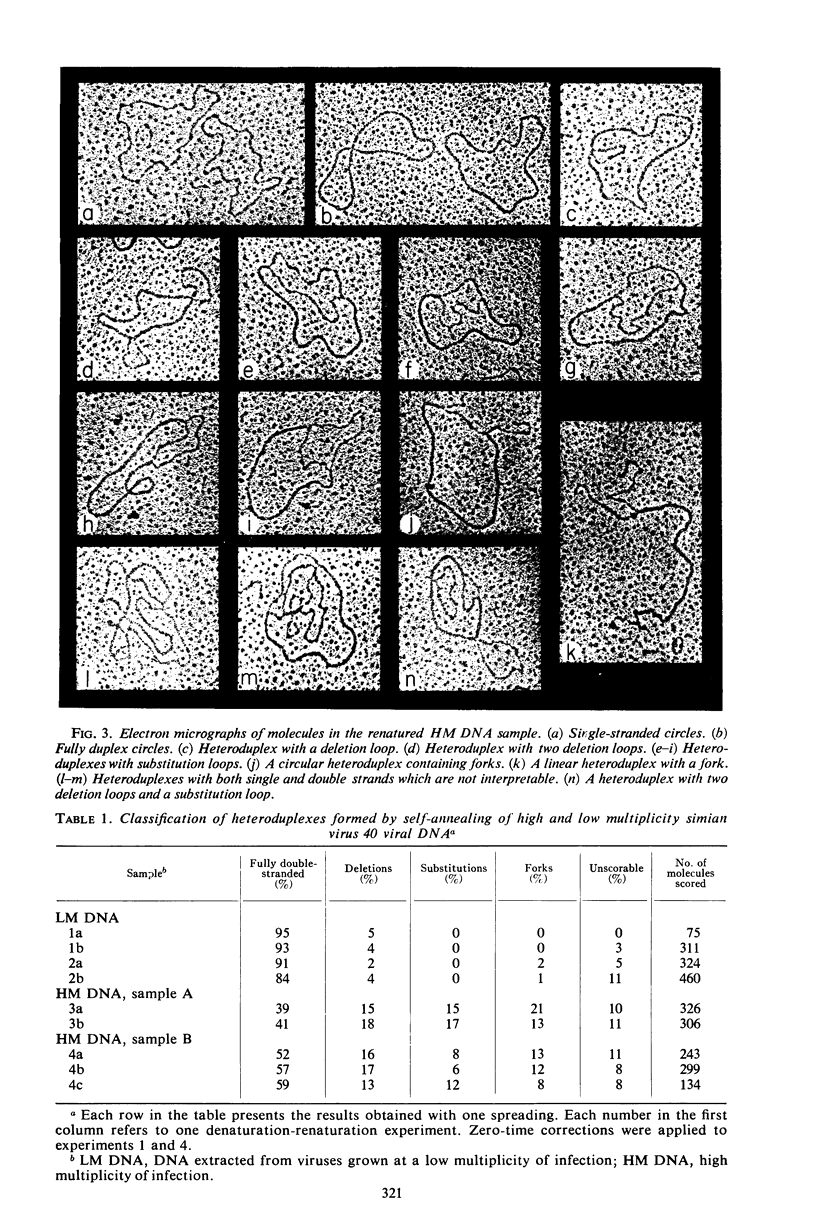
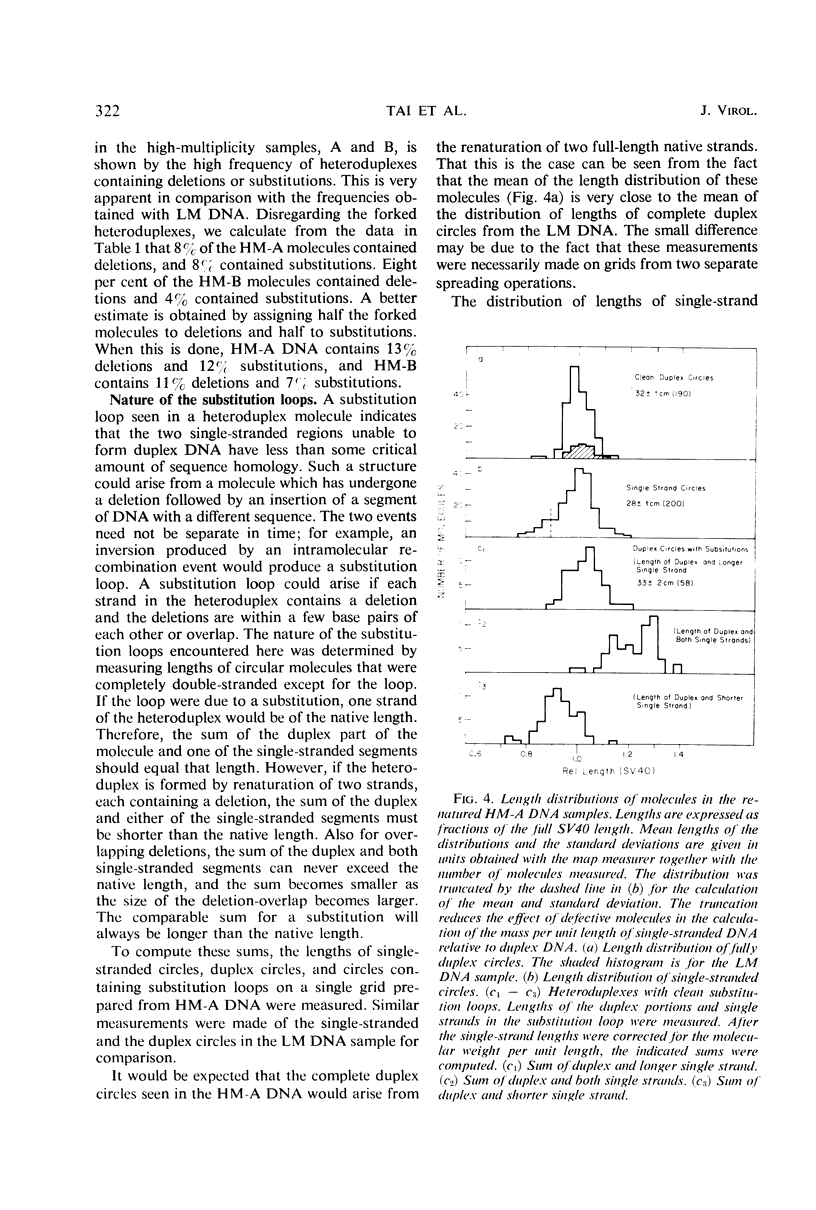
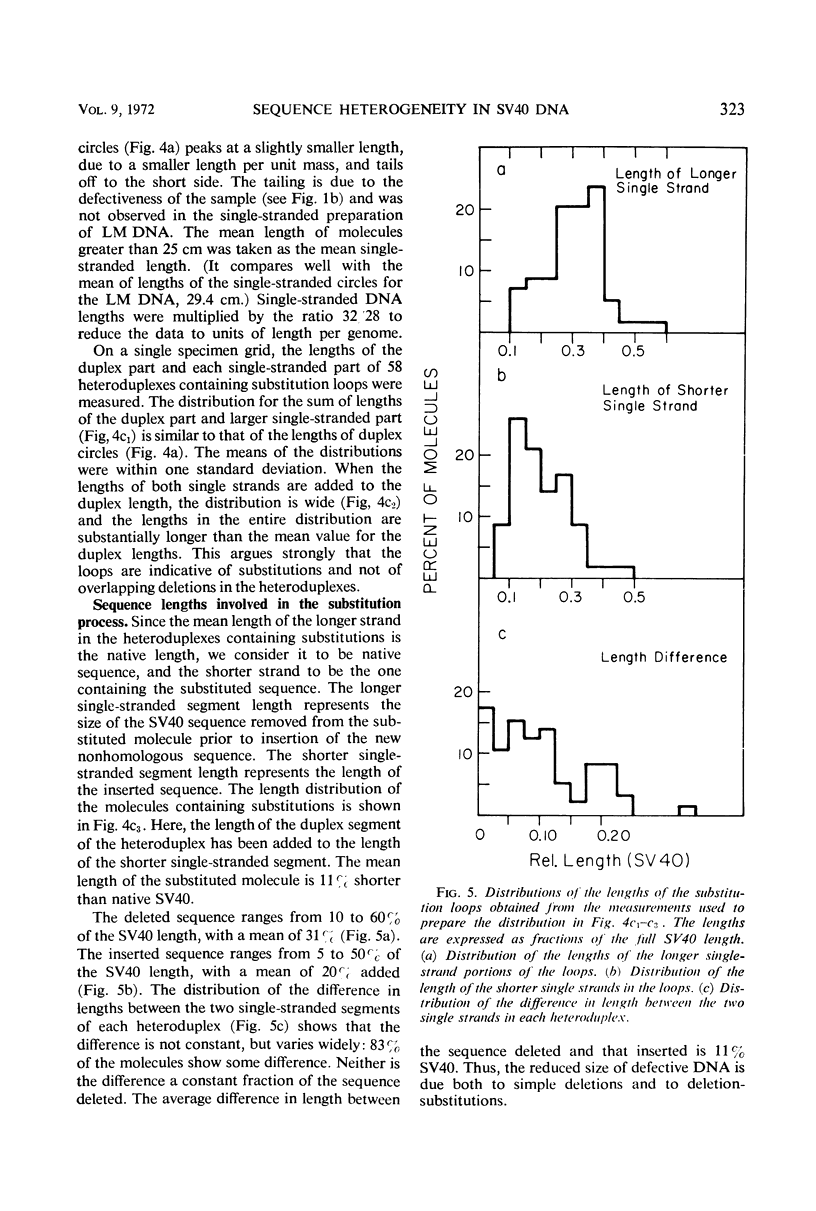
The heteroduplex molecules formed by self-annealing of denatured, singly nicked simian virus 40 (SV40) deoxyribonucleic acid (DNA) prepared from closed viral DNA were examined by formamide-protein film electron microscopy to test the DNA for sequence homogeneity. Sequence inhomogeneity appears in the heteroduplexes as single-strand loops. These result from sequence deletion or from sequence substitution, if regions greater than 50 nucleotides are involved. The undenatured DNA from viruses passaged twice at multiplicities of infection much less than 1 plaque-forming unit (PFU) per cell appeared to be homogeneous in size. The heteroduplexes formed by this DNA indicated that approximately 2% of the molecules carried deletions, but that substitutions were below the level of detection. In contrast, undenatured DNA from viruses grown by passaging undiluted lysates seven times or by infection with stock virus at a multiplicity of infection of 5 PFU per cell contained a large frequency of molecules shorter than the full length. The heteroduplex samples indicated that 12 and 7% of the undenatured material contained base substitutions, and 13 and 11% contained deletions. The deletions and substitutions appear to occur in separate molecules. Length measurements on heteroduplexes displaying the loop characteristic of substitutions have established that these molecules are from true sequence substitutions, and not from adjacent or overlapping deletions. More than 80% of the molecules carrying substitutions are shorter than the native SV40 length. On the average, the substituted sequence is about 20% of the length of SV40, but it replaces a sequence about 30% of the native length. The substituted sequences may be host cell nuclear DNA, possibly arising from integration of SV40 into the chromosome followed by excision of the SV40 DNA together with chromosomal DNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aloni Y., Winocour E., Sachs L., Torten J. Hybridization between SV40 DNA and cellular DNA's. J Mol Biol. 1969 Sep 14;44(2):333–345. doi: 10.1016/0022-2836(69)90179-x. [DOI] [PubMed] [Google Scholar]
- Gelb L. D., Kohne D. E., Martin M. A. Quantitation of Simian virus 40 sequences in African green monkey, mouse and virus-transformed cell genomes. J Mol Biol. 1971 Apr 14;57(1):129–145. doi: 10.1016/0022-2836(71)90123-9. [DOI] [PubMed] [Google Scholar]
- Jensen F. C., Koprowski H. Absence of repressor in SV40-transformed cells. Virology. 1969 Apr;37(4):687–690. doi: 10.1016/0042-6822(69)90290-6. [DOI] [PubMed] [Google Scholar]
- Knowles B. B., Jensen F. C., Steplewski Z., Koprowski H. Rescue of infectious SV40 after fusion between dfferent SV 40-transformed cells. Proc Natl Acad Sci U S A. 1968 Sep;61(1):42–45. doi: 10.1073/pnas.61.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski H., Jensen F. C., Steplewski Z. Activation of production of infectious tumor virus SV40 in heterokaryon cultures. Proc Natl Acad Sci U S A. 1967 Jul;58(1):127–133. doi: 10.1073/pnas.58.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A. J., Teresky A. K. Deoxyribonucleic acid replication in simian virus 40-infected cells. II. Detection and characterization of simian virus 40 pseudovirions. J Virol. 1970 Apr;5(4):451–457. doi: 10.1128/jvi.5.4.451-457.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. A., Axelrod D. SV40 gene activity during lytic infection and in a series of SV40 transformed mouse cells. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1203–1210. doi: 10.1073/pnas.64.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M. R., Hirt B., Weil R. Mouse cellular DNA enclosed in polyoma viral capsids (pseudovirions). Proc Natl Acad Sci U S A. 1967 Oct;58(4):1381–1388. doi: 10.1073/pnas.58.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K., Dulbecco R. Regulation of transcription of the SV40 DNA in productively infected and in transformed cells. Proc Natl Acad Sci U S A. 1968 Jun;60(2):525–532. doi: 10.1073/pnas.60.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich P. R., Black P. H., Weissman S. M. Nucleic acid homology studies of SV 40 virus-transformed and normal hamster cells. Proc Natl Acad Sci U S A. 1966 Jul;56(1):78–85. doi: 10.1073/pnas.56.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush M. G., Eason R., Vinograd J. Identification and properties of complex forms of SV40 DNA isolated from SV40-infected African Green monkey (BSC-1) cells. Biochim Biophys Acta. 1971 Feb 11;228(3):585–594. doi: 10.1016/0005-2787(71)90723-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Westphal H., Srinivasan P. R., Dulbecco R. The integrated state of viral DNA in SV40-transformed cells. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1288–1295. doi: 10.1073/pnas.60.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winocour E. On the apparent homology between DNA from polyoma virus and normal mouse synthetic RNA. Virology. 1967 Jan;31(1):15–28. doi: 10.1016/0042-6822(67)90003-7. [DOI] [PubMed] [Google Scholar]
- Yoshiike K. Studies on DNA from low-density particles of SV40. I. Heterogeneous defective virions produced by successive undiluted passages. Virology. 1968 Mar;34(3):391–401. doi: 10.1016/0042-6822(68)90059-7. [DOI] [PubMed] [Google Scholar]




