Abstract
Purpose
Although many lung cancers express the epidermal growth factor receptor and the vascular endothelial growth factor, only a small fraction of patients will respond to inhibitors of these pathways. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MS) has shown promise in biomarker discovery, potentially allowing the selection of patients who may benefit from such therapies. Here, we use a matrix-assisted laser desorption/ionization MS proteomic algorithm developed from a small dataset of erlotinib-bevacizumab treated patients to predict the clinical outcome of patients treated with erlotinib alone.
Methods
Pretreatment serum collected from patients in a phase I/II study of erlotinib in combination with bevacizumab for recurrent or refractory non-small cell lung cancer was used to develop a proteomic classifier. This classifier was validated using an independent treatment cohort and a control population.
Result
A proteomic profile based on 11 distinct m/z features was developed. This predictive algorithm was associated with outcome using the univariate Cox proportional hazard model in the training set (p = 0.0006 for overall survival; p = 0.0012 for progression-free survival). The signature also predicted overall survival and progression-free survival outcome when applied to a blinded test set of patients treated with erlotinib alone on Eastern Cooperative Oncology Group 3503 (n = 82, p < 0.0001 and p = 0.0018, respectively) but not when applied to a cohort of patients treated with chemotherapy alone (n = 61, p = 0.128).
Conclusion
The independently derived classifier supports the hypothesis that MS can reliably predict the outcome of patients treated with epidermal growth factor receptor kinase inhibitors.
Keywords: Lung cancer, Biomarkers, Proteomics
Recent advances in our understanding of cancer biology have led to the development of therapeutics that target pathways important for tumor growth and survival. One such pathway involves epidermal growth factor receptors (EGFRs). High EGFR expression is often observed in non-small cell lung cancer (NSCLC) and has been associated with a poor prognosis. Targeting this pathway with the EGFR tyrosine kinase inhibitor (EGFR-TKI) erlotinib confers a survival benefit for patients with advanced lung cancer.1 Another pathway important for tumor growth involves the vascular endothelial growth factor (VEGF), a major regulator of angiogenesis, and thus represents an ideal target for therapeutic intervention. VEGF expression is up-regulated in many solid tumors and is an independent predictor of poor prognosis in patients with NSCLC.2 A survival benefit is observed when bevacizumab, a humanized monoclonal antibody targeting VEGF, is combined with chemotherapy in patients with advanced lung cancers.3 Given their vital roles in tumor growth and survival, targeting the EGFR and vascular endothelial growth factor receptor pathways represents a rational approach toward the treatment of NSCLC.
Recent results from a randomized phase II trial evaluating the combination of bevacizumab and erlotinib without chemotherapy demonstrated that approximately 50% of patients derived clinical benefit from this treatment.4 The other 50% were exposed to toxicities whereas deriving no benefit. EGFR mutations, increased EGFR gene copy number, k-ras mutations, and overexpression of the EGFR protein have been explored as predictive markers for the response to treatment response with EGFR-TKIs. To date, EGFR mutations, copy number, and EGFR expression levels have been predictive of the response or the survival in some studies.5 EGFR gene copy number was also predictive for the EGFR-TKI response in the second and third line settings.6 These biomarkers require tumor tissue analysis and are not sufficiently conclusive for routinely selected patients who would derive benefits from therapy with EGFR-TKI. In addition, although there are candidate markers to predict response to erlotinib treatment, no markers are available to predict benefit from bevacizumab. Despite considerable evidence for the association of intratumoral and/or plasma VEGF levels with tumor progression and/or poor prognosis, pretreatment VEGF levels are not predictive of response to bevacizumab therapy.7 Thus, better prediction tools are needed to maximize treatment benefits while minimizing toxicity.
Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS) can be used to generate protein signatures from biologic specimens such as tissue, urine, and serum. The technique also offers the advantages of rapidity and sensitivity. Unfortunately, previous studies with serum MS proteomics as biomarkers have suffered from the lack of reproducibility and validation. These problems have led to general skepticism about this technology and its use in the development of cancer biomarkers.8 Recently, utilizing serum MALDI-TOF MS, Taguchi et al.9 reported a proteomic signature that independently classified patients according to their clinical outcome after treatment with EGFR-TKI therapy, but not with chemotherapy. This finding suggests that MALDI-TOF MS may still be useful for biomarker development and eventual clinical utility. In the present study, we developed another independent proteomic signature obtained from patients treated with erlotinib and bevacizumab that can not only accurately classify this group of patients based on clinical outcome in a leave-one-out analysis, but also can be used to independently classify outcome in patients treated with erlotinib alone. Furthermore, despite the small training set, the variability of signals between obtained spectra was small, suggesting that data generated from MS are reliable and reproducible. This study thus lends further support to the use of serum MALDI-TOF in biomarker discovery.
METHODS
Patients and Samples
MS was performed on pretreatment serum samples from patients who were treated with erlotinib and bevacizumab in an open-label, phase I/II study. Forty patients were enrolled in this study. All were diagnosed with histologically proven stage IIIB (with pleural effusion) or stage IV, recurrent, nonsquamous NSCLC. Pretreatment patient samples were available for 37 of 40 patients in the clinical trial. Further details regarding the patient population and the clinical trial were described previously.4 The validation cohort (n = 82) comprised of patients enrolled in Eastern Cooperative Oncology Group (ECOG) 350. The Vanderbilt University control group patients were comprised of unselected patients treated under various institutional review board approved chemotherapy protocols at Vanderbilt University Medical Center.9 These patients were treated in both the first and second line settings. None were treated with EGFR-TKI at time of relapse.
Sample Preparation and Mass Spectrometry
The sera were thawed on ice and diluted 1:20 in a saturated sinapinic acid solution (35 mg/ml sinapinic acid (Sigma, St. Louis, MO), 50% acetonitrile (Burdick & Jackson, Muskegon, MI), and 0.1% trifluoroacetic acid (Sigma, St. Louis, MO)). To avoid confounding variables associated with the run order, samples were randomly spotted in triplicate, placed on gold 64-well sample plates, and allowed to dry at room temperature. Mass spectra for all samples were generated in linear mode using a Voyager-DE STR workstation. The results from 500 to 525 independent spectrum acquisitions per sample were averaged to generate each spectrum. To avoid day-to-day bias, the spectrometry of all triplicate samples was repeated on two other days (for a total of 3 days). All mass spectra were output as column text files of intensity versus mass/charge (m/z). Mass spectra from ECOG 3503 and the Vanderbilt control cohort were obtained from Dr. David P. Carbone.9
Spectral Preprocessing
Raw spectra were analyzed with Wave-spec software developed at the Vanderbilt University High Dimensional Data Analysis Center. Detailed information on preprocessing was published previously.10 Briefly, preprocessing comprised of internal calibration, smoothing, baseline correction, normalization to the total ion current, feature selection based on signal-to-noise ratio, and binning of features. Internal calibration was performed using the Apo-C1 (m/z = 6631), hemoglobin (m/z = 15127), hemoglobin β chain (m/z = 15127), and two unidentified, common peaks m/z = 9423 and m/z = 13746. A total of 174 bins were selected from m/z ratios between 3000 and 20,000. The preprocessing parameters were optimized based on the training set and were fixed when applied to the preprocessing of the independent test set.
Experimental Reproducibility and Variability Determination
Experimental variability and reproducibility (both analytical and biologic) were first tested on 277 spectra generated from 37 available serum samples. We used Coefficients of Variation (CV) as a measure of the relative variability between the expression levels of each peak or feature. We also used a nested, random-effect model, testing the variability not only between patients, but also with the acquisition day factor nested within each patient. The intraclass correlation coefficient (ICC), a measure of correlation, consistency, or conformity for a dataset when it contains multiple groups, is defined by the expression:
where is the variance between patients and is the pooled variance within a patient and between days. The distribution across peaks for CV, variance components, and ICC are presented in quantiles and box-plots.
Risk Prediction Model
Serum sample were available from 37 of 40 patients enrolled in the clinical trial. Of these, 35 provided both spectral data and associated clinical data used to develop the risk prediction model. Features or peaks associated with survival were selected based on a univariate Cox regression analysis, considering mean intensity for one feature at a time. A false discovery rate of <0.05 was used as cutoff for selecting important features. Once significant features were selected, the compound score (denoted as ci) for patient i was defined as
| (1) |
where xj,i is the peak intensity of the feature j for patient i, wj is the Wald statistic, and βj is the coefficient obtained with the univariate Cox regression analysis for the set of k selected significant features. The compound score (ci) is then used as prediction index. A prediction model was then developed based on the Cox model with the compound score as predictor,
| (2) |
The compound score for a new patient with the vector of intensity ( ) for the selected features can be calculated by replacing xj,i with in (1), which is then used to predict the survival time for that patient.
Validation of Prediction Model Using ECOG and VU Control Cohorts
For the validation of the predictive power of the compound score derived in (2), the Cox proportional hazards (CPH) model was first fitted with this compound score as the only covariate, whereas overall survival (OS) and progression-free survival (PFS) served as outcome variables, respectively. After the univariate analysis, baseline clinical factors were entered into the model along with the compound score to determine whether the compound score could serve as an independent predictor for survival.
For the VU control cohort, smoking status, stage, and compound score were prespecified in the multivariable CPH model. For the ECOG cohort, model building techniques were used in four steps relying first on univariate testing for each of the candidate variables. Only variables with a statistical significance of p < 0.2 were considered for the next step of model building. Backward elimination, forward selection, and stepwise selection (with p < 0.1) were then performed on the variables to select possible models. In the final stepwise step, two-way interactions under the hierarchical principle were also added for consideration in the model. The Akaike’s information criterion was adopted to make comparisons among a number of possible models. The smaller the value of this statistic, the better the model fits the data.
The proportional hazards assumption of each covariate was evaluated after model building using time-by-covariate interactions. Model fitting was assessed via the Cox-Snell generalized residuals plot and diagnostics for influential points. OS and PFS survival curve for the subgroups of categorized compound scores were estimated using the Kaplan-Meier method. Analyses were performed using SAS version 9.1 and R version 2.1.1.
RESULTS
Reproducibility of Data Processing
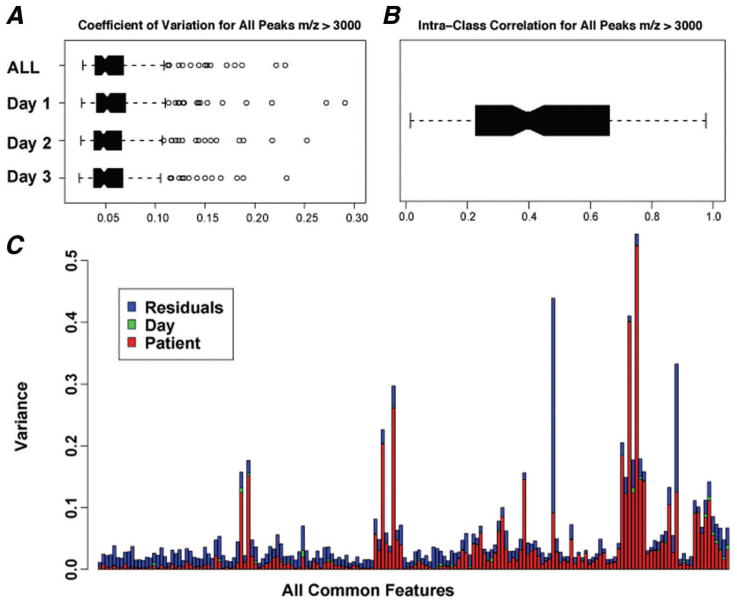
Patient characteristics from the three cohorts used for testing and validation are summarized in Table 1. We generated 277 spectra from the available 37 samples in the erlotinib/bevacizumab study. The average number of spectra per patient was 7.5 (range, 5–9). Across all sera, 139 peaks in the 3,000 to 20,000 m/z range were detected. To quantify the relative variability of the features or peaks, we generated Coefficients of Variation (CV) using 139 common peaks for all samples, and for samples from each of the 3 replicated days (Figure 1A). The comparable and low mean CV of <5% across the 3 days and comparable overall CV suggest that the spectra did not differ significantly and that our spectrometry was reproducible. We used intraclass correlation coefficients (ICC) to measure consistency and conformity among the multiple groups within our training set. The obtained mean ICC of 0.5192 indicates a relatively large intrasample variability (Figure 1B); therefore, nine replications of the same sample were measured to reduce the heterogeneity of the expression profiles in each tumor sample. Observed patient, day, and residual variance (a measure of variability distribution) for the 139 common peaks in our dataset are shown in Figure 1C.
TABLE 1.
Characteristics of the Non-Small Cell Lung Cancer Patients Cohorts
| Characteristics | Bevacizumab/Erlotinib (n =35a) | ECOG 3503 (n = 82) | VU Control (n = 61) |
|---|---|---|---|
| Sex (%) | |||
| Male | 14 (40) | 37 (45.1) | 42 (68.9) |
| Female | 21 (60) | 45 (54.9) | 19 (31.1) |
| Age (yr) | |||
| Median | 59 | 70 | 65 |
| Range | 36–72 | 41–93 | 40–84 |
| Stage | |||
| IIIB | N/Ab | 7 (8.5) | 28 (45.9) |
| VI | N/Ab | 59 (72.0) | 33 (54.1) |
| Recurrent | 16 (19.5) | ||
| RECIST (%) | |||
| Partial response | 8 | N/A | N/A |
| Stable disease | 21 | N/A | N/A |
| Progressive disease | 6 | N/A | N/A |
| Survival (mo) | |||
| Overall (95% CI) | 15.25 (7.75-inf) | 7.69 (5.39–10.05) | 14.63 (10.3–27.27) |
| Progression-free (95% CI) | 4 (2.25–12.5) | 3.38 (2.07–3.91) | |
The total number of patients with available clinical data for survival analysis.
The total number of patients in the bevacizumab/erlotinib trial was 40 (seven IIIb, 32 IV, one unknown).
CI, confidence interval; N/A, not applicable; RECIST, Response Evaluation Criteria in Solid Tumors.
FIGURE 1.
Reproducibility analysis of the bevacizumab/erlotinib training data set. A, Coefficient of variation (CV) using 139 common peaks across all sera in the 3000 to 20,000 m/z range. The results show low and comparable CV across all days, suggesting highly reproducible spectra. B, Intraclass correlation coefficient (ICC), a measure of reliability, is 0.5192, suggesting a good agreement at all common peaks. C, A summary of observed patient, day, and residual variance (a measure of variability distribution), for the 139 common peaks.
MALDI-MS Algorithm Accurately Classifies Outcomes for Patients Treated with Erlotinib-Bevacizumab
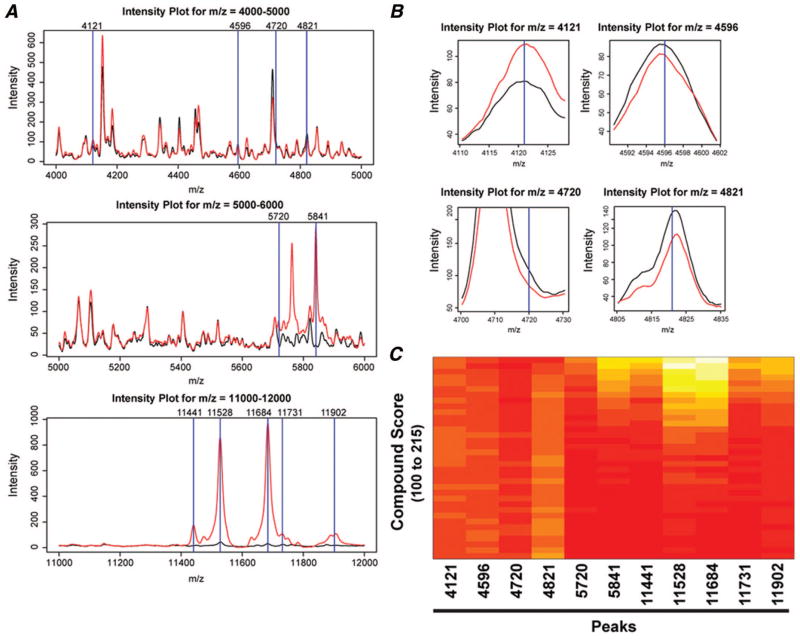
Features associated with survival were selected using a univariate CPH model, with peak intensity as covariate or associated with clinical outcome. Eleven features were selected according to their statistical significance, defined by a false discovery rate <0.05 (Table 2). Using the ±Wald statistic and peak intensities of these 11 features, a compound score for each patient was generated. To illustrate that the compound score can differentiate intensities of the 11 selected features, we generated an intensity plot comparing the mean peak intensity of the group with high intensity score, defined as those with compound score ≥ median compound score (114.7862), to that from the group with low intensity score (<114.7862). The difference in mean intensities between the two groups is easily observed in peaks with m/z values greater than 5000 (Figure 2A). At higher magnification, differences within peak regions 4121, 4596, and 4821 can also be detected (Figure 2B). Although the two average curves show negligible differences at peak 4710, some individual spectra from samples with high compound scores demonstrated disparate peaks at this value. Whether an identified peak is a product of technology or biology will ultimately be established by validation and perhaps protein identification. Figure 2C shows a heatmap to better visualize the relationship between the compound scores and peak intensities for the 11 selected features.
TABLE 2.
List of 11 Distinct m/z Features Used to Predict Clinical Outcome
| m/z | Lower m/z | Upper m/z | pFDR | |
|---|---|---|---|---|
| 1 | 4121 | 4112 | 4128 | 0.0482 |
| 2 | 4596 | 4581 | 4605 | 0.0857 |
| 3 | 4720 | 4717 | 4724 | 0.0476 |
| 4 | 4821 | 4804 | 4833 | 0.0445 |
| 5 | 5720 | 5714 | 5726 | 0.0362 |
| 6 | 5841 | 5832 | 5848 | 0.0447 |
| 7 | 11441 | 11423 | 11455 | 0.0396 |
| 8 | 11528 | 11515 | 11539 | 0.0483 |
| 9 | 11684 | 11673 | 11697 | 0.0368 |
| 10 | 11731 | 11717 | 11742 | 0.0345 |
| 11 | 11902 | 11890 | 11933 | 0.0381 |
pFDR, positive false discovery rate.
FIGURE 2.
The intensity plots comparing the mean spectra of samples from the group with high versus the group with low compound scores, respectively. A, Intensity plots for m/z regions 4000 to 5000, 5000 to 6000, and 11,000 to 12,000 demonstrating different intensities among 11 features (blue lines). The black line denotes mean intensity of peaks from patients with compound scores equal or less than the median. The red line represents mean intensity of peaks from patients whose compound scores were greater than the median. B, Magnification of intensity plots for features 4121, 4596, 4720, and 4821 showing a subtle difference in intensity between the mean spectrum with high and that with low compound score. C, A heatmap of the 11 peaks or features from the bevacizumab/erlotinib training data set, ranked according to compound score. The levels of peak intensity (features 4121, 5720, 5841, 11,441, 11,528, 11,684, 11,731, and 11,902) increase as compound scores increase, whereas peak intensities of features 4596, 4720, 4821 decrease.
MALDI-MS Algorithm Accurately Classifies Clinical Outcome in Patients Treated with Erlotinib-Containing Regimen
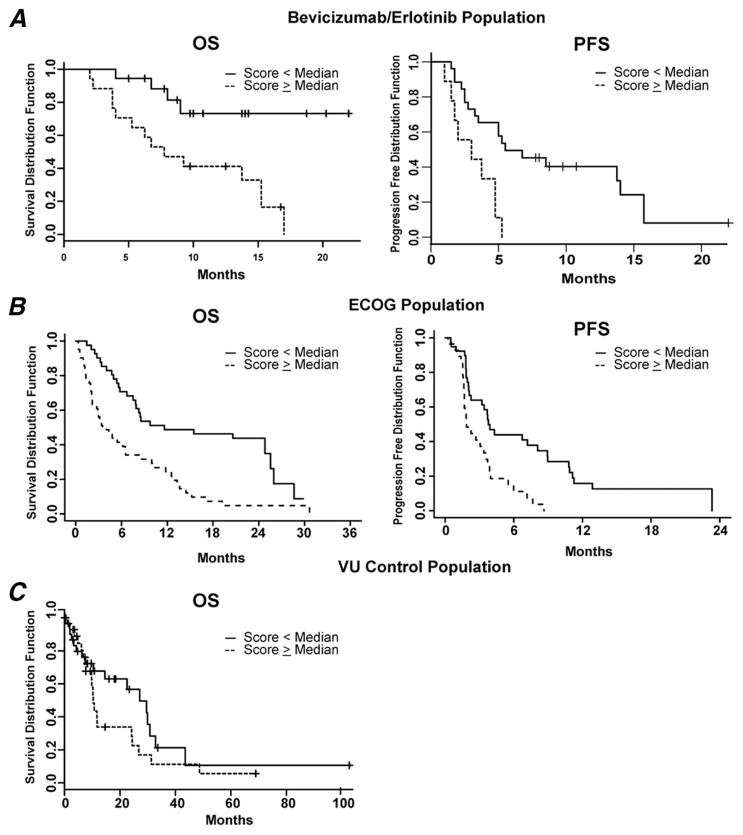
Using the univariate CPH model, the proteomic algorithm based on the 11 distinct m/z features was found to be associated with clinical outcome in both OS and PFS (p = 0.0006 for OS; p = 0.0012 for PFS). We used the median scores to generate binary data to improve data visualization. We found a tight relationship between compound scores and clinical outcome when we compared the survival of patients whose compound scores were less than the median (114.7862) to the survival of those whose scores were equal or greater. The Kaplan–Meier survival curves for the two groups are shown in Figure 3A. The group with the scores that were lower than the median shows a statistically significantly greater OS and PFS (log-rank test, p = 0.00284 and p = 0.014, respectively).
FIGURE 3.
Survival of patients from the training, testing, and control datasets sorted by median compound score using Kaplan-Meier analysis. Overall survival (OS) and progression-free survival (PFS) of patients, whose compound scores were less than the median were significantly better than those of patients whose scores are equal or greater than the median in bevacizumab/erlotinib (A) and ECOG 3503 (B) cohorts. (C) OS of VU control cohort based on median score.
We applied the prediction algorithm to a validation cohort from ECOG protocol E3503, a phase II trial of erlotinib. Each patient was given a compound score based on our classification algorithm, and the results were sent to the ECOG Statistical Office for correlation with the clinical data. The analysis is based on 82 patients, excluding ineligible patients and those without an available compound score. With the obtained proteomic signature, we were able to classify OS and PFS outcome for samples from the blinded test set of patients treated with erlotinib alone on ECOG 3503 (univariate CPH model, p < 0.0001 and p = 0.0018, respectively). When the compound scores were again sorted into two groups, one whose scores were equal or above the median (131.0321) and one below, patients with lower compound scores had better OS and PFS than those whose prediction scores were higher (log-rank test, p < 0.0001 and p = 0.003, respectively). Figure 3B shows a Kaplan-Meier plot for OS and PFS.
Because the analyses were based on survival, it is possible that the survival outcome classification derived from the MALDI-TOF MS algorithm merely indicates performance status or overall general health, and was not specific to treatment with the erlotinib-containing regimen. To test whether the profile was predictive or merely prognostic, we used a control cohort of 61 patients with advanced NSCLC who were treated with various chemotherapy regimens at Vanderbilt University Medical Center, and for whom clinical data and spectra were available.9 About 80% of these patients received carboplatin and paclitaxel. No difference in OS was noted between the two groups, one whose scores were equal or above the median (133.2427) and one below (univariate CPH, p = 0.128 and log-rank test, p = 0.181) (Figure 3C). These findings suggest that our MS algorithm is predictive of patients who would benefit from erlotinib, and not merely prognostic. However, further testing in a prospective, randomized setting is needed to ascertain that this observation is not merely an artifact due to the retrospective nature of our study.
To determine whether our profiles have independent predictive ability in outcome for patients treated with bevacizumab/erlotinib, we tested our proteomic algorithm, using the CPH model with the adjustment of common baseline factors. After adjusting for age, sex, and histology, our predictors remained significant for OS and PFS. In the ECOG cohort, the predictors held after adjustment for the number of meta-static sites and performance status. The results obtained using multivariate model for the different cohorts including that for VU Control are summarized in Table 3.
TABLE 3.
Multivariable Analysis for Patient Data Sets
| Factor | OS
|
PFS
|
||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | p | Hazard Ratio (95% CI) | p | |
| Erlotinib/bevacizumab | ||||
| Compound score | 1.024 (1.009–1.040) | 0.003 | 1.016 (1.005–1.028) | 0.004 |
| Age | 0.992 (0.933–1.056) | 0.809 | 0.979 (0.936–1.024) | 0.347 |
| Sex | 1.022 (0.328–3.185) | 0.97 | 0.893 (0.345–2.310) | 0.815 |
| Adeno vs. nonadeno | 1.679 (0.540–5.223) | 0.371 | 0.933 (0.318–2.736) | 0.899 |
| ECOG 3503 | ||||
| Compound score | 1.012 (1.003–1.021) | 0.012 | 1.014 (1.004–1.024) | 0.006 |
| Metastatic site | 1.414 (1.191–1.678) | p < 0.0001 | 1.298 (1.086–1.552) | 0.004 |
| PS = 0 vs. PS = 2 | 0.227 (0.109–0.473) | p < 0.0001 | NA | |
| PS = 1 vs. PS = 2 | 0.388 (0.209–0.791) | 0.003 | NA | |
| VU control | ||||
| Compound score | 1.011 (0.996–1.026) | 0.161 | NA | |
| Ex-smoker vs. current | 0.525 (0.233–1.181) | 0.119 | NA | |
| Nonsmoker vs. smoker | 0.879 (0.192–4.019) | 0.868 | NA | |
| Stage IV vs. IIIB | 0.714 (0.369–1.488) | 0.4 | NA | |
OS, overall survival; PFS, progression-free survival, CI, confidence interval; NA, not applicable.
DISCUSSION
In the present study, we developed a novel MALDI-TOF MS survival prediction model with which NSCLC patients can be identified who may benefit from treatment with an erlotinib-containing regimen. To develop this model, a training set of 37 patients treated with bevacizumab and erlotinib was used. Despite the small sample size, the model retained its predictive abilities on multivariate testing against the ECOG 3503 validation cohort. By contrast, we could not classify patients treated with chemotherapy alone, suggesting that it is not merely prognostic, but moreover predictive for outcome in patients treated with EGFR inhibition with or without bevacizumab. Though our findings are encouraging, we cannot rule out that potential confounders and selection bias due to the retrospective nature of our study may result in an incorrect estimate of the association between the algorithm and clinical outcome.
There is a dearth of practical and clinically useful biomarkers for diagnosis, prognosis, and disease monitoring in lung cancer. Early studies using MS to develop proteomic profiles from patient sera, coupled with advanced data mining algorithms had triggered hopes that this technology could be used for cancer diagnosis, prognosis, and disease/treatment monitoring.11 The enthusiasm for this technology has diminished due to the lack of reproducibility, procedural bias, and virtually no independent confirmation.8,12 Although the two current proteomic technologies MALDI-TOF MS and surface-enhanced laser desorption and ionization MS may differ in their approaches and sensitivity, the potential pitfall involving data reproducibility remains a significant universal concern. To avoid systematic or procedural bias in our study, the positions on the plates were randomized, and the sample spectrometry was replicated a total of nine times and on multiple days. Despite the relatively small dataset, the low and consistent coefficient variation among peaks, low variance, and high intraclass correlation affirm the potential for reproducibility of proteomic approaches.
Interestingly, while the patients in our cohort were treated with bevacizumab and erlotinib, our predictors could accurately classify patients from ECOG 3503 who were treated with erlotinib alone. Furthermore, of the 11 mass spectral features identified in our study, five overlap with those from another independent, proteomic algorithm developed by Taguchi et al.9 Their model was based on patients treated with erlotinib alone. Because there has been no demonstrated clinical efficacy for bevacizumab alone in human lung cancer, it is reasonable to propose that the predictive power of the classifier is the result of direct augmentation of the erlotinib activity by bevacizumab. It is also possible that the addition of bevacizumab to the erlotinib treatment in our training set, compared with the erlotinib-only population from Taguchi et al., may account for the incomplete overlap of mass spectral features. These hypotheses will need to be tested against patients treated with erlotinib/bevacizumab and erlotinib alone. Unfortunately, such samples are not yet available to us. Nevertheless, validation of our predictors using the ECOG independent dataset and the identification of similar discriminant mass spectral features in another independent algorithm suggest that our observations most likely reflect biology and not simply data overfitting.
Finally, although we have not yet identified the proteins represented by our 11 selected features, our proteomic signature is unable to predict outcome in the chemotherapy-treated cohort. This finding suggests that the selected features represent proteins that are not just stress-related. Of note, a number of current biomarkers used for cancer screening and treatment monitoring such as CA-125, prostate-specific antigen, and CA-19-9 have no direct role in cancer pathogenesis. Thus, it is highly possible that the proteins we have identified as predictive will not give insights into pathways or mechanisms of disease or treatment response. Clearly, only further rigorous testing in a prospective clinical trial setting can determine whether any candidate biomarker will have clinical utility regardless of its identification.
In summary, we have demonstrated that a small dataset can be used to develop a prediction model using MALDI-TOF MS from the sera of patients with NSCLC. With sample randomization and replication, we were able to avoid extreme sample variations and systemic biases. Furthermore, our model could accurately and independently identify patients who may benefit from treatment with an erlotinib-containing regimen. Although further validation with independent, larger cohorts is needed, our findings affirm that MS can be a useful tool in biomarker discovery.
Acknowledgments
Supported by SPORE in Lung Cancer P50 CA090949.
Footnotes
Disclosure: The authors declare no conflicts of interest.
References
- 1.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 2.Herbst RS, Onn A, Sandler A. Angiogenesis and lung cancer: prognostic and therapeutic implications. J Clin Oncol. 2005;23:3243–3256. doi: 10.1200/JCO.2005.18.853. [DOI] [PubMed] [Google Scholar]
- 3.Sandler A, Gray R, Perry MC. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Johnson DH, Mininberg E. Phase I/II trial evaluating the anti-vascular endothelial growth factor monoclonal antibody bevacizumab in combination with the HER-1/epidermal growth factor receptor tyrosine kinase inhibitor erlotinib for patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:2544–2555. doi: 10.1200/JCO.2005.02.477. [DOI] [PubMed] [Google Scholar]
- 5.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 6.Zhu CQ, da Cunha Santos G, Ding K. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR 21. J Clin Oncol. 2008;26:4268–4275. doi: 10.1200/JCO.2007.14.8924. [DOI] [PubMed] [Google Scholar]
- 7.Stinchcombe TE, Socinski MA. Bevacizumab in the treatment of non-small-cell lung cancer. Oncogene. 2007;26:3691–3698. doi: 10.1038/sj.onc.1210366. [DOI] [PubMed] [Google Scholar]
- 8.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–356. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 9.Taguchi F, Solomon B, Gregorc V. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 10.Yildiz PB, Shyr Y, Rahman JS. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. J Thorac Oncol. 2007;2:893–901. doi: 10.1097/JTO.0b013e31814b8be7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petricoin EF, Ardekani AM, Hitt BA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 12.Baggerly KA, Morris JS, Edmonson SR, Coombes KR. Signal in noise: evaluating reported reproducibility of serum proteomic tests for ovarian cancer. J Natl Cancer Inst. 2005;97:307–309. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]