Abstract
Targeting and inhibiting CMG2 (Capillary Morphogenesis Gene protein 2) represents a new strategy for therapeutic agents for cancer and retinal diseases due to CMG2’s role in blood vessel growth (angiogenesis). A high throughput FRET (Förster Resonance Energy Transfer) assay was developed for the identification of CMG2 inhibitors as anti-angiogenetic agents. Bioassay-guided separation led to the isolation and identification of two new compounds (1 and 2) from CR252M, an endophytic fungus Coccomyces proteae collected from a Costa Rican rainforest, and one known compound (3) from CR1207B (Aurapex penicillata). Secondary in vitro assays indicated anti-angiogenic activity. Compound 3 inhibited the endothelial cell migration at 52 µM, but did not show any endothelial cell antiproliferative effect at 156 µM. The structure of the two new compounds, A (1) and B (2), were elucidated on the basis of extensive spectroscopic analysis, including 1D and 2D NMR experiments.
Keywords: Fungus, Coccomyces proteae, Aurapex penicillata, CMG2, Phenolic
Anthrax Protective Antigen (PA) binds to two cell surface receptors, CMG2 and Tumor Endothelial Marker 8 (TEM8), which are responsible for allowing entry of anthrax toxin into host cells. PA and PASSSR, a mutated form of protective antigen, are anti-angiogenic, likely as a result of inhibition of CMG2-ECM (extracellular matrix) interactions.2 Thus, small molecules that bind to CMG2 and TEM8 have potential applications as both anthrax toxin antidotes and as agents for angiogenic diseases.2 Because PA is believed to use the natural ligand binding site on CMG2, compounds that inhibit the interaction of PASSSR with CMG2 are likely to inhibit tumor angiogenesis. In the course of identifying small molecules that can inhibit the interaction of PASSSR with the anthrax toxin receptor CMG2, we screened our sample libraries at Harvard Medical School’s high throughput screening facility (ICCB-L, Institute of Chemistry and Cell Biology at Longwood) using a high-throughput FRET assay. Our focus on natural products was based on an observation in a pilot screen that small molecule natural products, including those from endophytic fungi collected at Costa Rica, were frequently active for inhibition of the CMG2/PASSSR interaction. The crude extracts of the fungi CR252M3 and CR1207B3 inhibited the interaction of PASSSR with CMG2 and were selected for bioassay-guided separation. CR252M was first fractionated over a C18 SPE column, and fraction II was further separated by phenyl-hexyl prep-HPLC and then C8 semi-prep-HPLC to yield compounds 1 and 2.4 Compound 3 was obtained from CR1207B, after C18 SPE and C18 HPLC separation.4
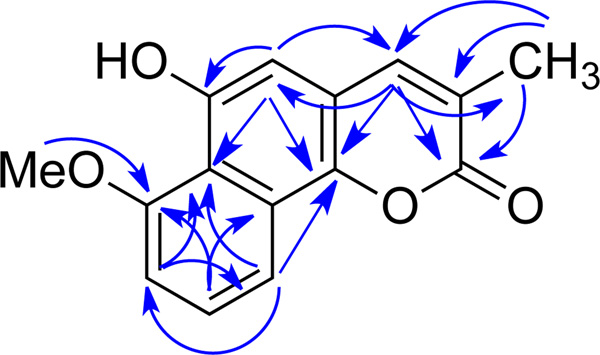
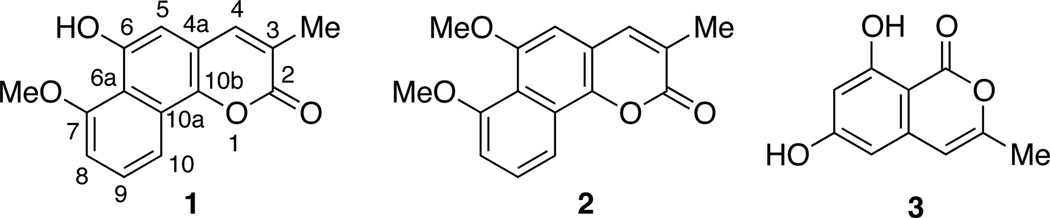
Compound 1 was obtained as a yellow solid. Its UV absorptions in MeOH, with λmax (log ε) 213 (3.16), 227 (3.24), 277 (3.28), 287 (3.31), 315 (sh), 390 (2.54) nm, indicated the presence of an extended coumarin moiety similar to that of neolambertellin.5,6 The IR spectroscopic data of compound 1, which showed absorption at 1700 cm−1, confirmed the existence of the C=O functional group. The positive ion HREIMS of 1 revealed a quasimolecular ion peak at m/z 257.0813 [M + H]+ corresponding to C15H13O4, (calculated for C15H13O4: 257.0814). The proton NMR spectrum of 1 broad singlet (δH 7.52, br s, H-4), one aromatic broad singlet (δH 6.76, br s, H-5), an ABC spin system (δH 8.14, d, J = 8.4 Hz, H-10; 7.49, t, J = 8.4 Hz, H-9; 6.97, d, J = 8.4 Hz, H-8), one methoxy (δH 4.10, s, 6-OMe), and one methyl singlet (δH 2.26, s, 3-Me). In the HMBC (Figure 1) spectrum, the 3-methyl group (δH 2.26) exhibited correlations to C-2 (δC 164.1, a lactone carbonyl), C-3 (δC 126.9), and C-4 (δC 141.4). The olefinic proton H-4 (δH 7.52) correlated to C-2, C-10b (δC 143.6), C-5 (δC 106.1), and 3-CH3 (δC 17.1); while H-5 (δH 6.76) demonstrated a 2J HMBC correlation to C-6 (δC 152.0, an oxygenated aromatic carbon) and 3J HMBC correlations to C-4, C-10b, and C-6a (δC 117.6), indicating a coumarin moiety with a methyl group at 3-position and a hydroxyl group at 6-position. Besides, the following HMBC correlations were also observed: from the 7-methoxy (δH 4.10) to C-7 (δC 157.2, an oxygenated aromatic carbon), H-8 (δH 6.97) to C-6a and C-10 (δC 115.8), H-9 (δH 7.49) to C-7 and C-10a (δC 126.4), and H-10 (δH 8.14) to C-6a, C-10b, and C-8 (δC 108.0), which indicated that the second aromatic ring was connected to the coumarin at 6a- and 10a-positions. Hence, the structure of 1 (7-O methyl neolambertellin) was determined as shown.7
Figure 1.
Key HMBC correlation of 1
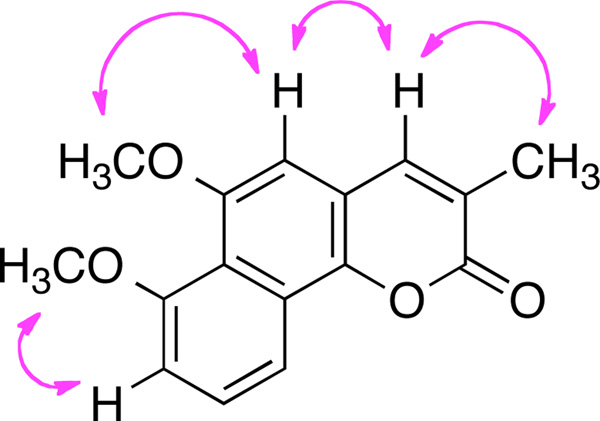
Compound 2 was also obtained as a yellow solid, and had a molecular formula of C16H14O4, which was 14 mass units more than 1. The proton NMR spectrum of 2 was similar to that of 1 except for the presence of the 6-O methyl group. The proton signals included one olefinic broad singlet (δH 7.92, br s, H-4), an aromatic singlet (δH 7.02, br s, H-5), an ABC spin system (δH 7.14, d, J = 7.8 Hz, H-8; 7.59, t, J = 7.8 Hz, H-9; 7.86, d, J = 7.8 Hz, H-10), two methoxy groups (δH 3.87, s, 6-OMe; 3.88, s, 7-OMe), and one methyl singlet (δH 2.14, s). Hence, it was readily deducted that 2 was the methylated product of compound 1. The ROESY (Figure 2) correlations between the 3-methyl (δH, 2.14, s) and H-4 (δH 7.92), H-4 and H-5 (δH 7.02), the 6-methoxy (δH 3.87) and H-5, and the 7-methoxy (δH 3.88) and H-8 (δH 7.14) conformed the structure of 2 as shown.8
Figure 2.
Key ROESY correlations of 2
Compound 3 from CR1207B was identified to be 6,8-dihydroxy-3-methyl-1H-isochromen-1-one.9–11
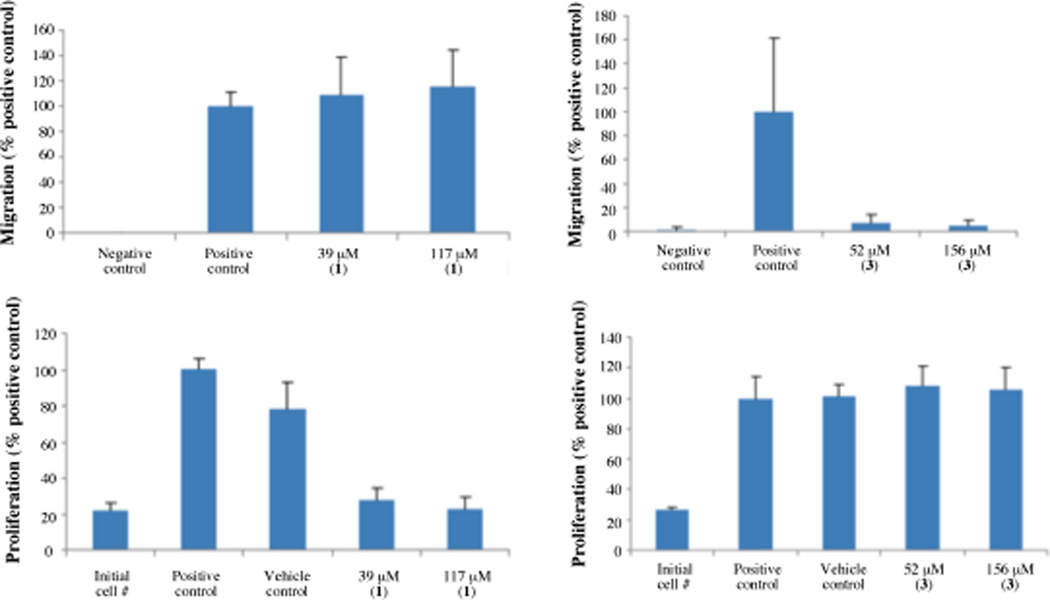
Both 1 and 2 are new compounds, and 1 and 3 were active in the CMG2 FRET assay12,17 with IC50 values of 0.5 and 0.6 µM, respectively, while compound 2 was inactive. Compound 2 did not inhibit CMG2 in the FRET assay, even though it was structurally similar to compound 1, which did inhibit. Compound 1, at 39 and 117 µM, did not inhibit endothelial cell migration,2,13,17 but significantly inhibited endothelial cell proliferation.2,14 On the other hand, compound 3 did not inhibit endothelial cell proliferation at 52 and 156 µM, but inhibited endothelial cell migration by ~96.0% and ~93.1%, respectively (Figure 3). Upregulation of CMG2 during endothelial tubule formation in collagen gels suggests it may have an important functional role in the angiogenic process.15 Reeves et al have demonstrated that expression of CMG2 regulates the capacity of macrovascular endothelial cells to proliferate and form capillary networks in vitro.16 PASSSR has been shown to inhibit the migration of endothelial cells at CMG2-inhibitory concentrations, with minimal inhibition of the second anthrax receptor, TEM8.2 Endothelial cells overexpressing CMG2 migrate much faster than the regular endothelial cells.2 Inhibition of their migration means inhibition of angiogenesis. Since endothelial cell migration is a commonly used predictor of antiangiogenic activity, it is interesting that compound 3 inhibited not only CMG2 but also endothelial cell migration without any obvious anti-proliferative activity. Further studies of its effect on angiogenesis in animals may confirm it as a candidate for therapy of angiogenic diseases.
Figure 3.
Effect of 1 and 3 on endothelial cell migration (top panel) and cell proliferation (bottom panel). For the migration assay panel, the positive control is full serum media with vehicle (DMSO) and the negative control is serum free media with 0.1% BSA, which is unable to support endothelial cell migration; In the proliferation assay panel, initial cell number refers to the readout from wells fixed in 100% ethanol at the start of the assay, thus being unable to proliferate further. The positive control is full serum media, and the vehicle control full serum media with DMSO vehicle; n = 3 for 1 and n = 2 for 3, p < 0.01.
Supplementary Material
Chart 1.
Structures of compounds 1 to 3
Acknowledgments
This work was generously supported by NIH U01 TW007404 (J.C. & G. T.), NIH U54 AI057159, and Department of Defense BC074070P1-02 (M.R and J.C.). We also thank Thomas Böttcher for photographing CR252M and reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at xxxxxx
References and notes
- 1.Discovery of Natural Product Based Drugs and Bioenergetic Materials from Costa Rica Biota, recent publication, see Cao S, Ross L, Tamayo G, Clardy J. Org. Lett. 2010;12:4661. doi: 10.1021/ol101972g. Cao S, Clardy J. Tetrahedron Lett. 2011;52:2206. doi: 10.1016/j.tetlet.2010.11.159. Ymele-Leki P, Cao S, Sharp J, Lambert KG, McAdam AJ, Husson RN, Tamayo G, Clardy J, Watnick PI. PLos ONE. 2012;7(2):e31307. doi: 10.1371/journal.pone.0031307. Cao S, McMillin DW, Tamayo G, Delmore J, Mitsiades CS, Clardy J. J. Nat. Prod. 2012;75:793. doi: 10.1021/np2009863.
- 2.(a) Rogers MR, Christensen KA, Birsner AE, Short SM, Wigelsworth DJ, Collier RJ, D’Amato RJ. Cancer Res. 2007;60:9980. doi: 10.1158/0008-5472.CAN-07-0829. [DOI] [PubMed] [Google Scholar]; (b) Cryan LM, Rogers MS. Targeting the anthrax receptors, TEM-8 and CMG-2, for anti-angiogenic therapy. Front Biosci. 2011;16:1574. doi: 10.2741/3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collection of fungal strains. CR252M was obtained as an endophytic isolate from the leaves of Disterigma humboldtii, collected in Braulio Carrilo National Park (collection permit R-012-2005-OT-CONAGEBIO) in January, 2006. CR 1207B was isolated from the petiole of Geonoma hoffmanniana, in the summit of Cacao Volcano in October, 2006 (collection permit R-CM-INBio-03-2006-OT). Vouchers of both, fungal isolates and host plants, are kept in the INBio’s Herbarium. Sequencing and species identification. For identification by internal transcribed spacer (ITS) sequencing, CR252M and CR1207B were cultured in the below-mentioned rich media (in the “Culturing” section) for 6 days. The mycelia were then retrieved by filtration and ground to a fine powder in liquid N2. Genomic DNA of CR252M was extracted using the SurePrep RNA/DNA/protein purification kit (Fisher Bioreagents), and large subunit rDNA was amplified by PCR using primers ITSF1 (ACAAGGTTTCCGTAGGTGA), LR5 (5′-TCCTGAGGGAAACTTCG-3′) and LROR (5′-ACCCGCTGAACTTAAGC-3′). Genomic DNA of CR1207B was extracted using the Wizard Genomic DNA Purification Kit (Promega), and large subunit rDNA was amplified by PCR using primers ITS4 (TCCTCCGCTTATTGATATGC) and ITS5 (GGAAGTAAAAGTCGTAACAAGG). PCR products were sequenced at Genewiz (http://www.genewiz.com/). The DNA sequence data obtained from the fungal strains CR252M and CR1207B have been deposited at GenBank with accession numbers JX089385 and JQ864529, respectively. >252M TACAGAGACCTTGCACCCACACAACCGAAGTAGAAGGAAAATCACGGGCTCCCGTGATCACCTACAGCGAACCCCCCCGGGATCGTCTACAGACTAAGTGGTTGTGGGCGGGGCCTGGAGCCCTGCTTAAGATATAGTCGATACCCACGCCTCGCGGCCTGGGATAGCTAGTCCGTAGGTGAACCTGCGGAAGGATCATTACTGAGATTTGTCCTCCGGGACGGACCTCCAACCCTATGTTTACTGTACCATGTTGCTTCGGCAGGCCGGCCTCACGGCTACCAGCCCCTCCTAAGGGGGCTGGCCAGTGCCTGCCAGAAGCCCCACGAAACTCGTTTTGTCAGTGTCGTCTGAGTAAATTTTAATAATTAAAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCACCCTCTGGTATTCCGGAGGGTATGCCTGTTCGAGCGTCATTAAACACATTCAAGCCTGGCTTGGTATTGGGACGCGCCTGCCCGGCGCCCCCTAAAATCAGTGGTCGCCGCAGCACAACTTCCAGCGTAGTTACCTTTCTCGCTGGGGAGTTGGAGCCTGTGGTCCGCCAGAATAGTAGCTCCTTAGAGTTTGACCTCGGATCAGGTAGGGATACCCGCTGAACTTAAGCATATCAATAAGCGGAGGAAAAGAAACCAACAGGGATTGCCTTAGTAACGGCGAGTGAAGCGGCAACAGCTCAAATTTGAAATCTGGCTTCGGCCCGAGTTGTAATTTGTAGAGGATGCTTAGGGTGTGGTTCCGGTCTAAGTTCCTTGGAACAGGACGTCATAGAGGGTGAGAATCCCGTATGTGACCGGCCACCTTCGCCCGTGTTAAGCTCCTTCGACGAGTCGAGTTGTTTGGGAATGCAGCTCTAAATGGGTGGTAAATTTCATCTAAAGCTAAATACTGGCCAGAGACCGATAGCGCACAAGTAGAGTGATCGAAAGATGAAAAGCACTTTGGAAAGAGAGTTAAATAGTACGTGAAATTGTTGAAAGGGAAGCGCTTGCAACCAGACTTGCGCGCAGTTGATCATCCGGTGTTCTCACCGGTGCACTCGGTTGCGCTCAGGCCAGCATCAGTTCTGGTGGTTGGATAAAGGCCCAGGGAATGTGGCTCCTCTCGGGGAGTGTTATAGCCCTGGGTGCAATGCAGCCTACCGGGACTGAGGACCGCGCTTCGGCTAGGATGCTGGCGTAATGGTTGTAAGCGACCCGTCTTGAAACACGGACCAAGGAGTCTAACATCTATGCGAGTGTTTGGGTGTCAAACCCATACGCGTAATGAAAGTGAACGGAGGTGAGAGCCCTTAAGGGTGCATCATCGACCGATCCTGATGTCTTCGGATGGATTTGAGTAAGAGCATAGCTGTTGGGACCCGAAAGATGGTGAACTATGCGTGATTAGGGTGAAGCCAGAGGAAACTCTGGTGGAGGCTCGTAGGGGTTCTGACGTGCAAATCGATCTCaAAAATTGCGTATAGGGGCGAAAGACTAATCGAACCATCTAAACCGACTGCTGGATGGAATATTACAGCGTAGTCTGGCCGCGGCCAGGCAACACTACCACTGTGCGAGTATGCCGAGAG GCTACCTGCAGCGGGGCCCAGCGCCCCGTCCACAGACTAAAC GGTAGTGGCCCCCCGGGGTTCTGATATAGCCGATTCCCACTGGCAAGTG >1207B GATATGCTTAAGTTCAGCGGGTATTCCTACCTGATCCGAGGTCAAATTTTCAGAATATTAGGGGTTTTTACGGCAAGAAGCAACCGCTAATCCTTCCAAAGCGAGGTTGAAAAAACTACTACGCTCAGAGTCTTAGCGAGCCCGCCACTAAATTTCAGGGCCTACCGTTTTACGGGTAGTGCCCCAACACCAAGCTAGGCTTGAGGGTTGAAATGACGCTCGAACAGGCATGCCCGCTGGAATTCCAGCGGGCGCAATGTGCGTTCAAAGATTCGATGATTCACTGAATTCTGCAATTCACATTACTTATCGCATTTCGCTGCGTTCTTCATCGATGCCAGAACCAAGAGATCCGTTGTTGAAAGTTTTGATTCATTTGTTTATTTATTAACTCAGAGGAGATACGTTATCAAAACAAAAGAGTTTAATGGGCCGCCGGCGGGCCTGCTCCAACACCGTTTGCACGGTGAGGGAGGGGGAGGCGGACAAAAAAAAGAATTTGTCCAAGCCTCCCCCCCGGGCTCGGCGCCGAGGCAACGATAAAAAAGGTATAAGTTCACATAGGGTATCTGGGGTTGTGCCCGTGAGGGCACAGTTCCAGCAATGATCCCTCCGCTGGTTCACCAACGGAGACCTTGTTACATA
- 4.General Experimental Procedures. IR and UV spectra were measured on Bruker Alpha-P and Amersham Biosciences Ultrospec 5300 pro, UV/visible spectrophotometers, respectively. 7-O-methyl neolambertellin (1) and 6,7-O,O-dimethyl neolambertellin (2) were purified from CR252M on an Agilent 1100 series HPLC (Agilent Technologies) using a preparative Phenomenex Luna Phenyl-hexyl column (250 × 21.2 mm, 5 µm particle size; flowrate: 10 mL/min) and an Eclipse XDB-C8 HPLC column (250 × 9.4 mm, 5 µm particle size; flowrate: 2 mL/min). Compound 3 was purified from CR1207B using a Phenomenex Luna C18 column (250 × 21.2 mm, 5 µm particle size; flowrate: 10 mL/min). All NMR experiments were carried out on a Varian INOVA 600 MHz spectrometer. HREIMS data were obtained on a Waters Micromass 70-VSE. Culturing. Agar plugs of CR252M were initially grown at 25 °C on yeast malt agar plates supplemented with 30 µg/mL streptomycin and 12 µg/mL chlortetracycline. After one week, 3 macerated agar plugs from this plate were placed in 75 mL media [potato dextrose broth (24 g/L)] in a 125 mL erlenmeyer at a pH of 6.1. It was grown at 28 °C and 150 rpm for 6 days. Then the grains of barley (200 g) and wheat (200 g) were ground with the aid of a mixer and subsequently the rest of components [1 mL of magnesium chloride (20 mg/mL solution stock), 2 grams of peptone, and 250 mL of distilled water] were added. The flasks containing 24.4 g of solid media, each, were autoclaved for 30 minutes at 121 °C. Each flask was inoculated with 15 mL of seed medium; and it was incubated at 25 °C for 21 days. Agar plugs of CR1207B were initially grown at 25 °C on yeast malt agar plates supplemented with 30 µg/mL streptomycin and 12 µg/mL chlortetracycline. After one week, agar plugs from this plate were placed in 75 mL of rich seed media [5 g peptone, 10 g dextrose, 3 g yeast extract, 10 g malt extract per 1 L water (pH 6.2)] in a 125 mL Erlenmeyer at a pH of 6.2. They were grown at 25 °C and 150 rpm for 6 days. 150 mL of 0.66% (w/v) malt extract and 5 g HP-20 resin were then added to each flask with 15 mL of the seed media in a 250 mL erlenmeyer, and the fungi were cultured under the same conditions for 16 days. The fungal cultures were then incubated at 25 °C without shaking for 5 days. Extraction and separation. The solid media (24.4 g × 4) of CR252M was extracted with 100 mL 90% EtOH three times, and the extract was concentrated under vacuum. The crude extract of CR252M in 90% H2O/MeOH was passed through a C18 SPE wand then eluted with methanol. The methanol eluent was dried with rotary evaporator. The methanol extract was fractionated using a phenyl-hexyl column on a HPLC. The active fraction SC1-101-8 (tR 34.5 min) after phenyl-hexyl HPLC was further separated with 50% MeCN/H2O on an Eclipse XDB-C8 HPLC column to yield compound 1 (tR 16.5 min, 1 mg/ 100g); while compound 2 (tR 18 min, 0.3 mg/100 g) was obtained with 45% MeCN/H2O. Flash chromatography of the crude CR1207B from 1.05 L (150 mL × 7) was dissolved in MeOH and filtered over SPE C18 to yield three fractions. The main fraction, fraction II, was subjected to a C18 prep-HPLC column (20–100% MeCN/H2O in 30 minutes), and five sub-fractions were collected. Sub-fraction four yielded the known compound 3 (tR 25.2 min, 3 mg/L), 6,8-dihydroxy-3-methyl-1H-isochromen-1-one.
- 5.Murakami T, Hashimoto M, Okuno T. Biorg. Med. Chem. Lett. 2005;15:4185. doi: 10.1016/j.bmcl.2005.06.082. [DOI] [PubMed] [Google Scholar]
- 6.Schuetz A, Murakami T, Takada N, Junker J, Hashimoto MM, Griesinger C. Angew. Chem. Int. Edit. 2008;47:2032. doi: 10.1002/anie.200705037. [DOI] [PubMed] [Google Scholar]
- 7.7-O-methyl Neolambertellin (1): yellow powder; UV (MeOH) λmax (log ε) 213 (3.16), 227 (3.24), 277 (3.28), 287 (3.31), 315 (sh), 390 (2.54) nm; IR ν 3384, 2931, 2919, 1700, 1592, 1402, 1390, 1352, 1074, 1061 cm−1; 1H NMR (600 MHz, CD3OD): δH 2.21 (brs, 3-CH3), 4.11 (s, 7-OCH3), 6.81 (s, H-5), 7.15 (d, J = 8.4 Hz, H-8), 7.56 (t, J = 8.4 Hz, H-9), 7.76 (brs, H-4), 8.00 (d, J = 8.4 Hz, H-10); 1H NMR (600 MHz, CDCl3): see Table 1; 13C NMR (150 MHz, 50% CD3OD/CDCl3): see Table 1; HREIMS m/z 257.0813 ([M+H]+, calcd for C15H13O4, 257.0814).
- 8 6,7-O,O-dimethyl Neolambertellin (2): yellow powder; UV (MeOH) λmax (log ε) 212 (2.95), 226 (3.03), 277 (3.01), 286 (3.03), 314 (sh), 385 (2.28) nm; IR ν 1700, 1590, 1400, 1391, 1350, 1075, 1060 cm−1; 1H NMR (600 MHz, DMSO-d6): see Table 1; 1H NMR (600 MHz, 90% CD3OD/CDCl3): δH 2.23 (d, J = 1.2 Hz; 3-CH3), 3.95 (s, 6-OCH3), 3.96 (s, 7-OCH3), 6.95 (s, H-5), 7.14 (d, J = 7.8 Hz, H-8), 7.58 (t, J = 7.8 Hz, H-9), 7.84 (brs, H-4), 8.03 (d, J = 7.8 Hz, H-10); 13C NMR (150 MHz, DMSO-d6): see Table 1; HREIMS m/z 271.0968 ([M+H]+, calcd for C16H15O4, 271.0970).
- 9.Hill RA, Carter RH, Staunton J. Chem Comm. 1975:380. [Google Scholar]
- 10.Kendall JK, Fisher TH, Schultz HP, Schultz TP. J. Org. Chem. 1989;54:4218. [Google Scholar]
- 11.6,8-dihydroxy-3-methyl-1H-Isochromen-1-one (3): spectroscopic data consistent with literature.
- 12.CMG2 FRET assay:17 Briefly, for high throughput screening, 30 µL of a solution of 17 nM CMG2R40C*AF546 in HBST was added to the wells of a barcode labeled Corning 3710 384-well-plate using a WellMate liquid handling robot (Matrix Technologies) with integrated stacker. Next, 0.3 µL of test compound (5 – 10 mg/mL) diluted in DMSO was added by pin transfer using a custom Epson robot to duplicate plates. Following a 1–3 h incubation, 10 µl of a 30 nM PAE733C*AF488 solution in 50 mM HEPES pH 7.4, 150 mM NaCl, 0.1 mM CaCl2 was then added to all wells using the Wellmate and plates were incubated for 3–4 h. Final CMG2 concentration (13 nM) and PA concentration (7.5 nM) were sufficient to promote quantitative binding of CMG2 in the absence of effective inhibitors, based on the previously reported Kd (≥ 170 pM). Incubation lengths varied minimally between individual wells, as a function of the time required for delivery of library compounds to individual positions in the well-plate. Following incubation, plates were read on an Envision (PerkinElmer) plate reader using a 485/14 excitation filter, with 535/25 and 595/60 emission filters incorporating a barcode reader to correlate fluorescence measurements with plates. For each plate, 32 positive control wells were generated by adding 10 mM EDTA to the CMG2 solution; 32 negative control wells were generated by addition of 10 mM NaCl to the CMG2 solution.
- 13.Endothelial cell migration assay:2,18 Human microvascular endothelial cells (Lonza) were maintained in EGM-2 media (Lonza) according to the vendor's instructions and used before passage 8. Polycarbonate transwell inserts, 6.5 mm diameter with 8.0 µm pores (Corning), were coated with 20 µg/mL fibronectin (Sigma) overnight at 4 °C. Cells were harvested and resuspended in EBM media (Lonza) containing 0.1% bovine serum albumin (Sigma). Cells (10,000–20,000 per well) were plated onto wells and placed within wells containing full serum EGM-2 medium alone or FS EGM-2 medium containing the molecule to be tested. Cells were allowed to migrate for 4 h at 37 °C, 5% CO2. Membranes were then fixed and processed using Diff-Quick staining solution (Dade Diagnostics). Cells on the top of the membrane were removed using cotton-tipped applicators. Membranes were removed from the insert using a scalpel and mounted on slides, and the number of cells in 4 10× microscopic fields were counted.
- 14.Endothelial cell proliferation assay:2a Endothelial cells were plated in a 96-well plate with 2,000 cells/well in full serum EGM-2 media (Lonza) overnight at 37 °C. After 16–24 h, media was removed and cells were treated with full serum media +/− compound treatment or vehicle control at various concentrations in triplicate. For three additional wells, cells were fixed with 100% EtOH, to allow measurement of initial cell number. After incubation for either 24 h or 72 h plates were washed with 200 µL PBS and placed in an −80 °C freezer. Cyquant (Invitrogen) was added to each well according manufacturer’s instructions and fluorescence at 485/545 nm was measured on a Wallac platereader in order to quantify relative cell number.
- 15.Bell SE, Mavila A, Salazar R, Bayless KJ, Kanagala S, Maxwell SA, Davis GE. J. Cell Sci. 2011;114:2755. doi: 10.1242/jcs.114.15.2755. [DOI] [PubMed] [Google Scholar]
- 16.Reeves CV, Dufraine J, Young JA, Kitajewski J. Oncogene. 2010;29:789. doi: 10.1038/onc.2009.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wigelsworth DJ, Krantz BA, Christensen KA, Lacy DB, Juris SJ, Collier RJ. J. Biol. Chem. 2004;279:23349. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- 18.QCM™ Endothelial Cell Migration Assay: http://www.millipore.com/catalogue/item/ECM202
Table 1.
| 1H | 13C | |||
|---|---|---|---|---|
| # | 1 | 2 | 1 | 2 |
| 2 | 164.1, C | 164.6, C | ||
| 3 | 126.9, C | 128.6, C | ||
| 4 | 7.52, br s | 7.92, br s | 141.4, CH | 143.1, CH |
| 4a | 116.7, C | 117.0, C | ||
| 5 | 6.76, br s | 7.02, br s | 106.1, CH | 105.3, CH |
| 6 | 152.0, C | 156.5, C | ||
| 6a | 117.6, C | 120.4, C | ||
| 7 | 157.2, C | 160.2, C | ||
| 8 | 6.97, d (8.4) | 7.14, d (7.8) | 108.0, CH | 112.1, CH |
| 9 | 7.49, t (8.4) | 7.59, t (7.8) | 128.5, CH | 131.5, CH |
| 10 | 8.14, d (8.4) | 7.86, d (7.8) | 115.8, CH | 116.2, CH |
| 10a | 126.4, C | 128.9, C | ||
| 10b | 143.6, C | 146.2, C | ||
| 3-Me | 2.26, s | 2.14, s | 17.1, CH3 | 19.9, CH3 |
| 6-OMe | 3.87, s | 59.4, CH3 | ||
| 7-OMe | 4.10, s | 3.88, s | 56.9, CH3 | 59.4, CH3 |
δ (ppm) 600 MHz; multiplicities; J values (Hz) in parentheses; 1 in CDCl3; 2 in DMSO-d6.
δ (ppm) 150 MHz; 1 in 50% CDCl3/CD3OD; 2, in DMSO-d6, chemical shifts from gHSQC and gHMBC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.