Abstract
Studies integrating clinicopathological and genetic features have revealed distinct patterns of genomic aberrations in Melanoma. Distributions of BRAF or NRAS mutations and gains of several oncogenes differ among melanoma subgroups while 9p21 deletions are found in all melanoma subtypes.
In the study, status of genes involved in cell cycle progression and apoptosis were evaluated in a panel of 17 frozen primary acral melanomas.
NRAS
mutations were found in 17% of the tumours. In contrast, BRAF mutations were not found. Gains of AURKA gene (20q13.3) were detected in 37.5% of samples, gains of CCND1 gene (11q13) or TERT gene (5p15.33) in 31.2% and gains of NRAS gene (1p13.2) in 25%. Alterations in 9p21 were identified in 69% of tumours. Gains of 11q13 and 20q13 were mutually exclusive; and 1p13.2 gain was associated with 5p15.33.
Our findings showed that alterations in RAS related pathways are present in 87.5% of acral lentiginous melanomas.
Keywords: Acral lentiginous melanoma, melanoma, MLPA, NRAS, AURKA
Introduction
Molecular studies have revealed the existence of different biological subsets of melanomas based on the patterns of alterations identified (1–3), some of which correlated with degree of chronic sun-induced damage and site of origin (3). Furthermore, such molecular differences could result in clinical and histopathological differences among lesions (4, 5).
Melanomas classified as acral lentiginous melanomas (ALM) develop on volar skin, usually unexposed to UV radiation and are characterized by the presence of an atypical lentiginous proliferation. ALM carry a high number of genomic alterations compared to other melanoma subtypes and most of them account for a smaller proportion of genome (1, 3). The molecular hallmarks of ALM are CCND1 amplifications (1, 6, 7) or somatic mutations in c-KIT (8).
Deletions in the 9p21 region where the CDKN2A gene is located are widely detected (9–11). However, other genes from this region could be implicated in melanoma since retention of the CDKN2A locus has been found in tumours with deletions at one or both sides of CDKN2A (10). Other reported aberrations include large amplifications of 12q (1, 3), 7q or 20q and gains localized at 5p15, 11q13, 11q14 (3) and 22q11-13 (1).
Questions addressed
To characterize acquired molecular genomic alterations in a set of ALM from Spanish patients. The study was focused on specific chromosomal regions where genes involved in signalling pathways, cell cycle progression and apoptosis are located.
Experimental design
Seventeen fresh-frozen histopathologically confirmed primary ALMs based on Clark’s classification were included. Sampling was guided by ex vivo dermoscopy (12) and documented by photography without altering the specimen, immediately fixed in formalin and embedded in paraffin (FFPE) for conventional histopathological diagnosis following the step sectioning protocol for melanoma. Clinical data is described in Table S1. Genomic characterization of the BRAF, NRAS, CDKN2A and MC1R genes was performed by PCR-direct sequencing. Deletions of the 9p21 region and gains of regions wherein oncogenes of interest are localized were carried out by the Multiplex Ligation-dependent Probe Amplification (MLPA) approach (Data S1). Significance of AURKA gain into protein was evaluated by inmunohistochemistry method (Data S1).
Unsupervised hierarchical clustering of amplified regions detected was carried out using Cluster 3.0 developed by Eisen Lab (University of California, Berkeley, California, USA) using Pearson’s correlations distance and average linkage clustering. The study was approved by the institutional review board of Hospital Clinic of Barcelona (Spain) and tumours were from the sample collection of Melanoma Unit at the Hospital Clínic of Barcelona.
Results
MC1R variants were found in 62.5% of patients (Table S1). NRAS missense mutations were detected in three out of 17 ALMs. In contrast, activating BRAF mutations were not present (Table S1).
9p21 alterations were detected in 69% of tumours, one sample failed to yield a result (Table S1). Whilst five tumours carried focal deletions of CDKN2A, CDKN2B and/or MTAP, 6 showed loss of the 9p21 region. No mutations were detected in CDKN2A.
Copy number gains were detected in 12 different loci (Table 1). All melanomas showed at least one altered locus; 43.7% of tumours presented two and 25% presented ≥ three loci. Four alterations were detected in at least 25% of samples: 1p13.2 (NRAS gene), 5p15.33 (TERT gene), 20q13.3 (AURKA gene) and 11q13 (CCND1 and contiguous genes).
Table 1.
List of oncogenes analyzed by the MLPA approach
| Chromosomic Region | Oncogenes | Frequency of tumours with amplifications |
|---|---|---|
| r01p13.2 | NRAS | 25% |
| r01p22.1 | BCAR3 | 0 |
| r01q32 | MDM4 | 0 |
| r03q27 | BCL6 | 0 |
| r05p15.33 | TERT | 31.2% |
| r05q13 | BIRC1 | 12.5% |
| r06p21 | CCND3 | 0 |
| r07q21.3 | CDK6 | 6.2% |
| r08q11 | MOS | 18.8% |
| r11q13 | RELA, GSTP1, CCND1, EMS1, FGF3, | 31.2%* |
| r11q22 | BIRC3 | 6.2% |
| r12p13.32 | CCND2 | 0 |
| r12p13.2 | BCLG | 0 |
| r12q14 | CDK4 | 12.5% |
| r12q14.3 | MDM2 | 12.5% |
| r13q12.3 | CCNA1 | 0 |
| r14q32.33 | AKT1 | 0 |
| r17q25 | BIRC5 | 6.2% |
| r18q21.3 | BCL2 | 0 |
| r19q12 | CCNE1 | 6.2% |
| r20q11.1 | BCL2L1 | 0 |
| r20q13.1 | PTPN1 | 0 |
| r20q13.3 | AURKA | 37.5% |
| r20q13.33 | FLJ20517 | 0 |
| Xq25 | BIRC4 | 0 |
Frequency regarding gains of the CCND1 gene
Unsupervised hierarchical clustering was carried out to analyze the distribution of these recurrent alterations. Although the analysis was not statistically significant, it suggested the presence of different patterns (Fig. 1). Any sample with 20q13.3 gain (37.5%) presented 11q13 gain (31.25%). Furthermore, gains of 1p13.2 (25%) were more associated with 5p15.33 gains. Deletions in 9p21 were associated with any specific gained region.
Figure 1.
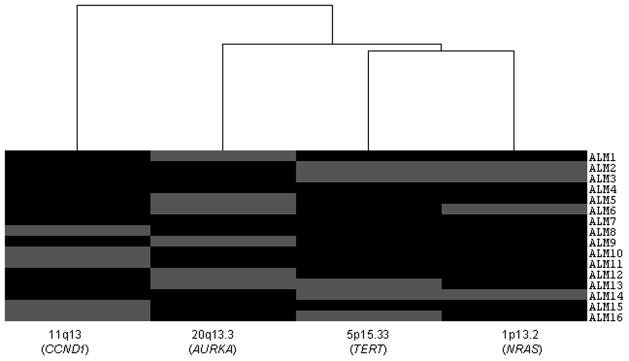
Hierarchical Clustering Subgroups of Acral lentiginous melanoma: Distribution of recurrent gains (11q13, 20q13.3, 5p15.33, 1p13.2). Amplification of a given region is indicated by grey box.
Aurora A protein was evaluated in the corresponding FFPE biopsies of 12 samples. Among samples harbouring AURKA gains (6 tumours) inmunohistochemistry failed in 2. Protein expression was detected in 3 out of 4 tumous in a range of 10% of cells up to 75% of cells (Figure S1). In contrast; Aurora A expression was not detected in samples without alteration.
Conclusion
Although the study showed a high proportion of patients harbouring germinal MC1R variants, the frequency of them did not differ from the frequency detected in the control population (data not shown). Activating NRAS mutations were detected in ALM as previously described (7, 13–15). BRAF mutations were not present in any sample which could be explained by the large proportion of samples carrying other deregulated genes located downstream of the MAPK pathway (CCND1 or CDK4) as described by Curtin et al. (3).
Focal amplifications have been described as molecular markers of ALM (1, 3). The most frequently gained loci were at 1p13.2, 5p15.33, 11q13 and 20q13.3. Our data suggests the existence of different ALM subgroups based on distribution of these alterations. Although a large set of tumours should be analyzed to obtain statistical power to detect such profiles, there is plausible biological evidence to support the existence of such patterns. A group of tumours carried 20q13.3 gains (AURKA gene) which has been reported previously in melanoma (16). AURKA overexpression which has been closely related to gene amplification or genetic instability (17, 18), could be implicated in promoting cancer cell survival activating Akt and stimulating the PI3K pathway (19). Interestingly, AURKA may converge upon oncogenic Ras signalling through the RALGEF pathway (20, 21). Another subgroup presented 11q13 gains including CCND1 which is a frequent initial molecular event in ALM (2, 6, 7, 22). Gains of AURKA and CCND1 were mutually exclusive, suggesting that both genes would lead to deregulation of cell proliferation in the same way. Revalidation of these results has been carried out by FISH methodology in a large set of ALM obtaining concordant findings (manuscript in preparation).
The third subgroup presented 1p13.2 gains (NRAS), frequently associated with 5p15.33 (TERT) alterations. Increased copy number of NRAS has been described previously (9, 23, 24).
In summary, based on these results, we hypothesize that alterations in cell progression genes (NRAS, AURKA or CCND1) could play similar roles as driver alterations in ALM. Further studies have to be performed on a large set of Acral lentiginous melanoma in order to elucidate the existence of these patterns and also to evaluate the cross-talk between downstream NRAS pathways and melanoma development.
Supplementary Material
Acknowledgments
S.P and J.M designed the research study. C.C and P.A collected tumour biopsies and obtained the clinical data. J.A.P.B and Z.O performed the research. J.A.P.B and C.B analyzed the results, J.A.P.B wrote the paper. This work was supported by a grant from Fondo de Investigaciones Sanitarias (03/0019). Melanoma Unit in Barcelona is partially funded by Grants from Fondo de Investigaciones Sanitarias (09/01393) Spain, by the AGAUR 2009 SGR 1337 of the Catalan Government, Spain; by the CIBER de Enfermedades Raras, ISCIII, Madrid, Spain: by the European Commission under the 6th Framework Programme, Contract nr: LSHC-CT-2006-018702 (GenoMEL) and by the National Cancer Institute (NCI) of the US National Institute of Health (NIH) (CA83115).
Footnotes
Conflict of interest: The authors declare no conflict of interest
References
- 1.Bastian BC, Kashani-Sabet M, Hamm H, et al. Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res. 2000;60:1968–1973. [PubMed] [Google Scholar]
- 2.Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163:1765–1770. doi: 10.1016/S0002-9440(10)63536-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 4.Wurm EM, Lin LL, Ferguson B, et al. A blueprint for staging of murine melanocytic lesions based on the Cdk4 (R24C/R24C) :Tyr-NRAS (Q) (61K) model. Exp Dermatol. 2012;21:676–681. doi: 10.1111/j.1600-0625.2012.01543.x. [DOI] [PubMed] [Google Scholar]
- 5.Whiteman DC, Pavan WJ, Bastian BC. The melanomas: a synthesis of epidemiological, clinical, histopathological, genetic, and biological aspects, supporting distinct subtypes, causal pathways, and cells of origin. Pigment Cell Melanoma Res. 2011;24:879–897. doi: 10.1111/j.1755-148X.2011.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauter ER, Yeo UC, von Stemm A, et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Res. 2002;62:3200–3206. [PubMed] [Google Scholar]
- 7.Takata M, Goto Y, Ichii N, et al. Constitutive activation of the mitogen-activated protein kinase signaling pathway in acral melanomas. J Invest Dermatol. 2005;125:318–322. doi: 10.1111/j.0022-202X.2005.23812.x. [DOI] [PubMed] [Google Scholar]
- 8.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 9.Jonsson G, Dahl C, Staaf J, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26:4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz A, Puig S, Lynch M, Castel T, Estivill X. Retention of the CDKN2A locus and low frequency of point mutations in primary and metastatic cutaneous malignant melanoma. Int J Cancer. 1998;76:312–316. doi: 10.1002/(sici)1097-0215(19980504)76:3<312::aid-ijc4>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 11.Kumar R, Smeds J, Lundh Rozell B, Hemminki K. Loss of heterozygosity at chromosome 9p21 (INK4-p14ARF locus): homozygous deletions and mutations in the p16 and p14ARF genes in sporadic primary melanomas. Melanoma Res. 1999;9:138–147. doi: 10.1097/00008390-199904000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Scope A, Busam KJ, Malvehy J, et al. Ex vivo dermoscopy of melanocytic tumors: time for dermatopathologists to learn dermoscopy. Arch Dermatol. 2007;143:1548–1552. doi: 10.1001/archderm.143.12.1548. [DOI] [PubMed] [Google Scholar]
- 13.Reifenberger J, Knobbe CB, Sterzinger AA, et al. Frequent alterations of Ras signaling pathway genes in sporadic malignant melanomas. Int J Cancer. 2004;109:377–384. doi: 10.1002/ijc.11722. [DOI] [PubMed] [Google Scholar]
- 14.Takata M, Lin J, Takayanagi S, et al. Genetic and epigenetic alterations in the differential diagnosis of malignant melanoma and spitzoid lesion. Br J Dermatol. 2007;156:1287–1294. doi: 10.1111/j.1365-2133.2007.07924.x. [DOI] [PubMed] [Google Scholar]
- 15.Chernoff KA, Bordone L, Horst B, et al. GAB2 amplifications refine molecular classification of melanoma. Clin Cancer Res. 2009;15:4288–4291. doi: 10.1158/1078-0432.CCR-09-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koynova DK, Jordanova ES, Milev AD, et al. Gene-specific fluorescence in-situ hybridization analysis on tissue microarray to refine the region of chromosome 20q amplification in melanoma. Melanoma Res. 2007;17:37–41. doi: 10.1097/CMR.0b013e3280141617. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka E, Hashimoto Y, Ito T, et al. The clinical significance of Aurora-A/STK15/BTAK expression in human esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11:1827–1834. doi: 10.1158/1078-0432.CCR-04-1627. [DOI] [PubMed] [Google Scholar]
- 19.Yao JE, Yan M, Guan Z, et al. Aurora-A down-regulates IkappaBalpha via Akt activation and interacts with insulin-like growth factor-1 induced phosphatidylinositol 3-kinase pathway for cancer cell survival. Mol Cancer. 2009;8:95. doi: 10.1186/1476-4598-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim KH, Brady DC, Kashatus DF, et al. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol. 2009;30:508–523. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim KH, Baines AT, Fiordalisi JJ, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 22.Glatz-Krieger K, Pache M, Tapia C, et al. Anatomic site-specific patterns of gene copy number gains in skin, mucosal, and uveal melanomas detected by fluorescence in situ hybridization. Virchows Arch. 2006;449:328–333. doi: 10.1007/s00428-006-0167-8. [DOI] [PubMed] [Google Scholar]
- 23.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 24.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95:1878–1890. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


