Abstract
Eukaryotic genomes are extensively transcribed, forming both messenger (m) and noncoding (nc) RNAs. ncRNAs made by RNA polymerase II (Pol II) often initiate from bidirectional promoters (nucleosome-depleted chromatin) that synthesise mRNA and ncRNA in opposite directions. We demonstrate that actively transcribed mRNA encoding genes by adopting a gene loop conformation, restrict divergent transcription of ncRNAs. Since gene loop formation depends on a protein factor (Ssu72) that co-associates with both promoter and terminator, its inactivation leads to increased synthesis of promoter-associated divergent ncRNAs, referred to as Ssu72 restricted transcripts (SRT). Similarly, inactivation of individual gene loops by gene mutation enhances SRT synthesis. We demonstrate that gene loop conformation enforces transcriptional directionality on otherwise bidirectional promoters.
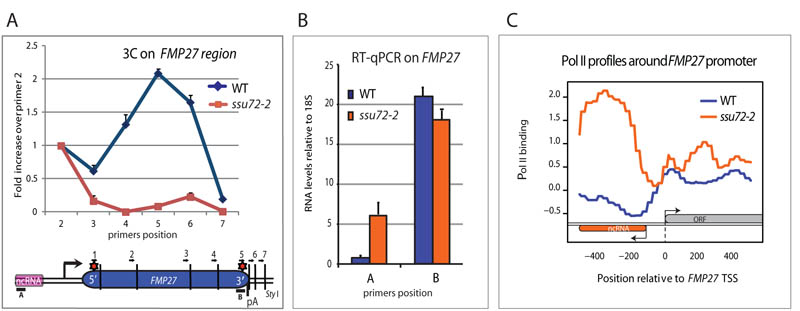
Eukaryotic genomes are ubiquitously transcribed, generating an extensive network of ncRNAs (1, 2). Most ncRNAs are made by Pol II, which can initiate transcription non-specifically and bidirectionally on nucleosome-depleted chromatin (3-5). While this promiscuous transcription is partly restricted by rapid transcript degradation (6, 7), we demonstrate that actively transcribed genes adopt a gene loop conformation that reduces aberrant transcription by focusing Pol II into productive mRNA synthesis. Gene loop formation depends on both promoter-associated transcription factors as well as polyadenylation complex (pAC) factors (8-11) such as Ssu72, localised at the 5′ and 3′ ends of genes (12, 13). We initially confirmed that its mutation (ssu72-2) prevents gene loop formation across FMP27, based on quantitative 3C analysis (Fig. 1). We also detected an increase in promoter associated antisense ncRNA and increased Pol II density over the FMP27 promoter region in ssu72-2 (Fig. 1). Furthermore we observed unanticipated genetic interactions between either Ssu72 or pAC associated Pta1 and the nuclear exosome component Rrp6, responsible for degradation of many ncRNAs, especially cryptic unstable transcripts (CUTs) (Fig. S1; (6, 7). Taken together our initial results indicated that loss of gene loop formation by inactivation of Ssu72 results in the production of aberrant ncRNAs that are stabilized in Δrrp6 mutant cells.
Figure 1. Ssu72 inactivation abrogates FMP27 gene loop and enhances antisense transcription.
A) Graphical representation of 3C interaction levels in ssu72-2 vs WT across FMP27. Red stars show significant 3C interaction. Positions of 3C primers are indicated as are RT-qPCR amplicons. For 3C analysis primer 1 (anchor) was combined sequentially with downstream primers 2-7. Error bars represent SEM. B) RT-qPCR analysis of FMP27 mRNA and ncRNA in ssu72-2 vs WT. Error bar represent SEM. C) Pol II profile (ChIP-seq) across FMP27 promoter region (12) in ssu72-2 vs WT.
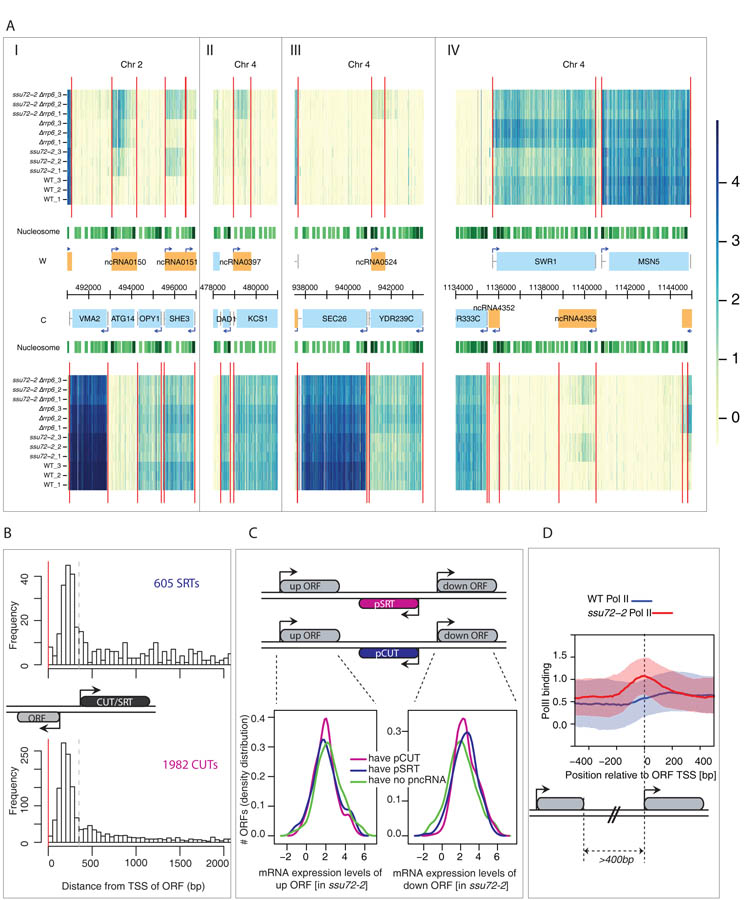
We next compared the effect of mutating RRP6 and SSU72 alone or together on the genomic profile of coding and ncRNAs. Total RNA from wild type (WT), ssu72-2, Δrrp6 and double ssu72-2Δrrp6 strains grown at 32oC (semi-permissive conditions) was hybridized to strand-specific S. cerevisiae tiling arrays. The profiles obtained confirmed that loss of Rrp6 causes accumulation of CUTs, especially from bidirectional promoters (7). However ssu72-2 mutation alone or in combination with Δrrp6 gave rise to many additional ncRNAs (Fig. 2A). Ssu72 is involved in the transcription termination of snoRNAs as is clearly revealed by the widespread appearances of extended transcripts for these genes in ssu72-2 ((14); Fig. S2AI). The profiles also unveil a role of Ssu72 in transcriptional termination of CUTs as many show 3′ extensions in the ssu72-2Δrrp6 double mutant as compared to Δrrp6 (Fig. S2AII). Ssu72 inactivation also leads to increased initiation of new cryptic transcripts. Ssu72-restricted transcripts (SRTs), which like CUTs, often run in a divergent orientation from bidirectional promoters. Some SRTs were detected in the single ssu72-2 mutant strain and others only in combination with RRP6 deletion (Fig. 2A). The array data demonstrated the presence of 605 SRTs in addition to the expected 1982 CUTs (Fig. 2B) as validated in specific cases (Fig. S2B).
Figure 2. Ssu72 inactivation leads to novel ncRNA transcription (SRTs).
A) I-IV. Genomic transcription across 28 kb of chromosomes 2 and 4 (x axis) for the Watson (W, top) and the Crick (C, bottom) strands. For the whole genome see http://steinmetzlab.embl.de/proudfoot_lab/index.html. Normalized signal intensities shown for profiled samples (y axis). Triplicate data shown for WT (1–3), ssu72-2 (1–3), Δrrp6 (1-3) and ssu72-2Δrrp6 (1–3) strains. Red vertical lines represent inferred transcript boundaries. Nucleosome positions (green tracks, darker for more significant scores; (25) and genome annotations are shown in the centre: annotated ORFs (blue boxes), ncRNAs (orange boxes), and TSS (arrows). (I) ncRNA0151, (II) ncRNA0397, (III) ncRNA0524 and (IV) ncRNA4353 represent Ssu72 repressed transcripts (SRTs) that are promoter-associated. B) Distribution of relative distances of 605 SRTs (upper panel) versus 1982 CUTs (lower panel) to their nearest ORF TSS (red line). Dashed lines (350 bp) indicate cut-off position used to define intergenic cryptic transcription sharing ORF promoters. C) Distributions of gene expression levels are shown for downstream (down) and upstream (up) ORFs of tandem genes (3 categories: downstream promoter pSRT (blue) or pCUT (purple) or no pncRNA (green). Downstream ORFs with pncRNAs are significantly higher expressed than those without (p=0.001 [pSRT] and p<2.2e-16 [pCUTs]) No significant association was found for upstream ORFs. D) Pol II profiles for ssu72-2 (red) and wild type (blue) around ORF promoter in tandem genes more than 400bp apart. Solid lines indicate median Pol II-occupancy and shaded areas 25-75 percentiles. Pol II occupancy increases upstream of TSS in ssu72-2.
CUT and SRT initiation is associated with mRNA transcription start sites (TSS; Fig. 2B; (6, 7). To focus on promoter-associated ncRNAs (pncRNA) we selected CUTs and SRTs that are positioned between tandem open reading frames (ORFs). 678 pCUTs and 135 pSRTs initiate antisense between tandem ORFs. Promoters that generate a divergent pncRNA tend to express more mRNA (down ORF) (Fig. 2C). In contrast we found no correlation in mRNA expression level (up ORF) with downstream positioned ncRNAs. We further showed that SRT expression is not due to loss of NRD dependent CUT termination or differential RNA stability effects (Fig. S3A and S3B). Finally, a genome-wide Pol II occupancy profile of the ssu72-2 mutant (12) revealed a distinct peak upstream of the TSS, which is absent in the wild type (Fig. 2D) as well as a higher Pol II occupancy over SRT transcript regions in the mutant as compared to WT (Fig. S3C). Overall these results established that loss of Ssu72 promotes de novo initiation of pSRTs.
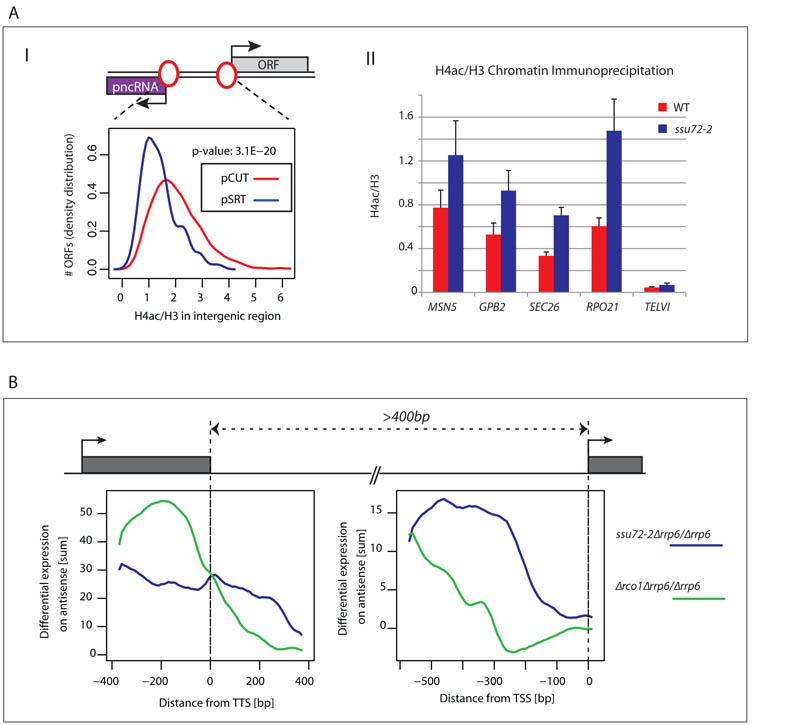
Publicly available genome-wide data revealed that pSRT-associated promoters are especially depleted of histone H4 acetylation (15), implying a more repressed transcriptional state also shown in 4 selected pSRT-producing promoters (Fig. 3A). Loss of Ssu72 seems to relax this repressed chromatin structure by promoting histone acetylation and consequent pSRT expression. A genome-wide analysis of S. cerevisiae nascent transcripts reported a potentially similar connection between ncRNA levels and histone deacetylation (16). Loss of histone deacetylase Rco1 (in Rpd3S complex) known to contribute to H4 deacetylation in gene 3′ regions also increased antisense transcription suggesting its potential role in promoter directionality. However, antisense transcripts may derive from antisense initiation at gene 3′ ends (17). We compared pSRTs to antisense ncRNA induced in Δrco1 (Rco1-restricted transcripts or RRTs) by generating transcriptome profiles for Δrco1 and Δrco1Δrrp6 matching our ssu72-2 profiles. To distinguish between transcripts arising from gene 5′ or 3′ ends, we selected tandem genes separated by either more or less than 400bp (Fig. S4A). We showed that RRT expression (in regions where pSRTs are also detected) in Δrco1Δrrp6 vs Δrrp6 is clearly greater in close tandem gene configuration than in distant ones. This argues that RRTs are produced from gene terminator regions. We therefore performed a metagene analysis on tandem gene pairs more than 400bp apart that have a pncRNA arising between them. SRTs peak near the TSS, whereas RRTs align with the TTS (Fig 3B) also validated for specific tandem and divergent gene pairs (Fig. S4B). The terminator association of RRTs fits with the known gene 3′ end association of Rpd3S (18, 19). Collectively we show that, contrary to previous interpretation (16), antisense RRTs are terminator-derived, while SRTs are promoter-derived. Ssu72 thus enforces promoter directionality. We also detected a small but significant trend of decreased expression in ssu72-2 for tandem genes that generate pSRTs (Fig. S5). This suggests that loss of promoter directionality results in decreased genic transcription. Since Ssu72 is required for gene loop formation we tested if other gene loop associated factors similarly act to restrict pncRNA synthesis. Inactivation of TFIIB (a.k.a. Sua7) or other pAC components, Pta1, Rna14 and Rna15 have all been shown to restrict gene loop formation (10, 11). Similarly we show that their inactivation caused an increase in pncRNA in a range of S. cerevisiae genes (Fig. S6).
Figure 3. pSRTs initiate from bidirectional promoters.
A) I. Histone H4 acetylation (as a ratio with H3) compared over intergenic region between ORF TSS and divergent pSRT (blue) or pCUT (purple) TSS in wild type strains. Intergenic regions of pCUTs show higher H4 acetylation levels than those of pSRTs. II. ChIP analysis across the promoter regions of the indicated loci with WT and ssu72-2 derived chromatin using anti-H4ac. Ssu72 inactivation caused H4 acetylation increase at all 4 loci. Telomeric region (TELV1) used as negative control. Error bars represent SEM. B) Metagene analysis of Δrco1Δrrp6 vs Δrrp6 (green) and ssu72-2Δrrp6 vs Δrrp6 (blue) differential expression levels for all antisense ncRNAs that initiate between tandem genes in relative position to upstream gene TTS and the downstream gene TSS.
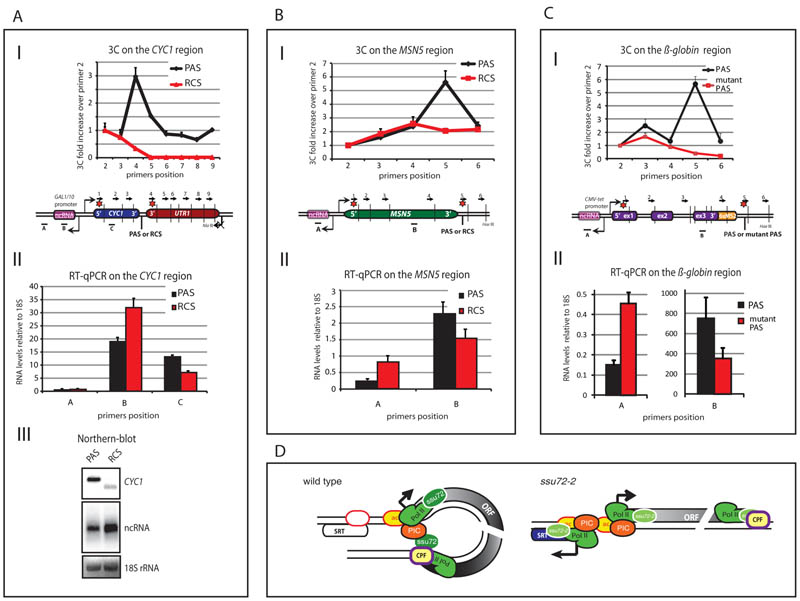
Since gene loops require both an active promoter and functional polyA signals (PAS) (20), we tested the effect of terminating transcription on pSRT formation by directly replacing the PAS with an Rnt1 cleavage signal (RCS) that promotes efficient termination but not mRNA polyadenylation (21). Plasmid constructs containing CYC1 with transcription initiated on a GAL1 promoter and terminated by either a PAS or RCS were transformed into Δrrp6 strain. Following galactose induction, chromatin was subjected to 3C analysis (Fig. 4AI). A clear peak of interaction between the promoter and PAS but not RCS was evident confirming that RCS-mediated Pol II termination prevents gene looping. Next we measured transcript levels of CYC1 mRNA and pncRNA in transformed Δrrp6 strains (Fig. 4AII, III). The GAL1 promoter associated pncRNA was enhanced in level when the CYC1 PAS was converted into an RCS due to loss of the PAS-dependent gene loop. In a genomic context conversion of MSN5 (which generates a pSRT; Fig. 2A, S2B) PAS into an RCS showed loss of gene looping and a 3-fold increase in pSRT production, mimicking the effect of Ssu72 inactivation (Fig. 4B). Finally, an integrated β-globin gene construct with an SV40 late PAS or mutated version (22) in human embryonic kidney cells (HEK293) displays a gene loop conformation with the wild type, but not mutant PAS construct. Similarly we observed a 3-fold increase in levels of divergent pncRNA with the mutated PAS (Fig. 4C) indicating a similar effect in a mammalian system.
Figure 4. Gene loop disruption by PAS mutation enhances divergent transcription.
A-C) I. Graphical representation of 3C interaction analyses for CYC1 plasmid (A), MSN5 (B) and mammalian β-globin gene construct (C) as in Fig. 1A. II, RT-qPCR analyses of mRNA versus pncRNAs. AIII. Northern blot for CYC1 transcripts. Note that the CYC1 mRNA is smaller with RCS replacement due to lack of pA tail. Also UTR1 downstream of CYC1 is inactive due to promoter deletion, denoted by crossed arrow. Error bars represent SEM. D) Gene loop defines transcription unit and promotes transcription directionality. Loss of gene loop in ssu72-2 or with PAS mutation increases antisense pncRNA transcription. Gene loops involving Ssu72 (green oval) may act to maintain nucleosome (yellow bubble; ac denotes H4Ac) positions, preventing association of a second PIC (red) leading to divergent transcription initiation.
Our results indicate that gene loops act to maintain the directionality of transcription. Indeed loss of a mammalian gene’s PAS can directly influence recruitment of transcription factors, with consequent reduction in gene expression (22). PAS mutation has also been shown to increase levels of divergent transcripts (23). Based on our results, such effects are directly explicable by the loss of gene loop formation and the potential to recycle factors from the terminator back to the promoter (see model, Fig. 4D). The role of Rpd3S in restricting antisense terminator transcripts (Fig. 3) clearly illustrates the importance of histone deacetylation in preventing inappropriate ncRNA synthesis. We predict that gene loops may similarly act to influence the recruitment of 5′ localised HDACs such as Set3 (24). This would maintain promoters in a deactylated, inactive state until gene activation selectively promotes transcription of genes rather than divergent pSRTs. We postulate that gene looping contributes to determining which transcription units are fully productive.
Supplementary Material
Acknowledgments
Thanks to Bernard Dichtl and Joanna Kufel for strains and Hashanthi Wijayatilake for FMP27 and β-globin 3C reagents. Supported by the Wellcome Trust (NJP), NIH and DFG (LMS), EMBL (JBZ, NML, LMS) and the SNF and EMBO (JC). Genomic data is deposited at http://steinmetzlab.embl.de/proudfoot_lab/index.html.
References
- 1.Mattick JS, Taft RJ, Faulkner GJ. Trends Genet. 2010 Jan;26:21. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P. Nature. 2009 Feb 19;457:974. doi: 10.1038/457974b. [DOI] [PubMed] [Google Scholar]
- 3.Core LJ, Lis JT. Science. 2008 Mar 28;319:1791. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preker P, et al. Science. 2008 Dec 19;322:1851. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 5.Seila AC, et al. Science. 2008 Dec 19;322:1849. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neil H, et al. Nature. 2009 Feb 19;457:1038. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 7.Xu Z, et al. Nature. 2009 Feb 19;457:1033. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Sullivan JM, et al. Nat Genet. 2004 Sep;36:1014. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 9.Ansari A, Hampsey M. Genes Dev. 2005 Dec 15;19:2969. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh BN, Hampsey M. Mol Cell. 2007 Sep 7;27:806. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Medler S, et al. J Biol Chem. 2011 Sep 30;286:33709. doi: 10.1074/jbc.M110.193870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang DW, et al. J Biol Chem. 2012 Mar 9;287:8541. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pappas DL, Jr., Hampsey M. Mol Cell Biol. 2000 Nov;20:8343. doi: 10.1128/mcb.20.22.8343-8351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinmetz EJ, Brow DA. Mol Cell Biol. 2003 Sep;23:6339. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokholok DK, et al. Cell. 2005 Aug 26;122:517. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 16.Churchman LS, Weissman JS. Nature. 2011 Jan 20;469:368. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray SC, et al. Nucleic Acids Res. 2011 Nov 28; [Google Scholar]
- 18.Carrozza MJ, et al. Cell. 2005 Nov 18;123:581. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Keogh MC, et al. Cell. 2005 Nov 18;123:593. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 20.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Mol Cell. 2008 Jan 18;29:56. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Mol Cell. 2009 Oct 9;36:88. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Mol Cell. 2010 Nov 12;40:410. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Preker P, et al. Nucleic Acids Res. 2011 May 19; doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim T, Buratowski S. Cell. 2009 Apr 17;137:259. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavrich TN, et al. Genome Res. 2008 Jul;18:1073. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Cell. 2007 Nov 16;131:706. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Wach A. Yeast. 1996 Mar 15;12:259. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Genes Dev. 2009 Nov 15;23:2610. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinmetz EJ, et al. Mol Cell. 2006 Dec 8;24:735. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Mischo HE, et al. Mol Cell. 2011 Jan 7;41:21. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto I, Ware DE, Hampsey M. Cell. 1992 Mar 6;68:977. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- 32.Torchet C, et al. Mol Cell. 2002 Jun;9:1285. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 33.Li B, et al. Genes Dev. 2007 Jun 1;21:1422. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venters BJ, Pugh BF. Genome Res. 2009 Mar;19:360. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Nature. 2007 Dec 13;450:1031. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.