Abstract
Viral nanoparticles (VNPs) based on plant viruses such as Cowpea mosaic virus (CPMV) can be used for a broad range of biomedical applications because they present a robust scaffold that allows functionalization by chemical conjugation and genetic modification, thereby offering an efficient drug delivery platform that can target specific cells and tissues. VNPs such as CPMV show natural affinity to cells; however, cellular uptake is inefficient. Here we show that chemical modification of the CPMV surface with a highly reactive, specific and UV-traceable hydrazone linker allows bioconjugation of polyarginine (R5) cell penetrating peptides (CPPs), which can overcome these limitations. The resulting CPMV–R5 particles were taken up into a human cervical cancer cell line (HeLa) more efficiently than native particles. Uptake efficiency was dependent on the density of R5 peptides on the surface of the VNP; particles displaying 40 R5 peptides per CPMV (denoted as CPMV–R5H) interact strongly with the plasma membrane and are taken up into the cells via an energy-dependent mechanism while particles displaying 10 R5 peptides per CPMV (CPMV–R5L) are only slowly taken up. The fate of CPMV–R5 versus native CPMV particles within cells was evaluated in a co-localization time course study. It was indicated that the intracellular localization of CPMV–R5 and CPMV differs; CPMV remains trapped in Lamp-1 positive endolysosomes over long time frames; in contrast, 30–50% of the CPMV–R5 particles transitioned from the endosome into other cellular vesicles or compartments. Our data provide the groundwork for the development of efficient drug delivery formulations based on CPMV–R5.
Introduction
A variety of nanoparticle platform technologies are currently being developed as the basis for image-guided therapies.1 Nanomaterials have many favorable properties that allow us to overcome some of the limitations of current diagnostic reagents and drugs, such as short plasma circulation times, poor solubility in aqueous media and nonspecific biodistribution.2 For example, nanomaterials have a large surface-to-volume ratio compared to traditional delivery vehicles, which offers a greater capacity for the carriage of drugs and/or imaging reagents. Furthermore, nanoparticles containing imaging reagents and/or drugs can be modified with hydrophilic molecules and polymers such as polyethylene glycol to enhance solubility and increase the plasma circulation time, and they can also be conjugated with ligands that target specific cells and tissues.3 Multifunctional delivery platforms can also be assembled by combining the principles described above.4
In the past few years, the potential biomedical applications of nanoparticles have been studied extensively. Nanoparticle formulations must be taken up into target cells efficiently in order for the drug payload to be effective, and many cancer drugs also need to be delivered to specific subcellular compartments (e.g. doxorubicin must be delivered to the nucleus as it intercalates into nuclear DNA, and mitotic inhibitors such as paclitaxel are effective only when they are delivered to mitochondria).5 Cell penetrating peptides (CPPs), also known as protein transduction domains (PTDs), promote translocation across membranes and are therefore potentially valuable in nanomedicine because they allow the efficient uptake of various cargos. Cationic CPPs contain clusters of primarily arginine and lysine residues, e.g. the transcription factor TAT and polyarginine peptides such as the R5 peptide used in this study. CPPs have been used to facilitate the transport of cargos ranging from small imaging molecules such as dyes to macromolecules such as avidin. Efficient cellular delivery has been observed for several CPPs.6
In addition to the delivery of small molecules, peptides and macromolecules, TAT and arginine-rich peptides such as polyarginine have also been used successfully for the delivery of nanoparticles. These range from 2 nm gold nanoparticles to 150 nm liposomes, demonstrating the versatility of CPPs in different nanomedical applications.7 Interactions between cells and CPP-nanoparticles depend predominantly on electrostatic forces, particularly interactions between positively charged peptides on the particle surface and negatively charged proteoglycans such as heparin sulfate, which are densely clustered on the surface of the cell.8 Arginine-rich peptides such as TAT and polyarginine promote cellular uptake of nanoparticles via mechanisms that are under active investigation.7d,9
We used nanoparticles derived from the plant virus Cowpea mosaic virus (CPMV) to evaluate the design and engineering principles needed for an efficient cellular delivery platform. CPMV has been studied extensively in the context of biomedical applications; it can be regarded as the gold standard. However, many other VNP systems are currently being developed for drug delivery and imaging applications. These include Cowpea chlorotic mottle virus (CCMV), Hibiscus chlorotic ringspot virus HCRV), Red clover necrotic mottle virus (RCNMV), and MS2.10 Similarly, nanoparticles based on CPMV have been engineered and developed as vaccines, drug delivery and imaging.11 CPMV interacts with surface-exposed vimentin, a type III filament protein that is predominately expressed in the cytosol of mesenchymal cells but is also found on the surface of endothelial cells and certain cancer cell lines, thus representing a potential tumor marker.12 Although this interaction with vimentin means that CPMV is targeted naturally to cancer cells, surface-vimentin expression on cells is low, which means that vimentin-mediated cellular uptake of CPMV is inefficient.13
To overcome these limitations, we developed a strategy to decorate CPMV particles with CPPs. Surprisingly, despite the nearly ubiquitous use of CPPs as cellular delivery agents, their use with viral nanoparticles has not been extensively investigated.7f First, we established optimized hydrazone ligation reactions on the CPMV surface by labeling the VNP coat protein with benzaldehydes and coupling with a hydrazinopyridine-modified peptide conjugate. The resulting bisarylhydrazone bond is UV-traceable and therefore allows in situ and real time quantitation of labeling efficiency. We analyzed the attachment of cationic arginine-rich CPPs to the anionic virus surface, evaluating the resulting particles in tissue culture by confocal microscopy and flow cytometry.
Results and discussion
Bioconjugation using hydrazone chemistry
CPMV particles were purified from infected black-eyed pea plants yielding 0.3–0.6 mg CPMV per 1 g infected leaves. The purity of the virus was confirmed based on the A260 : A280 ratio (1.8–1.9), which indicated a pure virus preparation based on the RNA:protein ratio. Particle integrity was also confirmed by size-exclusion chromatography (SEC).
Hydrazone chemistry has recently been applied to the CPMV platform and was shown to be a versatile strategy allowing the decoration of CPMV with targeting ligands specific for vascular endothelial growth factor receptor-1.11f We optimized the ligation reaction further using the catalyst aniline, which accelerates the rate of hydrazone bond formation by two orders of magnitude, allowing the reaction to proceed rapidly even at neutral and basic pH14 (the optimum of pH for hydrazone chemistry is 4.5).
The ligation of peptides to the particle surface via hydrazone bonds was carried out using a two-step protocol. First, we used N-hydroxysuccinimide chemistry to modify lysine side chains exposed on the particle surface with 4-formylbenzamide (4FB).15 Second, the aldehyde functional group of the peptide was conjugated to the resulting hydrazide to form a stable hydrazone bond (Fig. 1). The efficiency of 4FB decoration was determined using the aromatic 2-hydrazinopyridine-dihydrochloride hydrazine, which reacts specifically with 4FB-modified groups to form a UV-traceable and quantifiable hydrazone bond. The 4FB/ particle ratio was 280 ± 20 4FB groups per CPMV as determined by UV/vis spectrophotometry using the molar extinction coefficient ε350 nm = 18 000M−1 cm−1. CPMV displays 300 addressable lysine side chains;15 thus, maximum coverage with aldehyde groups was achieved. The reactivity of the 4FB group and the optimal aniline concentration were determined using FLAG-tag peptide. At neutral pH, hydrazinopyridines react readily with benzaldehyde groups. With no aniline present, up to 80 hydrazone bonds were formed after 2 h and up to 240 bonds were formed after 24 h. When aniline was added to a final concentration of either 10 mM or 100 mM, the maximum coverage of all 300 addressable benzaldehyde groups was achieved after just 1 h. (The number of hydrazone bonds per particle was determined in situ and in real time by UV/vis spectrometry using the absorption coefficient of the hydrazone bond (ε354 nm = 29 000 M−1 cm−1) and the molar extinction coefficient of CPMV (ε260 nm = 8.1 g−1 L cm−1) (Fig. S1†).) The yields were comparable to the labeling efficiencies achieved using the more common Cu-catalyzed azide–alkyne cycloaddition (click chemistry) protocols.16 However, an advantage of the hydrazone protocol is that the number of labels can be determined in situ in real time.14a
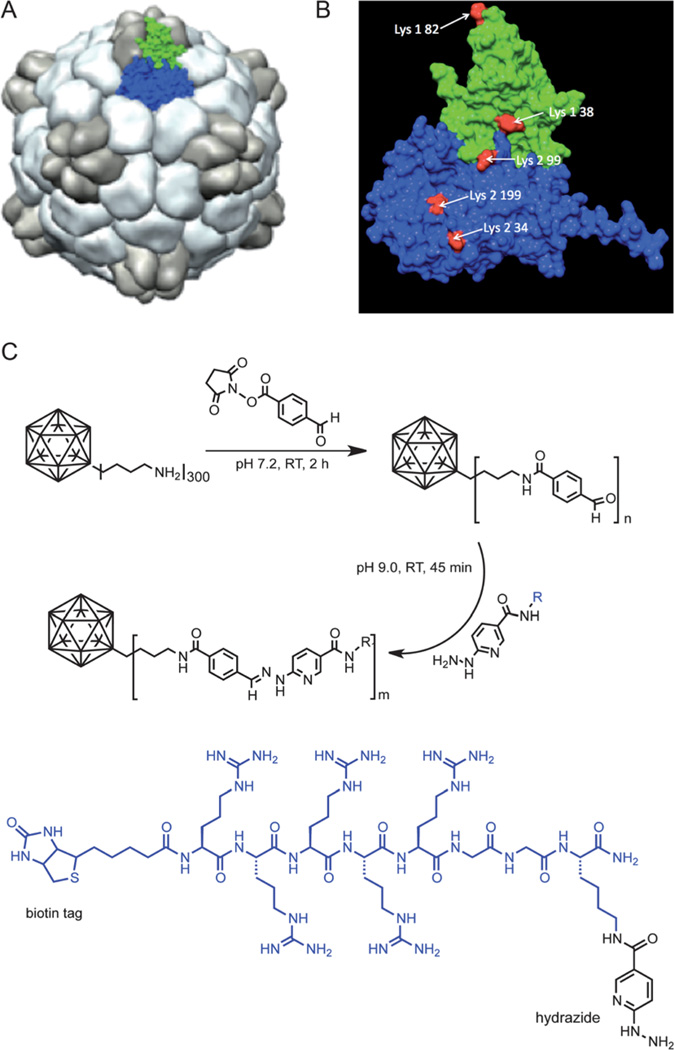
Fig. 1.
(A) Structure of CPMV; surface rendered model highlighting the asymmetric unit consisting of the S protein (green) and L protein (blue). (B) CPMV asymmetric unit; reactive Lys side chains are highlighted in red. (C) Bioconjugation of CPMV with 4FB followed by reaction with R5 peptide.
Attachment of the R5 peptide
Conjugation of the positively charged R5 peptide to the overall negatively charged CPMV particles was found more challenging. We initially reacted CPMV–4FB at 1 mg mL−1 with the R5 peptide using a 2 : 1 molar excess of hydrazinopyridine groups over 4FB groups, 10 mM aniline and incubation for 2 h, in 0.1 M phosphate buffer at pH 7.0. Visible precipitation of the particles occurred after 15 min and only 8% of the sample could be recovered after filtration. UV/vis spectrometry indicated successful hydrazone bond formation (peak = 354 nm) but also particle aggregation (elevated baseline), reflecting bridging interactions between the positively charged peptides and the CPMV particles, which have a net negative surface charge (detailed structural information on CPMV can be found at http://www.viperdb.scripps.edu). Consistent with a surface neutralization cause of aggregation, we found that mixing native CPMV particles with the R5 peptide (non-covalent) also resulted in the formation of aggregates.
We explored a range of alternative protocols to optimize the solubility of the VNPs during conjugation of positively charged peptides: (1) a 4-PEG spacer (PFB linker from Solulink) was introduced to shield the particle surface and extend the peptide away from the surface; (2) varying peptide/CPMV ratios and concentrations were tested; (3) varying incubation times were evaluated; and (4) a range of buffers with varying ionic strengths and pH values were explored. Details of the conditions we tested are provided in the ESI (Table S1†). We found that adjustment of the buffer, pH and salt concentration (0.1 M Tris, 10 mM NaCl, pH 9.0) enabled the production of soluble CPMV conjugates displaying up to 40 R5 peptides per particle. Adjustment of the reaction buffer to increase the pH and adding salt significantly decelerated the charge-dependent aggregation process, thereby allowing controlled conjugation of the R5 peptide to CPMV. By varying the excess of R5 peptide or linker used the density of peptides per CPMV could be controlled. Two formulations were made and analyzed: CPMV–R5H denotes CPMV displaying 40 R5 peptides (H = high density) and CPMV–R5L particles with only 10 R5 peptides per particle (L = low density) (Fig. 2).
Fig. 2.
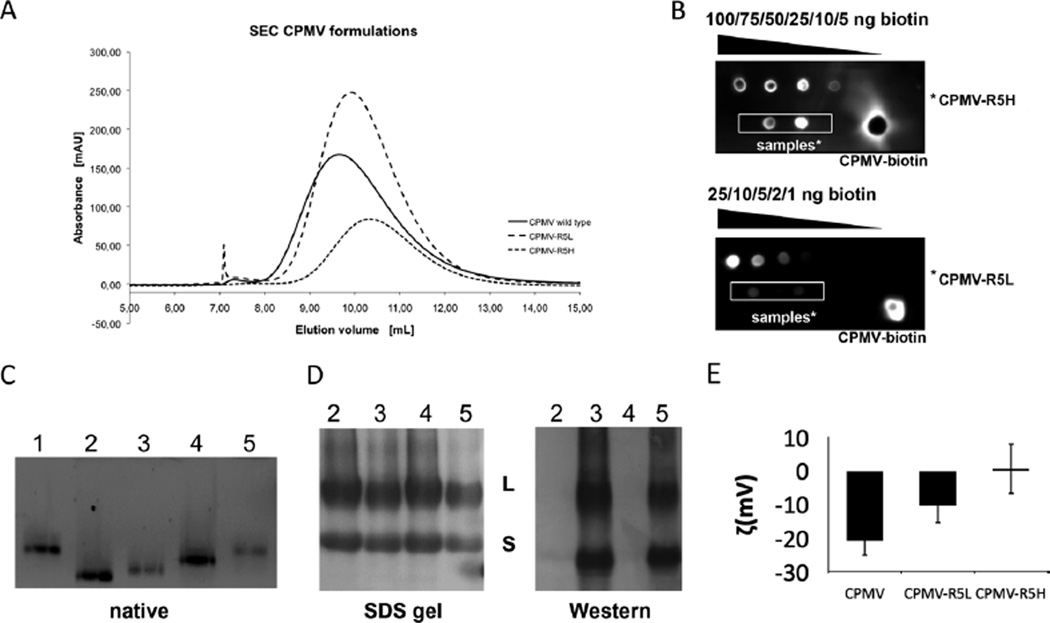
Characterization of CPMV labeling with the biotinylated R5 peptide. (A) Size exclusion chromatography of wild-type CPMV, CPMV–R5L and CPMV–R5Hat 280 nm. (B) ECL dot blot of purified CPMV particles. The number of biotin labels per particle was determined using standardized biotin concentrations and Chemidoc XRS software. (C) Native gel electrophoresis of intact CPMV particles (10 µg) using a 0.8% (w/v) agarose gel. Particles were visualized under UV light. Lane 1 = CPMV, 2 = CPMV–4FB, 3 = CPMV–R5H, 4 = CPMV–PFB, 5 = CPMV–R5L. (D) SDS–PAGE of CPMV particles (10 µg) using a 4–12% Bis-Tris gel and western blotting using streptavidin–alkaline phosphatase to detect the N-terminal biotin tag of the R5 peptide. (E) Zeta potential of CPMV wild type, CPMV–R5L and CPMV–R5H formulations.
The integrity of CPMV–R5L and CPMV–R5H was confirmed using SEC (A260 : A280 = 1.8–1.9) (Fig. 2A). CPMV–R5L/H eluted from the column at a slightly larger elution volume compared to native CPMV; this may indicate that the R5 peptide interacts with the column material, thus slowing down the migration of the particles. We found that the CPMV–R5L/H formulation remained structurally sound over at least one month when stored in 0.1 M potassium phosphate buffer at pH 7.0.
The labeling efficiency was quantified using the R5-biotin tag followed by quantitative electrochemiluminescence (ECL) dot blot analysis, confirming the presence of 40 attached peptides for CPMV–R5H and 10 for CPMV–R5L (Fig. 2B). Conjugation was also supported by SDS gel electrophoresis and western blotting (Fig. 2D). Both the L and S subunits of the CPMV coat protein were successfully labeled. Further characterization by agarose gel electrophoresis (Fig. 2C) indicated that the R5-modified CPMV particles have a slightly retarded mobility compared to CPMV–4FB, confirming the addition of the cationic peptide. The mobility toward the anode increases upon introduction of the non-charged 4FB group to surface lysine side chains, and then decreases upon labeling with positively R5 peptides. Zeta potential measurements indicated that CPMV–R5H particles had a neutral to positive zeta potential value 0.60 mV, Fig. 2E), whereas CPMV–R5L particles had a negative zeta potential value (−10.47 mV, Fig. 2E). As expected, the zeta potential values of native CPMV particles were even lower −20.77 mV, Fig. 2E). In agreement with the UV and ECL data, the zeta potential measurements correlated with the number of R5 peptides conjugated to the surface of the particles.
Cellular uptake of CPMV–R5 formulations
The efficiency of CPMV–R5 particle internalization (and the impact of peptide density on this process) was determined using a combination of confocal microscopy and flow cytometry to study the uptake of native CPMV, CPMV–R5L and CPMV–R5H particles into HeLa cells, a human cervical cancer cell line. HeLa cells are an ideal model for this analysis because previous studies have shown that CPMV binds to vimentin displayed on the surface of HeLa cells and is then taken up by endocytosis.13 The efficiency of particle internalization was determined by fluorescence-activated cell sorting (FACS), which showed that CPMV–R5H particles bound and/or internalized eight times more efficiently than native CPMV. There was only a marginal difference in efficiency between CPMV–R5L and wild type CPMV particles (Fig. 3A), thus indicating that the ratio of the CPP per nanoparticle carrier is relevant and uptake is more efficient at higher peptide densities (10 vs. 40 peptides per VNP).
Fig. 3.
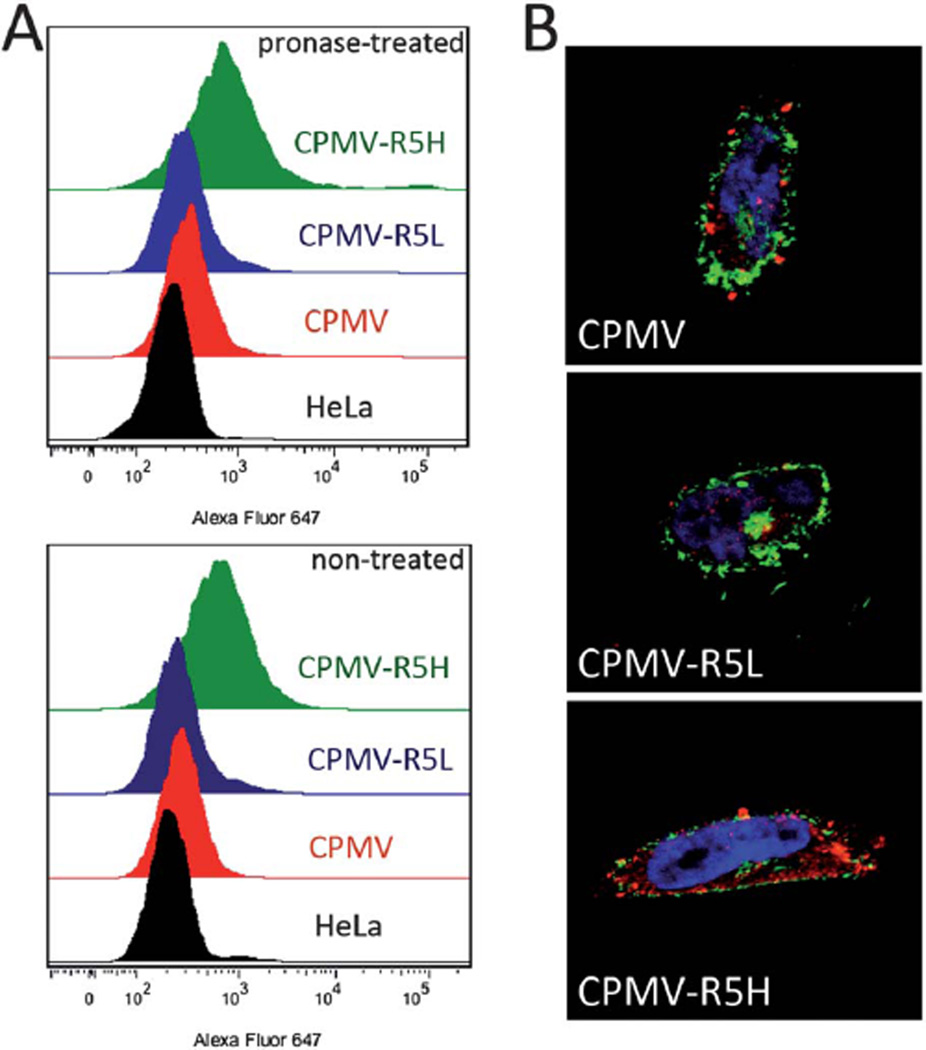
Uptake of VNPs into human cervical cancer cells (HeLa). (A) Evaluation of particle uptake by flow cytometry. Cells were either untreated or treated with 0.1% (w/v) pronase for 3 h. Experiments were conducted using 105 CPMV particles per cell and repeated at least twice; triplicate samples were analyzed, such that 10 000 events were recorded. (B) Fluorescence confocal microscopy of HeLa cells and CPMV formulations (red). Cell membrane is labeled with wheat germ agglutinin (green), and nucleus is labeled with DAPI (blue). Single plane images were analyzed using ImageJ.
The ratio of surface-bound and internalized VNPs was determined by treating cells previously exposed to CPMV and CPMV–R5 formulations with the enzyme pronase (a proteolytic enzyme and alternative to trypsin17), to remove particles bound to the external membrane surface. The fluorescence signal from treated and untreated samples was comparable in both formulations, suggesting that CPMV nanoparticles and R5-conjugates were internalized. Images obtained by confocal microscopy further supported that the CPMV conjugates were indeed taken up by the cells (Fig. 3B). Together these studies suggest that the CPMV–R5 particles are primarily internalized into cells. It should be noted that using native CPMV only about 10% of the cells showed a signal; this is consistent with findings by Koudelka et al., who showed that CPMV uptake in mammalian cells is dependent on surface vimentin interaction and that CPMV uptake into HeLa occurs in 6% of the cell population.12 This suggests that CPMV–R5 formulations use a vimentin-independent internalization mechanism.
The energy dependency of VNP internalization
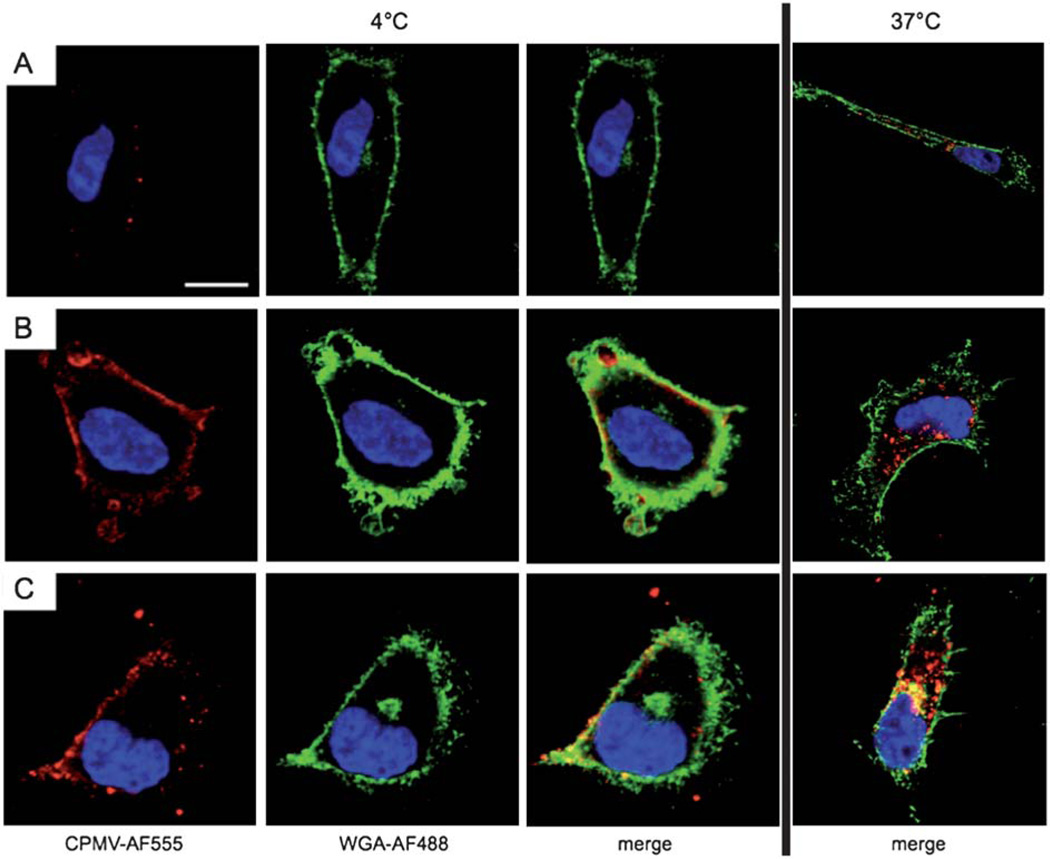
To dissect the mechanism of CPMV–R5 particles in more detail, we carried out energy-dependency and co-localization studies. To determine whether the uptake of CPMV, CPMV–R5L and CPMV–R5H relies on energy-dependent internalization pathways, HeLa cells were incubated with CPMV–R5 particles at 4 °C for 3 h and compared to control cells incubated at 37 °C for the same duration. The cells were then fixed, stained and analyzed by confocal microscopy. Lowering the temperature in this manner did not prevent any of the three formulations interacting with the cell membrane, but none of the VNPs were internalized, indicating that uptake is indeed energy-dependent.
Wild type CPMV particles were shown to be distributed in isolated patches across the cell surface (Fig. 4A). The patchy membrane binding of wild type CPMV to the cell membrane can be explained by the polarized expression pattern of surface vimentin on HeLa cells as previously reported.12,13 In contrast, CPMV–R5 particles were uniformly and ubiquitously distributed, indicating strong interactions with abundant anionic surface proteins, heparin, and phospholipids (Fig. 4B and C). It is interesting to note that CPMV–R5L and CPMV–R5H both interact strongly with cell membranes, but only CPMV–R5H particles are taken up efficiently (Fig. 4). These data taken together indicate that the presentation of R5 peptides on the CPMV surface can enhance binding to the cell surface, and CPMV–R5H particles with their abundant cationic ligands bind and internalize the most efficiently. CPMV particles with and without CPPs were unable to be internalized into the cell at 4 °C. Temperature-dependent ATP depletion inhibits all endocytotic pathways (clathrin-mediated endocytosis, caveolin-mediated endocytosis, macropinocytosis, and phagocytosis), and this suggests that native CPMV and the CPMV–R5 formulations are reliant on endocytosis for internalization. Our data support the conclusions of earlier studies involving a range of CPP-conjugated nanoparticles, such as TAT quantum dots and gold particles,7d,9 and CPP-conjugated macromolecules such as TAT– streptavidin.18 In all cases, internalization relied on energy-dependent endocytosis mechanisms.
Fig. 4.
Temperature/energy-dependent uptake of CPMV formulations. Confocal microscopy of HeLa cells and CPMV formulations (red). The cell membrane is labeled with wheat germ agglutinin (green), and the nucleus is labeled with DAPI (blue). Scale bar = 10 µm. Cells were incubated for 3 h with 106 VNPs per cell at 4 °C (left) or 37 °C (right). (A) CPMV, (B) CPMV–R5L, (C) CPMV–R5H. Imaging was performed using a Biorad 2100 confocal microscope with a 60× oil objective. Images were analyzed using ImageJ.
Cellular localization of CPMV and CPMV–R5 formulations
The internalization mechanism and pathway for nanoparticles tagged with CPPs are highly controversial, with different studies producing inconsistent and conflicting data. These studies implicate different endocytic pathways depending on many factors e.g. cargo size,19 cargo charge,20 peptide density21 and cell line.22 Studying the fate of nanoparticles in cells also remains technologically challenging.
We sought to determine the intracellular fate of each VNP formulation. This is an important challenge in the field of platform design because many therapeutic compounds including cytotoxic drugs and siRNAs are only active when they accumulate in the appropriate subcellular compartments.7f The fate of CPMV–R5 following internalization therefore affects the potential therapeutic efficacy of any drug compound conjugated to or encapsulated within it. We analyzed the subcellular fate of the different VNP formulations by carrying out confocal microscopy co-localization studies using the following specific intracellular markers: (1) transferrin, a marker of clathrin-mediated endocytosis; (2) the early endosomal marker EEA1; and (iii) the lysosomal marker Lamp-1. Previous studies have shown that native CPMV particles become trapped in the endolysosomal compartments and co-localize with Lamp-1.23 Our experiments showed that the VNPs were not co-localized with either EEA1 or transferrin (Fig. S2†), but provided clear evidence for co-localization with Lamp-1 (Fig. 5). Previous studies on CPP-mediated cell delivery suggest that arginine-rich peptides internalize via lipid rafts but not via clathrin-dependent endocytosis,7h which is in good agreement with our observation that CPMV–R5 particles do not co-localize with transferrin.
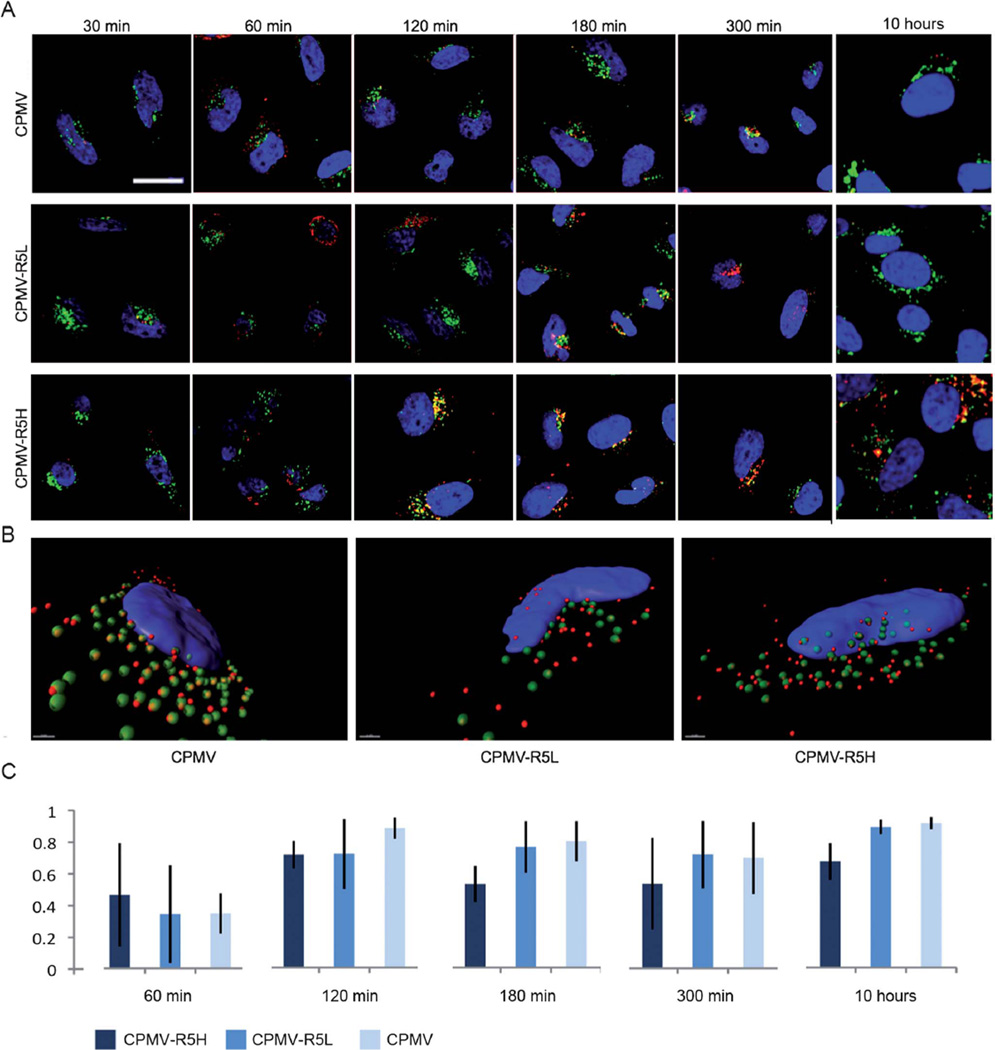
Fig. 5.
Subcellular fate of CPMV formulations in HeLa cells studied by confocal microscopy. CPMV formulations (red), endolysosomes are labeled with Lamp-1 (green), and nuclei are labeled with DAPI (blue). The overlay of endolysosomes and VNP signals is shown in yellow. (A) Time course and translocation of CPMV formulation over 10 hours. Scale bar = 10 µm. Images were analyzed using ImageJ. (B) Three-dimensional reconstruction of z-sectional data at time point 120 min. Images were recorded at a step size of 0.3 µm. Data were reconstructed using Imaris software. Scale bar = 1 µm. (C) Colocalization analysis of eight representative cells for each CPMV formulation showing the percentage of total internalized CPMV particles colocalizing with the endolyosomes, error bars represent the averages values of standard deviations of all analyzed cells. Co-localization analysis was performed using LSM Examiner software.
It is unclear which specific endocytic pathways are responsible for the uptake of CPMV, and multiple pathways may be involved. Clathrin-coated vesicles, caveosomes and macropinosomes can fuse with early endosomes for endosomal sorting. As endocytic compartments mature, a number of proteins in addition to EEA1 are recruited in a time-dependent manner.24 Because early endosomes are not the final cellular destination of the particles, the minimal co-localization with EEA1 may indicate that the particles pass through early endosomes rapidly.
Native CPMV particles have been shown to co-localize with Lamp-1 over long time frames,23 and our data confirmed that both CPMV and CPMV–R5L behaved in the same manner; CPMV–R5H showed faster internalization kinetics (Fig. 5). Z-stacked images were reconstructed and a detailed three-dimensional image analysis was performed using ImageJ and Imaris software. Statistical analysis using Student’s t-distribution test showed that 70% of the CPMV–R5H and CPMV–R5L particles and 90% of wild type CPMV particles (p < 0.05) co-localized with Lamp-1 (t = 120 min). The differences between the formulations were found to be statistically significant (fewer CPMV–R5 particles co-localized with the endolysosomal compartment). This phenomenon became more apparent at later time points. At t = 180 min and 300 min post-inoculation, only 50% of CPMV–R5H particles co-localized with Lamp-1 (Fig. 5C). Further characterization of CPMV–R5 translocation at late time points (t = 10 h post-inoculation) showed similar results. Significantly more CPMV–R5H particles were internalized compared to CPMV–R5L and native CPMV, and a lower proportion (20% fewer) of these particles were retained within the endolysosomes. The hazy signal of CPMV–R5H in near proximity to and around the endolysosomes may suggest endolyosomal escape; this is particularly apparent at late time points (Fig. 5A, CPMV–R5, 10 hours).
In summary, our data indicate that CPMV–R5H conjugates are efficiently taken up by HeLa cells via an energy-dependent mechanism, co-localize at least partially with Lamp-1 marker of endolysosomes, and are detectable within cells over long time periods. The intracellular fate of CPMV–R5H differs compared to native CPMV. 90% of native CPMV particles appear trapped within Lamp-1 endolysosomes, uptake is inefficient and only negligible amounts of particles are detectable only 10 hours post-inoculation.
It is understood that CPPs promote efficient cellular uptake via electrostatic binding to the negatively charged cell membrane. A combination of cellular entry mechanisms may be involved depending on the cell line, carrier system, and CPP used. Some studies have indicated endolysosomal escape mechanisms and translocation of the cargo into the cytoplasm.6 However, the study of these phenomena remains technologically challenging; fixatives such as methanol or paraformaldehyde can lead to artifacts.25 Further, the disappearance of CPP-delivered nanoparticles from one particular vesicle type (here Lamp-1 stained endolysosomes) does not exclude the probability that the nanoparticles could have been sorted into another type of vesicle (rather than translocated into the cytoplasm). However, the fluorescence signal of CPMV–R5H appears to be more diffused compared to native CPMV (Fig. 3 and 5); the hazy signal around the endolysosomes at late time points may indeed indicate partial endolyosomal escape. 3D image analysis indicated that a lower proportion (20% fewer) of CPMV–R5H were retained within the endolysosomes. At longer times following VNP addition, the fluorescence in the cells does become more diffuse which is consistent with some cytoplasmic delivery of the VNP (Fig. 5).
Studying of the fate of CPMV formulations in cells remains challenging. We explored the use of pH-sensitive dyes to gain further insights into the intracellular localization of CPMV, but the resulting data were inconclusive. Co-localization studies using Lamp-1 and confocal image analysis indicate that: (1) CPMV–R5H particles enter cells with high efficiency; (2) CPMV–R5H particles may enter the cell using an alternative internalization pathway that bypasses the endolysosomes such as caveolae-mediated endocytosis or macropinocytosis24 and reside in Lamp-1 negative vesicles; and (3) some CPMV–R5H particles may translocate into the cytoplasm or vesicles that do not contain Lamp-1. The observation that the particles reside inside the cells over long time frames in high concentration is interesting and may provide a platform for sustained release drug delivery system.
Conclusions
Protein-based nanoparticles such as CPMV have many properties that are beneficial for image-guided therapy, i.e. they are biocompatible, non-infectious in humans and extremely robust, and their structure is easily controlled by both chemical conjugation and genetic engineering. These features allow the creation of multifunctional platforms that combine targeting with sensitive imaging reagents and/or efficacious drugs for the treatment of cancer. A key challenge in the field of nanoparticle platform technology development is the control of intracellular fate once the particle has been taken up into the target cell. To address this challenge and design VNP formulations for efficient intracellular delivery, we made use of a CPP carrying a strong positive charge (the R5 peptide) by establishing hydrazone ligation chemistry as a simple and straightforward bioconjugation method. After careful optimization, this allowed CPMV particles to be loaded with up to 40 R5 peptides. Detailed in vitro studies using flow cytometry and confocal microscopy showed that VNPs with densely arrayed cationic peptides promoted intracellular targeting, increasing the number of particles taken up into HeLa cells by endocytosis. Quantitation of the colocalization of CPMV with endolysosomal markers suggests that the CPMV–R5H particles are escaping the endolysosomes, consistent with delivery to other cellular compartments or the cytoplasm. Our data provide the basis for the development of efficient drug delivery formulations that can be targeted to specific intracellular compartments.
Materials and methods
Peptide synthesis
The biotinylated 6-hydrazinopyridyl FLAG peptide was synthesized on a Waters Delta Prep 4000 preparative chromatography system using a Phenomenex Jupiter 10µ Proteo 90 Å (250 × 21.2 mm) column for separation as previously described.14a Analytical chromatography was carried out using a reversed-phase HP050 HPLC system with a Phenomenex Prodigy 4µ 90 Å (250 × 4.60 mm) column for separation. Electrospray ionization mass spectrometry (ESI-MS) was performed on a SCIEX API-I single quadruple mass spectrometer. The tBoc-R(Tos)(R(Tos))4Gly2K(Fmoc)-MBHA resin was synthesized manually using solid phase peptide synthesis (SPPS) and an in situ neutralization/1H-benzotriazolium-1-[bis(dimethylamino)methylene]-5-chloro-hexafluorophosphate-(1-),3-oxide (HCTU) activation procedure for tBoc chemistry on a p-methylbenzhydrylamine (MBHA) resin (0.59 meq. g−1).26 Biotinylated 6-hydrazinopyridyl-polyarginine peptide (0.1 mmol scale) was synthesized by removing the tBoc group of tBoc-R(Tos)(R(Tos))4Gly2K(Fmoc)-MBHA with trifluoroacetic acid (TFA) (2 × 1 min). The N-terminus was then biotinylated with 61 mg (0.25 mmol) biotin using the in situ neutralization/HCTU activation procedure (454 µL 0.5 M HCTU in DMF, 114 µL DIEA) (45 min). The resin was washed with DMF and the Fmoc side chain protecting group of the C-terminal lysine residue was removed on the resin by treatment with 20 vol% piperidine in DMF (4 × 3 min) followed by a DMF flow wash. Finally, 0.1 mmol (25 mg) 6-Boc-HNA was added to 0.1 mmol PyBOP® and 0.12 mmol (d = 0.742 kg L−1, 21 µL) DIEA in 1 mL of DMF and the mixture was added to the resin (45 min). The resin was washed with DMF and the tBoc group was removed with TFA (2 × 1 min). The resin was washed with DMF and DCM and dried under vacuum. The peptide was cleaved from the resin by HF using 4 vol% anisole as a scavenger. After lyophilization, biotinylated 6-hydrazinopyridyl-polyarginine peptide was purified by RP HPLC (gradient: 5–30% 9 : 1 v/v MeCN/H2O in H2O, 0.1 vol% TFA for 80 min; flow: 20 mL min−1). 2: ESI-MS calcd for C56H100N30O11S ([M + H]+): 1401.7, found 1401.0 ± 0.7.
Propagation and isolation of CPMV particles
Cowpea plants (Vigna unguiculata) were inoculated with 20 ng µL−1 CPMV in 0.1 M K-phosphate buffer (pH 7.0) and propagated for 18–20 days using established procedures.27 Virus concentration in plant extracts was determined by UV/vis spectrometry (ε = 8.1 mL mg−1 cm−1) and virus integrity was determined by size exclusion chromatography (see below).
Labeling CPMV lysine residues with 4-formylbenzamide
CPMV particles (final concentration 2 mg mL−1) were incubated with 4FB succinimidyl ester (Solulink) at a 10 : 1 molar excess per amine side chain of DMSO in 0.1MK-phosphate buffer (pH 7.0) for 2 h at room temperature, with agitation. The final DMSO concentration was adjusted to 10% of the final reaction volume. CPMV–4FB conjugates were purified using 10 kDa cutoff spin columns (Millipore) and stored at 4 °C.
Quantification of 4FB labeling efficiency
The 4FB labeling efficiency was determined using the Solulink 4FB molar substitution protocol (MSR), in which 0.1 mg of 4FB-modified CPMV particles were mixed with a 0.5 mM solution of 2-hydrazinopyridine-2-HCL (2-HP) prepared in 0.1 M phosphate buffer (pH 7.0). The reaction was incubated at room temperature for 2 h and analyzed by UV/vis spectrophotometry. The number of 4FB labels per particle was calculated using the bond-specific extinction coefficient (ε350 nm = 18 000 M−1 cm−1).
Modification of CPMV–4FB with the biotin-FLAG–hydrazide
CPMV particles modified with 4FB were diluted to a concentration of 1 mg mL−1 in 0.1 mM phosphate buffer and mixed with a 1.7-fold molar excess of the biotin-FLAG–hydrazide in a reaction containing 10 mM aniline catalyst. After incubating for 2 h at room temperature, the samples were purified using 10 kDa cut-off spin columns and stored at 4 °C. The resulting hydrazone bonds were quantified by UV/vis spectrometry using the bond-specific extinction coefficient (ε354 nm = 29 000 M−1 cm−1).
Modification of CPMV–4FB with the biotin-R5-hydrazide
CPMV particles modified with 4FB were diluted to a concentration of 0.5 mg mL−1 in 0.1 M Tris–HCl buffer (pH 9.0) containing 10 mM of the aniline catalyst, and mixed with the biotinylated R5-hydrazide prepared in water at a concentration of 10 mg mL−1 (1.2-fold molar excess). The reaction was carried out at room temperature for 35 min. The reaction mix was subsequently centrifuged at 14 000 rpm (Eppendorf 5400 microfuge) for 10 min in order to clear the reaction mix of aggregated particles. The supernatant was purified using a 10 kDa cut-off spin column and the particles were stored in 0.1 M phosphate buffer (pH 7.0) at 4 °C.
Size exclusion chromatography (SEC)
All labeled particles were analyzed by SEC using a Superose 6 column on the ÄKTA Explorer chromatography system (GE Healthcare). Concentrated samples (100 µL of 1 mg mL−1) were analyzed at a flow rate of 0.4 mL min−1, using 0.1 M potassium phosphate buffer (pH 7.0).
Gel electrophoresis
CPMV, CPMV–FLAG, CPMV–R5L and CPMV–R5H particles were analyzed by denaturing and native 0.8% agarose gel electrophoresis in 1× TBE buffer with 0.5× TBE as the running buffer (10 µg per lane). After separation the native gel was stained with Coomassie Blue and photographed using FluorChemSP imaging system. Protein subunits were analyzed on denaturing 4–12% NuPAGE gels (Invitrogen) using 1× MOPS buffer (Invitrogen). After separation, the gel was stained with Coomassie Blue and photographed using FluorChemSP imaging system or processed for western blotting (see below).
Western blotting
To detect biotinylated particles, CPMV, CPMV–FLAG, CPMV–R5L and CPMV–R5H were analyzed by western blot using electrochemiluminescent probes. We separated 5 µg samples on a 4–12% NuPAGE Bis-Tris gel using MOPS buffer, and the separated proteins were transferred onto methanol-activated PVDF membranes using NuPAGE Transfer Buffer (Invitrogen). The membranes were blocked at room temperature for 1 h using 0.1 M PBS (pH 7.4) containing 5% w/v skimmed milk powder, and the particles were detected with alkaline phosphatase-conjugated streptavidin (Sigma-Aldrich) (1 : 1000 in TBS containing 0.05% (v/v) Tween 20). Alkaline phosphatase activity was detected using the NCIP/NBT liquid substrate system (Sigma-Aldrich) according to the manufacturer’s instructions. To determine the number of biotin labels per particle, 1 µL samples were immobilized on nitrocellulose membranes, which were blocked at room temperature for 1 h in 0.1 M PBS (pH 7.4) containing 5% (w/v) skimmed milk powder. The biotin was detected using horseradish peroxidase-conjugated streptavidin (1 : 1000 in TBS containing 0.05% (v/v) Tween 20), and the horseradish peroxidase activity was detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce). The electrochemiluminescent signal was visualized and the number of biotin labels was calculated using biotin standard solutions on the BioRad ChemiDoc XRS system.
Zeta potential measurements
Wild-type CPMV (1.5 mL of a 0.04 mg mL−1 solution) and CPMV–R5H/R5L (1.5 mL of a 0.08 mg mL−1 solution) were dispersed in 100 mM potassium phosphate buffer (pH 7) and the zeta potential was measured using a 90 Plus zeta potential analyzer (Brookhaven Instruments Co., USA) with five measurements, each comprising 10 runs.
Cell cultures
Cells were grown and maintained in minimal essential medium (MEM) supplemented with 10% (v/v) heat inactivated fetal bovine serum (FBS), 1% (v/v) l-glutamine and 1% (v/v) penicillin–streptomycin at 37 °C in a 5% CO2 humidified atmosphere. All reagents were obtained from Gibco.
Confocal image analysis
Confocal images were obtained by seeding 100 µL of HeLa cells (1 × 106 cells per mL) on glass-bottomed Petri 35 mm dishes and adding wild-type CPMV, CPMV–R5H and CPMV–R5L particles prepared at a ratio of 1 × 106 particles per cell. The mixtures were incubated at 37 °C for different durations and then fixed/stained at room temperature as follows: (i) cells were fixed in 4% paraformaldehyde and 0.3% glutaraldehyde in DPBS (pH 7.2 for 5 min); (ii) cell membranes were stained with Alexa Fluor 488-conjugated wheat germ agglutinin (Invitrogen; 1 : 500 for 1 h); (iii) cells were permeabilized with 0.2% Triton-X 100 in DPBS for 2 min; (iv) the cells were blocked with 10% goat serum for 1 h; (v) CPMV was stained using a rabbit anti-CPMV antibody (1 : 500) for 1 h followed by a goat anti-rabbit Alexa Fluor 555 or Alexa Fluor 647-conjugated antibody (Invitrogen) for 1 h; (vi) early endosomes of endolysosomes were stained using mouse anti-EEA1 or mouse anti-Lamp 1 antibodies (Invitrogen; 1 : 500) for 1 h followed by Alexa Fluor 555 or Alexa Fluor 647 conjugated goat anti-mouse antibodies; and (vii) cell nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI) (1 : 9500) for 10 min. Slides were mounted using Vecta Shield mounting medium. Confocal images were obtained using a Biorad 2100 confocal microscope with a 60× oil objective. Images were created using ImageJ software. Colocalization analysis was performed using single plane images. 8 ROIs showing representative cells for each internalization study were analysed using a Zeiss LSM examiner. The weighted colocalization coefficient was used to quantify particle co-clustering with endolysosomal markers.
Fluorescence activated cell sorting (FACS)
To quantify the intracellular uptake of the different formulations, HeLa cells were collected using enzyme-free Hank’s based Cell Dissociation Buffer and distributed in 200 µL aliquots at a concentration of 5 × 106 cell per mL in V-bottom 96-well plates for FACS analysis. We added 1 × 105 VNPs per cell and incubated the preparations for 3 h at 37 °C. To confirm whether CPMV particles were internalized or merely bound to the surface, cells were exposed to 0.1% (w/v) pronase in DPBS for 15 min at 4 °C in order to strip off surface particles. The cells were washed three times in DBPS and fixed in 2% (v/v) formaldehyde for 10 min in DPBS at room temperature. CPMV particles were stained using a rabbit anti-CPMV antibody (1 : 1000 in FACS buffer), i.e. DPBS supplemented with 0.5% (v/v) saponin, 25 mM HEPES, 2 mM EDTA and 1% FBS followed by a goat anti-rabbit secondary antibody conjugated to Alexa Fluor 647 (1 : 2000 in FACS buffer). Cells were washed with FACS buffer between steps and centrifuged three times at 500 × g. Cells were resuspended and analyzed using a FACS LSM II instrument. At least 10 000 gated events were recorded. Experiments were repeated at least twice and triplicates of each sample were measured. Data were analyzed using FlowJo 8.6.3 software.
Energy dependency studies
We seeded 100 µL of HeLa cells (1 × 106 cell per mL) on 35 mm glass-bottomed Petri dishes, and added the three CPMV formulations at a concentration of 1 × 106 particles per cell. The mixture was incubated for 3 h at either 4 °C or 37 °C and differences in the efficiency of particle internalization were determined by immunostaining as described above.
Acknowledgements
This work was supported by NIH/NIBIB grants R00 EB009105 (to Steinmetz) and P30 EB011317 (to Steinmetz), a student fellowship from the Undergraduate Research Opportunities Program (UROP, to Wu) and the Adam-Schall-Gesellschaft (to Wu). We would like to thank Prof. Manchester (UCSD) for helpful discussions.
Footnotes
Electronic supplementary information (ESI) available: Experimental details and additional supporting data. See DOI: 10.1039/c2nr30366c
References
- 1.(a) Steinmetz NF. Nanomedicine. 2010;349 doi: 10.1016/j.nano.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huo Q, Liu J, Wang LQ, Jiang Y, Lambert TN, Fang E. J. Am. Chem. Soc. 2006;128:6447. doi: 10.1021/ja060367p. [DOI] [PubMed] [Google Scholar]; (c) Alexiou C, Schmid RJ, Jurgons R, Kremer M, Wanner G, Bergemann C, Huenges E, Nawroth T, Arnold W, Parak FG. Eur. Biophys. J. 2006;35:446. doi: 10.1007/s00249-006-0042-1. [DOI] [PubMed] [Google Scholar]; (d) Dutta T, Agashe HB, Garg M, Balakrishnan P, Kabra M, Jain NK. J. Drug Targeting. 2007;15:89. doi: 10.1080/10611860600965914. [DOI] [PubMed] [Google Scholar]; (e) Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Clin. Pharmacol. Ther. 2008;83:761. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 2.Jain KK. Med. Princ. Pract. 2008;17:89. doi: 10.1159/000112961. [DOI] [PubMed] [Google Scholar]
- 3.Mohanraj V, Chen Y. J. Pharm. Res. 2006;5:561. [Google Scholar]
- 4.(a) Sanvicens N, Marco MP. Trends Biotechnol. 2008;26:425. doi: 10.1016/j.tibtech.2008.04.005. [DOI] [PubMed] [Google Scholar]; (b) Torchilin VP. Adv. Drug Delivery Rev. 2006;58:1532. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Razis ED, Fountzilas G. Ann. Oncol. 2001;12:593. doi: 10.1023/a:1011108807105. [DOI] [PubMed] [Google Scholar]
- 6.(a) Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. J. Biol. Chem. 2001;276:5836. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]; (b) Lundberg P, Magzoub M, Lindberg M, Hallbrink M, Jarvet J, Eriksson LE, Langel U, Graslund A. Biochem. Biophys. Res. Commun. 2002;299:85. doi: 10.1016/s0006-291x(02)02595-0. [DOI] [PubMed] [Google Scholar]; (c) Hallbrink M, Floren A, Elmquist A, Pooga M, Bartfai T, Langel U. Biochim. Biophys. Acta. 2001;1515:101. doi: 10.1016/s0005-2736(01)00398-4. [DOI] [PubMed] [Google Scholar]; (d) Gupta B, Levchenko TS, Torchilin VP. Adv. Drug Delivery Rev. 2005;57:637. doi: 10.1016/j.addr.2004.10.007. [DOI] [PubMed] [Google Scholar]; (e) Schwarze SR, Ho A, Vocero-Akbani A, Dowdy SF. Science. 1999;285:1569. doi: 10.1126/science.285.5433.1569. [DOI] [PubMed] [Google Scholar]
- 7.(a) Khalil IA, Kogure K, Futaki S, Harashima H. Int. J. Pharm. 2008;354:39. doi: 10.1016/j.ijpharm.2007.12.003. [DOI] [PubMed] [Google Scholar]; (b) de la Fuente JM, Berry CC. Bioconjugate Chem. 2005;16:1176. doi: 10.1021/bc050033+. [DOI] [PubMed] [Google Scholar]; (c) Khalil IA, Kogure K, Futaki S, Hama S, Akita H, Ueno M, Kishida H, Kudoh M, Mishina Y, Kataoka K, Yamada M, Harashima H. Gene Ther. 2007;14:682. doi: 10.1038/sj.gt.3302910. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Ruan G, Agrawal A, Marcus AI, Nie S. J. Am. Chem. Soc. 2007;129:14759. doi: 10.1021/ja074936k. [DOI] [PubMed] [Google Scholar]; (e) Lewin M, Carlesso N, Tung CH, Tang XW, Cory D, Scadden DT, Weissleder R. Nat. Biotechnol. 2000;18:410. doi: 10.1038/74464. [DOI] [PubMed] [Google Scholar]; (f) Wei B, Wei Y, Zhang K, Wang J, Xu R, Zhan S, Lin G, Wang W, Liu M, Wang L, Zhang R, Li J. Biomed. Pharmacother. 2009;63:313. doi: 10.1016/j.biopha.2008.07.086. [DOI] [PubMed] [Google Scholar]; (g) Tseng YL, Liu JJ, Hong RL. Mol. Pharmacol. 2002;62:864. doi: 10.1124/mol.62.4.864. [DOI] [PubMed] [Google Scholar]; (h) Jones SW, Christison R, Bundell K, Voyce CJ, Brockband SM, Lindsay MA. Br. J. Pharmacol. 2005;145:1093. doi: 10.1038/sj.bjp.0706279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs SM, Raines RT. Biochemistry. 2004;43:2438. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Delehanty JB, Bradburne CE, Boeneman K, Susumu K, Farrell D, Mei BC, Blanco-Canosa JB, Dawson G, Dawson PE, Mattoussi H, Medintz IL. Integr. Biol. 2010;2:265. doi: 10.1039/c0ib00002g. [DOI] [PubMed] [Google Scholar]; (b) Nativo P, Prior IA, Brust M. ACS Nano. 2008;2:1639. doi: 10.1021/nn800330a. [DOI] [PubMed] [Google Scholar]
- 10.(a) Anderson EA, Isaacman S, Peabody DS, Wang EY, Canary JW, Kirshenbaum K. Nano Lett. 2006;6:1160. doi: 10.1021/nl060378g. [DOI] [PubMed] [Google Scholar]; (b) Hooker JM, Datta A, Botta M, Raymond KN, Francis MB. Nano Lett. 2007;7:2207. doi: 10.1021/nl070512c. [DOI] [PubMed] [Google Scholar]; (c) Hooker JM, O’Neil JP, Romanini DW, Taylor SE, Francis MB. Mol. Imaging Biol. 2008;10:182. doi: 10.1007/s11307-008-0136-5. [DOI] [PubMed] [Google Scholar]; (d) Kaiser CR, Flenniken ML, Gillitzer E, Harmsen AL, Harmsen AG, Jutila MA, Douglas T, Young MJ. Int. J. Nanomed. 2007;2:715. [PMC free article] [PubMed] [Google Scholar]; (e) Kovacs EW, Hooker JM, Romanini DW, Holder PG, Berry KE, Francis MB. Bioconjugate Chem. 2007;18:1140. doi: 10.1021/bc070006e. [DOI] [PubMed] [Google Scholar]; (f) Loo L, Guenther RH, Lommel SA, Franzen S. J. Am. Chem. Soc. 2007;129:11111. doi: 10.1021/ja071896b. [DOI] [PubMed] [Google Scholar]; (g) Loo L, Guenther RH, Lommel SA, Franzen S. Chem. Commun. 2008:88. doi: 10.1039/b714748a. [DOI] [PubMed] [Google Scholar]; (h) Ren Y, Wong SM, Lim LY. J. Gen. Virol. 2006;87:2749. doi: 10.1099/vir.0.81944-0. [DOI] [PubMed] [Google Scholar]; (i) Ren Y, Wong SM, Lim LY. Bioconjugate Chem. 2007;18:836. doi: 10.1021/bc060361p. [DOI] [PubMed] [Google Scholar]; (j) Suci PA, Berglund DL, Liepold L, Brumfield S, Pitts B, Davison W, Oltrogge L, Hoyt KO, Codd S, Stewart PS, Young M, Douglas T. Chem. Biol. 2007;14:387. doi: 10.1016/j.chembiol.2007.02.006. [DOI] [PubMed] [Google Scholar]; (k) Suci PA, Varpness Z, Gillitzer E, Douglas T, Young M. Langmuir. 2007;23:12280. doi: 10.1021/la7021424. [DOI] [PubMed] [Google Scholar]
- 11.(a) Phelps JP, Dang N, Rasochova L. J. Virol. Methods. 2007;141:146. doi: 10.1016/j.jviromet.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Porta C, Spall VE, Loveland J, Johnson JE, Barker PJ, Lomonossoff GP. Virology. 1994;202:949. doi: 10.1006/viro.1994.1417. [DOI] [PubMed] [Google Scholar]; (c) Brennan FR, Jones TD, Hamilton WD. Mol. Biotechnol. 2001;17:15. doi: 10.1385/MB:17:1:15. [DOI] [PubMed] [Google Scholar]; (d) Destito G, Yeh R, Rae CS, Finn MG, Manchester M. Chem. Biol. 2007;14:1152. doi: 10.1016/j.chembiol.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Nat. Med. 2006;12:354. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Brunel FM, Lewis JD, Destito G, Steinmetz NF, Manchester M, Stuhlmann H, Dawson PE. Nano Lett. 2010;10:1093. doi: 10.1021/nl1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Steinmetz NF, Hong V, Spoerke ED, Lu P, Breitenkamp K, Finn MG, Manchester M. J. Am. Chem. Soc. 2009;131:17093. doi: 10.1021/ja902293w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Nat. Protoc. 2010;5:1406. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Steinmetz NF, Ablack AL, Hickey JL, Ablack J, Manocha B, Mymryk JS, Luyt LG, Lewis JD. Small. 2011:1664. doi: 10.1002/smll.201000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koudelka KJ, Destito G, Plummer EM, Trauger SA, Siuzdak G, Manchester M. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000417. e1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinmetz NF, Cho CF, Ablack A, Lewis JD, Manchester M. Nanomedicine. 2011;6:351. doi: 10.2217/nnm.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Dirksen A, Yegneswaran S, Dawson PE. Angew. Chem. 2010;49:2023. doi: 10.1002/anie.200906756. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dirksen A, Hackeng TM, Dawson PE. Angew. Chem. 2006;45:7581. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 15.Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, Lin T. Chem. Biol. 2004;11:855. doi: 10.1016/j.chembiol.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 16.Hong V, Presolski SI, Ma C, Finn MG. Angew Chemie. 2009;48:9879. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foley JF, Aftonomos B. J. Cell. Physiol. 1970;75:159. doi: 10.1002/jcp.1040750204. [DOI] [PubMed] [Google Scholar]
- 18.Rinne J, Albarran B, Jylhava J, Ihalainen TO, Kankaanpaa P, Hytonen VP, Stayton PS, Kulomaa MS, Vihinen-Ranta M. BMC Biotechnol. 2007;7:1. doi: 10.1186/1472-6750-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Biochem. J. 2004;377:159. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gratton SE, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11613. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks H, Lebleu B, Vives E. Adv. Drug Delivery Rev. 2005;57:559. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Douglas KL, Piccirillo CA, Tabrizian M. Eur. J. Pharm. Biopharm. 2008;68:676. doi: 10.1016/j.ejpb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez MJ, Plummer EM, Rae CS, Manchester M. PLoS One. 2009;4:e7981. doi: 10.1371/journal.pone.0007981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Kerr MC, Lindsay MR, Luetterforst R, Hamilton N, Simpson F, Parton RG, Gleeson PA, Teasdale RD. J. Cell Sci. 2006;119:3967. doi: 10.1242/jcs.03167. [DOI] [PubMed] [Google Scholar]; (b) Kiss AL, Botos E. J. Cell. Mol. Med. 2009;13:1228. doi: 10.1111/j.1582-4934.2009.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green I, Christison R, Voyce CJ, Bundell KR, Lindsay MA. Trends Pharmacol. Sci. 2003;24:213. doi: 10.1016/S0165-6147(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 26.Schnolzer M, Alewood P, Jones A, Alewood D, Kent SB. Int. J. Pept. Protein Res. 1992;40:180. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 27.Wellink J. Methods Mol. Biol. 1998;81:205. doi: 10.1385/0-89603-385-6:205. [DOI] [PubMed] [Google Scholar]