Abstract
Two different groups, using ChIP-seq data, have recently published the genome-wide distribution of histones H3.1 and H3.3 in Arabidopsis thaliana. In one report, Stroud and colleagues determined that, whereas H3.1 was enriched in repetitive pericentromeric and silent chromatin, H3.3 was enriched in transcriptionally active regions. This work was performed using seedlings, which contained dividing and non-dividing cells. In a second report, Wollmann and colleagues found similar results analyzing dividing or non-dividing tissue. None of these reports addressed the analysis of telomeres or centromeres. Our group has recently described an experimental approach that allows the study of the epigenetic status of some Arabidopsis repetitive sequences by analyzing ChIP-seq data. By using this approach and the data generated by Stroud, Wollmann and colleagues, we found that telomeres are enriched in H3.3 with regard to the centromeric 178 bp repeats, whereas the centromeric repeats are enriched in H3.1 with regard to telomeres.
Histone variants influence important biological processes like cellular differentiation and development1,2,3. Recently, Stroud and colleagues as well as Wollmann and colleagues have mapped the genome-wide distribution of two histone H3 variants in Arabidopsis thaliana: H3.1 and H3.34,5. They found that H3.1 was enriched in repetitive pericentromeric and silent chromatin whereas H3.3 was enriched in transcriptionally active regions of the genome. These results are consistent with genome-wide analyses of H3.3 enrichment in Drosophila and mammalian cells6,7,8. Whereas H3.1 is incorporated during DNA replication, H3.3 can be incorporated outside of S phase, during processes like transcription. Interestingly, Stroud and colleagues also found that both histone variants were enriched in replication origins reflecting an overall high nucleosomal density, which represents an unique characteristic of plant replication origins and indicates differences in chromatin structure organization between plants origins and the origins of other eukaryotes. In addition, Wollmann and colleagues found that the transcriptional variations associated with leaf differentiation were accompanied by changes of H3.3 levels, but not of H3.1. Here, we analyze the ChIP-seq data released by Stroud, Wollmann and colleagues to study the distribution of H3.1 and H3.3 at telomeres and centromeres.
Results
The studies reported by Stroud, Wollmann and colleagues did not address the analysis of some repetitive sequences present at different genomic loci because it is actually challenging9. This was the case for centromeric sequences and for telomeric sequences, which in Arabidopsis are very abundant at interstitial telomeric loci and are denoted as ITSs10,11,12,13,14. These ITSs challenge the analysis of Arabidopsis telomeric chromatin structure by ChIP, ChIP-on-chip or ChIP-seq9.
We have recently described an experimental approach that allows the study of the epigenetic status of telomeres versus centromeres by analyzing ChIP–seq data15. This approach was designed to analyze telomeres independently of ITSs. Since ITSs in Arabidopsis are mostly composed of very short stretches of perfect telomeric repeats interspersed with degenerated repeats10,13,14,16, a short stretch of perfect telomeric repeats might essentially represent telomeres. Blast analyses of the Arabidopsis genome revealed that 98% of the (CCCTAAA)4 sequences are found at telomeres whereas only 2% of these sequences localize at ITSs15. Therefore, the DNA sequence (CCCTAAA)4 reveals Arabidopsis telomeres but not ITSs in ChIp-seq experiments15. For heterochromatic comparison, we selected the sequences CEN1 and CEN2, which are conserved regions of the 178 bp satellite repeats present at centromeres15. These sequences allowed us to analyze the chromatin organization of the Arabidopsis 178 bp satellite repeats, as an average, which are known to be heterochromatic17,18,19,20,21.
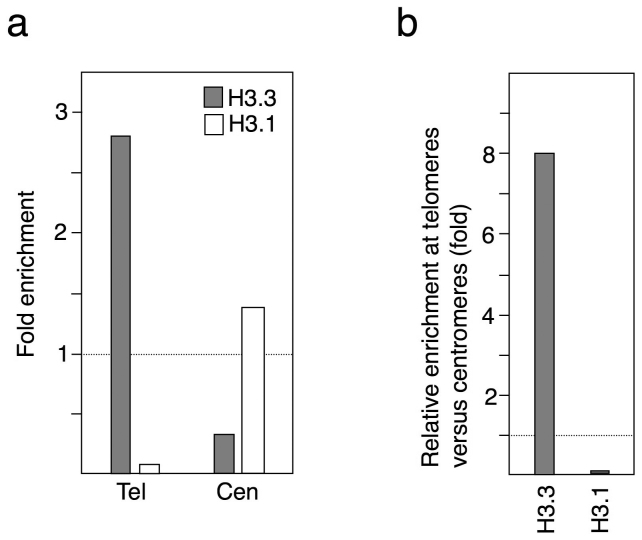
To analyze the data reported by Stroud and colleagues we determined the number of reads containing the sequence (CCCTAAA)6 for telomeres instead of (CCCTAAA)4, because the length of their reads allowed it. About 99% of the (CCCTAAA)6 sequences are found at telomeres. For centromeres, we determined the number of reads containing CEN1 and CEN2 (Supplementary Table 1). Then, we calculated the relative enrichment of H3.1 and H3.3 at telomeres and at centromeres. We found that telomeres were enriched in H3.3 and had low levels of H3.1. Conversely, centromeres had low levels of H3.3 and slightly higher than average genomic levels of H3.1 (Fig. 1a). Actually, whereas telomeres were enriched in H3.3 versus centromeres, centromeres were enriched in H3.1 versus telomeres (Fig. 1b). Thus, Arabidopsis telomeres are labeled with H3.3 and the 178 bp repeats are labeled with H3.1.
Figure 1. Relative enrichment of histones H3.1 and H3.3 at telomeres and centromeres in 10 days old seedlings.
Histone H3 variants levels were analyzed using study SRP010096 (GSE3840), as indicated in the Methods section. (a) Enrichment of histones H3.1 and H3.3 at telomeres and at centromeres versus the input samples. Tel indicates telomeres whereas Cen indicates centromeres. (b) Relative enrichment of histones H3.1 and H3.3 at telomeres versus centromeres.
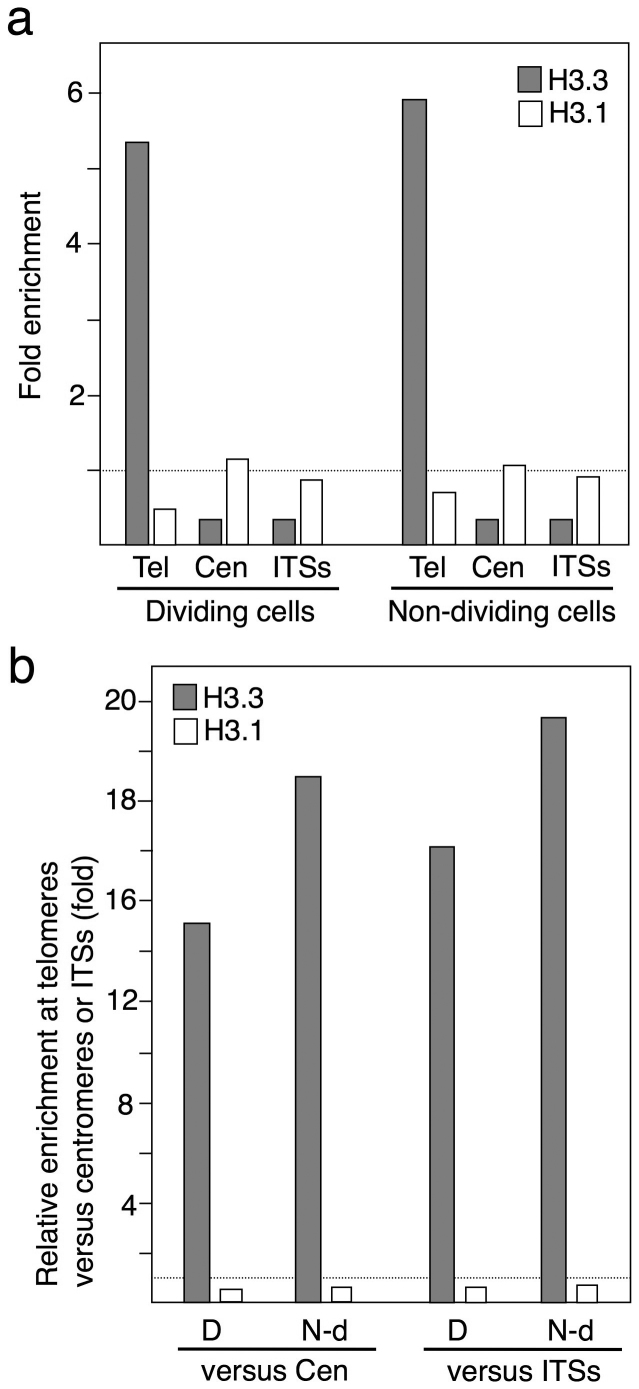
To analyze the data reported by Wollmann and colleagues we determined the number of reads containing the sequence (CCCTAAA)5 for telomeres and CEN1 and CEN2 for centromeres (Supplementary Table 2). We performed these analyses in dividing tissues (meristems, leaf primordia and young leaves) or in non-dividing tissues (mature, differentiated leaves) and found similar results in both kinds of tissues (Fig. 2). Whereas telomeres were enriched in H3.3 and had lower than average H3.1 levels, centromeres had low levels of H3.3 and average levels of H3.1 (Fig. 2a). In this study, telomeres were also enriched in H3.3 versus centromeres and centromeres were enriched in H3.1 versus telomeres (Fig. 2b). Therefore, these results confirm that Arabidopsis telomeres are labeled with H3.3 and that the 178 bp repeats associate with H3.1, independently of the proliferative capacity of the tissues and their state of differentiation.
Figure 2. Relative enrichment of histones H3.1 and H3.3 at telomeres, centromeres and ITSs in dividing and in non-dividing cells.
Histone H3 variants levels were analyzed using study SRP011591 (GSE36631), as indicated in the Methods section. (a) Enrichment of histones H3.1 and H3.3 at telomeres, centromeres and ITSs versus the control immunoglobulin G samples. Tel indicates telomeres whereas Cen indicates centromeres. (b) Relative enrichment of histones H3.1 and H3.3 at telomeres versus centromeres or ITSs. D indicates dividing cells whereas N-d indicates non-dividing cells.
As a control, we determined the levels of H3.1 and H3.3 enrichment at a pericentromeric region of Arabidopsis chromosome 1, which contains a high number of ITSs. We counted the number of reads for the sequence TCTAAACCCTAAACCGTACACC in dividing cells and in non-dividing cells (Supplementary Table 2). This sequence is found only 34 times in the Arabidopsis genome, within the pericentromeric region mentioned above (between coordinates 15106975 and 15431297; see for example the DNA surrounding the repeat located 4876 bp upstream of AT1G40131). We found that the enrichment levels of H3.3 and H3.1 at the selected sequence were similar to those found for centromeres in dividing and in non-dividing cells (Fig. 2a). Therefore, telomeres are enriched in H3.3 versus ITSs, as an average, and ITSs are enriched in H3.1 versus telomeres (Fig. 2b). These results agree with the heterochromatic nature of Arabidopsis ITSs22 and confirm the accuracy of our ChIP-seq analysis approach.
Discussion
Our study extends the conclusions reported by Stroud, Wollmann and colleagues4,5. It is known that part of the 178 bp repeats associate with another variant of H3, CENH3, and leads to the formation of the centromere1,2,3. These repeats are surrounded by additional 178 bp repeats that are pericentromeric and associate with H3 chromatin1,2,3,20,23. The data shown in Figures 1 and 2 indicate the association of H3.1 with pericentromeric 178 bp repeats.
Our analysis supports previous studies reporting that mammalian telomeres associate with H3.36,24,25,26, which were performed without differentiating between telomeres and ITSs. In addition, the presence of H3.3 at telomeres agrees with the fact that Arabidopsis telomeres have low levels of heterochromatic marks (like H3K9me2 or H3K27me) and exhibit euchromatic features15,22. H3.3 do not associate with H3K27me3, a euchromatic repressive mark, in the Arabidopsis genome4. However, H3.3 and H3K27me3 are both found at telomeres15, which might reflect the particular chromatin organization of Arabidopsis telomeres or the dynamic nature of their epigenetic marks.
It is known that Arabidopsis telomeres are transcribed leading to the so-called TERRA transcripts27. These transcripts are found in a wide variety of organisms and have been related to the stability of telomeres28. The presence of H3.3 at Arabidopsis telomeres in non-dividing leaf tissues agrees with the existence of TERRA transcripts being required for telomere stability. However, since old Arabidopsis leaves undergo endoreduplication29,30, the presence of H3.3 at differentiated leaf telomeres could also be related to TERRA transcripts being required for telomeres endoreduplication.
Methods
The enrichment levels of H3.1 and H3.3 at telomeres, centromeres and ITSs were calculated using different ChIP-seq runs from studies SRP010096 (Gene Expression Omnibus accession number GSE3840)4 and/or SRP011591 (GSE36631)5, availables at NCBI. First, in all cases, the number of reads was normalized against the number of spots for each run. In the case of SRP010096, telomeric and centromeric enrichments were calculated by dividing the reads corresponding to the immunoprecipitated samples between the reads corresponding to the input samples. For telomeres, we plotted the data obtained after counting the number of (CCCTAAA)6 reads. Similar results were obtained when the number of (CCCTAAA)4 reads were determined (data not shown). For centromeres, we plotted the mean data obtained for CEN1 (TTGGCTTTGTATCTTCTAACAAG) and for CEN2 (CATATTTGACTCCAAAACACTAA), which were very similar. In the case of SRP011591, telomeric, centromeric and ITSs enrichments were calculated by dividing the reads corresponding to the immunoprecipitated samples between the reads corresponding to the immunoglobulin G control, using the reads from repeat 2 runs. For telomeres, we plotted the data obtained after counting the number of (CCCTAAA)5 reads, although similar results were obtained when the number of (CCCTAAA)4 reads were determined (data not shown). For centromeres, we plotted the mean data obtained for CEN1 and for CEN2, which were also very similar. For ITSs, we determined the number of reads corresponding to the sequence TCTAAACCCTAAACCGTACACC. In all cases, the relative enrichment at telomeres versus centromeres or versus ITSs was obtained by dividing the telomeric enrichment values by the centromeric or the ITSs enrichment values.
Author Contributions
M.A.V. designed experiments and M.I.V. and M.A.V. performed analyses and wrote the manuscript.
Supplementary Material
Supplementary Tables
Acknowledgments
We acknowledge support from the Spanish Ministry of Economy and Competitivity (Grant BIO2011-24794).
References
- Hake S. & Allis C. Histone H3 variants and their potential role in indexing mammalian genomes: the "H3 barcode hypothesis". Proc. Natl. Acad. Sci. USA 103, 6428–6435 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L., Allis C. & Lewis P. Histone variants in metazoan development. Dev. Cell 19, 662–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. & Henikoff S. Histone variants – ancient wrap artists of the epigenome. Nat. Rev. 11, 264–275 (2010). [DOI] [PubMed] [Google Scholar]
- Stroud H. et al. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 109, 5370–5375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H. et al. Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 8, e1002658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C., Bell O. & Schübeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19, 1761–1766 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J. & Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37, 1090–1097 (2005). [DOI] [PubMed] [Google Scholar]
- Vaquero-Sedas M. & Vega-Palas M. On the chromatin structure of eukaryotic telomeres. Epigenetics 6, 1055–1058 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámez-Arjona F., López-López C., Vaquero-Sedas M. & Vega-Palas M. On the organization of the nucleosomes associated with telomeric sequences. Biochim. Biophys. Acta 1803, 1058–1061 (2010). [DOI] [PubMed] [Google Scholar]
- Richards E., Goodman H. & Ausubel F. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 19, 3351–3357 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. & Yan J. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutation Res. 658, 95–110 (2008). [DOI] [PubMed] [Google Scholar]
- Regad F., Lebas M. & Lescure B. ITSs within the Arabidopsis thaliana genome. J. Mol. Biol. 239, 163–169 (1994). [DOI] [PubMed] [Google Scholar]
- Uchida W., Matsunaga S., Sugiyama R. & Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 77, 63–67 (2002). [DOI] [PubMed] [Google Scholar]
- Vaquero-Sedas M. & Vega-Palas M. Analysis of the epigenetic status of telomeres by using ChIP-seq data. Nucleic Acids Res. 40, e163 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J., Depeiges A., White C. & Gallego M. ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 5, e1000380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L., Cao X. & Jacobsen S. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12, 1360–1367 (2002). [DOI] [PubMed] [Google Scholar]
- Lindroth A. et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080 (2001). [DOI] [PubMed] [Google Scholar]
- Lindroth A. et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23, 4286–4296 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Lee H., Koo D. & Jiang J. Epigenetic modification of centromeric chromatin: Hypomethylation of DNA sequences in the CENH3-associated chromatin in Arabidopsis thaliana and maize. Plant Cell 20, 25–34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P. et al. Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13, 867–876 (1998). [DOI] [PubMed] [Google Scholar]
- Vaquero-Sedas M., Gámez-Arjona F. & Vega-Palas M. Arabidopsis telomeres exhibit euchromatic features. Nucleic Acids Res. 39, 2007–2017 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Zapater J., Estelle M. & Somerville C. A high repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204, 417–423 (1986). [Google Scholar]
- Wong L. et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P., Elsaesser S., Noh K., Stadler S. & Allis C. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107, 14075–1480 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbsky J. et al. siRNA-mediated methylation of Arabidopsis telomeres. PloS Genet. 6, e1000986 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A. & Azzalin C. The telomeric transcriptome: From fission yeast to mammals. Int. J. Biochem. & Cell Biol. 44, 1055–1059 (2012). [DOI] [PubMed] [Google Scholar]
- Massonnet C. et al. New insights into the control of endoreduplication: Endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol. 157, 2044–2055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C. The Arabidopsis book Chap. 2 (ASPB Publications, 2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables