SUMMARY
Packaging of viral genomes into empty procapsids is powered by a large DNA-packaging motor. In most viruses, this machine is composed of a large (L) and a small (S) terminase subunit complexed with a dodecamer of portal protein. Here, we describe the 1.75 Å crystal structure of the bacteriophage P22 S-terminase in a nonameric conformation. The structure presents a central channel ~23 Å in diameter, sufficiently large to accommodate hydrated B-DNA. The last 23 residues of S-terminase are essential for binding to DNA and assembly to L-terminase. Upon binding to its own DNA, S-terminase functions as a specific activator of L-terminase ATPase activity. The DNA-dependent stimulation of ATPase activity thus rationalizes the exclusive specificity of genome-packaging motors for viral DNA in the crowd of host DNA, ensuring fidelity of packaging and avoiding wasteful ATP hydrolysis. This posits a model for DNA-dependent activation of genome-packaging motors of general interest in virology.
INTRODUCTION
Viral genome packaging is a complex, non-spontaneous reaction, catalyzed in many DNA viruses by a powerful genome-packaging motor (Casjens, 2011; Catalano, 2005; Rao and Feiss, 2008; Sun et al., 2010). This molecular machine consists of a dodecameric portal protein and an ATPase known as terminase that converts ATP hydrolysis into linear translation of DNA. In double stranded DNA (dsDNA) bacteriophages, the terminase is formed by a small and a large subunit (referred to as S- and L-terminase), assembled in a complex of unknown stoichiometry. The ATPase activity resides in the L-terminase subunit, which binds directly to the portal protein (Rao and Feiss, 2008). In contrast, the S-terminase subunit binds to packaging initiation sites (referred to as pac in P22 (Jackson et al., 1978)) to prepare for genome packaging (Casjens et al., 1992) and regulates the ATPase activity of L-terminase (Baumann and Black, 2003; Leffers and Rao, 2000). This function is likely very important in vivo to sustain the enormous rate of genome-packaging, which can be as high as ~2,000 bp/sec (Fuller et al., 2007).
All S-terminases characterized to date adopt an oligomeric quaternary structure (Rao and Feiss, 2008), but the exact stoichiometry of assembly varies in different viruses. In bacteriophage P22, S-terminase (gp3) consists of 162 amino acids (M.W. ~18.6 kDa) and self-assembles into a nonameric ring (Nemecek et al., 2007; Nemecek et al., 2008; Roy et al., 2011). In contrast, the S-terminase subunit of bacteriophage Sf6, a close relative of P22 assembles into octamers (Zhao et al., 2010). Although octamers were also observed in solution for bacteriophage T7 (gp18) (White and Richardson, 1987) and T4 (gp16) (Lin et al., 1997) S-terminases, a recent crystal structure of the T4-like phage 44RR S-terminase (gp16) revealed a mix of undecamers and dodecamers (Sun et al., 2012). Similarly, phage SPP1 S-terminase (gp1) was reported to form decameric rings in solution (Camacho et al., 2003), but the crystal structure of the SPP1-like Bacillus phage SF6 revealed a mix of nonamers and decamers, with the nonamer being the predominant conformation (Buttner et al., 2012). Finally, phage λ S-terminase (gpNu1) forms a hetero-trimer bound to a monomer of L-terminase (gpA1), and in vitro this protomer can further assemble into tetramers (Maluf et al., 2006). Thus, as previously observed for viral portal proteins (Cingolani et al., 2002; Lorenzen et al., 2008), S-terminases appear to be highly polymorphic in solution and in crystal.
Similar to S-terminases, L-terminases can also oligomerize: a pentameric quaternary structure was suggested for T4 L-terminase bound to procapsid (Sun et al., 2008). The N-terminal ATPase domain of L-terminase is thought to contact directly the portal vertex (Sun et al., 2008), while the C-terminus harbors a nuclease domain required to cleave DNA after encapsidation (Duffy and Feiss, 2002; Kanamaru et al., 2004). Regardless of the exact stoichiometry of assembly, the genes encoding L- and S-terminase subunits are essential in all tailed bacteriophages and herpesviruses (Rao and Feiss, 2008), underscoring their essential function in DNA packaging. Unfortunately, the molecular characterization of viral genome-packaging motors is in its infancy as compared to other multisubunit ATPases, like the F1-ATPase. Atomic-level structural information is presently available only for the full length L-terminase of T4 (Sun et al., 2008; Sun et al., 2007), and isolated L-terminase nuclease domains of SPP1 (Smits et al., 2009), P22 (Roy and Cingolani, 2012) and human cytomegalovirus (herpesvirus 5) (Nadal et al., 2010). No high resolution information exists for the terminase holoenzyme or in complex with portal protein. Likewise, it is unclear how L- and S-terminase assemble during packaging to form a functional holoenzyme and how DNA is recruited to initiate packaging.
We have studied the S-terminase subunit of bacteriophage P22 to provide an atomic description of a prototypical S-terminase subunit and to determine its functional role in viral DNA-packaging. Our results indicate that this protein is a dedicated DNA-dependent ATPase-activating factor within the genome-packaging motor.
RESULTS
Structure determination of the bacteriophage P22 S-terminase subunit
The S-terminase subunit of bacteriophage P22 assembles in solution (Nemecek et al., 2007) into a homo-nonamer. In vitro, this oligomer unfolds irreversibly and in a highly cooperative manner, with an apparent melting temperature (Tm) of ~85° C (Figure S1A). We crystallized the full length (fl) S-terminase in different space groups with one or two nonamers in the asymmetric unit (Roy et al., 2011). Although most crystals diffracted weakly to ~3.5 Å resolution, we found that omitting protease inhibitors during crystallization resulted in degradation of the last 23 C-terminal residues, which dramatically improved diffraction quality (Roy et al., 2011). We determined the crystal structure of P22 nonameric cleaved S-terminase (cl-S-terminase) to an Rfree ~21.65 %, at 1.75 Å resolution (Figure 1A,B, Table 1). The entire polypeptide chain, with the exception of C-terminal residues 140–162 which are not present in the cleaved crystal form, has been unambiguously traced. This structural model was then used to phase an orthorhombic crystal form of fl-S-terminase grown in the presence of protease inhibitors (Table 1). Unexpectedly, this structure also had no discernible electron density for the last 23 residues, which, although present in crystal, are disordered in the crystal structure (Figure S1B,C).
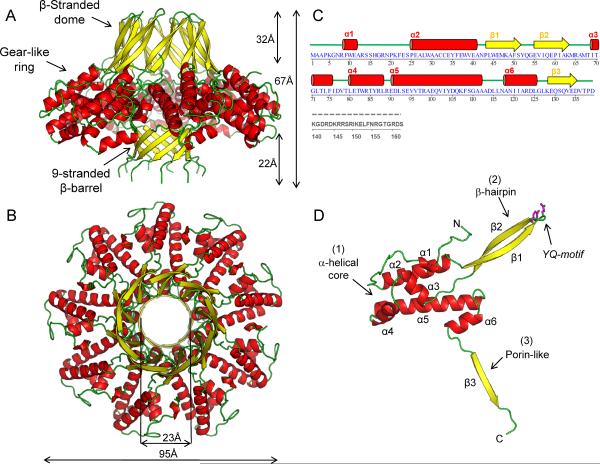
Figure 1. Quaternary structure of the nonameric S-terminase subunit of bacteriophage P22.
Ribbon diagram of S-terminase in side (A) and top (B) views. The oligomer is colored by secondary structure elements with α-helices, β-strands and loops in red, yellow and green, respectively. The overall diameter of S-terminase is ~95 Å with an internal hollow channel ~23 Å. (C) Secondary structure and amino acid sequence of bacteriophage P22 S-terminase subunit. Dashed in gray is the DNA-binding domain spanning residues 140–162, which is proteolytically cleaved in the 1.75 Å structure used for high resolution refinement and is disordered in the 3.35 Å structure of fl-S-terminase (Figure S1B). The illustration was generated using STRIDE (Heinig and Frishman, 2004). (D) Ribbon diagram of S-terminase protomer colored as in (A). The side chains for the YQ-motif on the tip of S-terminase are shown as sticks.
Table 1.
Crystallographic data collection and refinement statistics.
| Cleaved S-terminase | Full Length S-terminase | ||
|---|---|---|---|
| Native-gp3 | SeMet-gp3 | FL-gp3 | |
| Data collection | |||
| Space group | P21 | P21 | P21212 |
| Cell dimensions | |||
| a, b, c (Å) | 76.48, 100.90, 89.95 | 80.22, 101.18, 89.09 | 143.46, 144.93, 144.61 |
| α, β, γ (Å) | 90, 93.73, 90 | 90, 92.87, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.99 | 0.97 | 0.97 |
| Resolution (Å) | 15–1.75 (1.81–1.75) | 20–2.5 (2.54–2.50) | 20–3.35 (3.42–3.35) |
| Reflections (tot/unique) | 2,483,158 / 126,076 | 7,505,096 / 49,292 | 747,842 / 43,496 |
| R sym | 7.1 (67.4) | 9.6 (30.8) | 17.5 (64.6) |
| I / σI | 28.2 (1.8) | 36.8 (9.0) | 14.7 (3.3) |
| Completeness (%) | 93.5 (62.5) | 99.8 (100.0) | 99.9 (99.7) |
| Redundancy | 4.4 (3.3) | 8.3 (8.1) | 5.7 (5.6) |
| Refinement | |||
| Resolution (Å) | 15–1.75 | 20–3.35 | |
| No. reflections | 126,076 | 41,398 | |
| Rwork / Rfreeb | 17.73 / 21.65 | 23.24 / 26.45 | |
| No. atoms | |||
| Protein | 11,158 | 19,348 | |
| Water | 1,397 | 0 | |
| B-factors (Å2) | |||
| Protein | 36.5 | 59 | |
| Water | 40.1 | ||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.006 | 0.009 | |
| Bond angles (°) | 0.918 | 1.208 | |
Values in parentheses are for highest-resolution shells.
Rsym =Σi,h|I(i,h) − <I(h)>|/Σi,h|I(i,h)| where I(i,h) and <I(h)> are the ith and mean measurement of intensity of reflection h.
The Rfree value was calculated using 2,000 reflections selected randomly for native-gp3 and in thin resolution shells for Full Lenath S-terminase.
P22 S-terminase folds into a nonamer
The S-terminase subunit of bacteriophage P22 folds into a hollow nonameric ring of mixed α/β structure that resembles a jelly-fish (Figure 1A,B). The quaternary structure of S-terminase consists of a central gear-like ring ~95 Å in diameter, which is sandwiched by a 9-stranded β-barrel, similar to that found in β-porins (Galdiero et al., 2007), and a β-stranded dome, formed by nine slightly twisted β-hairpins (Figure 1A). The solvent-filled channel inside S-terminase varies between 20–25 Å in diameter (Figure 1B), large enough to accommodate B-DNA, which is ~20 Å in diameter in its hydrated form (Vlieghe et al., 1999). The nonamer is built by nine slightly non-identical subunits, symmetrically arranged around a central channel that has an overall height of ~67 Å (Figure 1A,B). Superimposition of S-terminase protomers reveals significant structural plasticity in the nine β-hairpins forming the β-dome (residues 42–65) that are rotated up ~8° with respect to each other. The body of each protomer is nearly parallel to the 9-fold axis running along the central channel (Figure 1A). The ring-shaped nonamer has a total solvent-accessible surface area of ~54,030 Å2 and ~1,980 Å2 of surface area is buried at the interface between two neighboring protomers. This interface is stabilized by a complex network of salt bridges between a largely positive face of one S-terminase protomer and the complementary largely negative face of its adjacent neighbor. Most notably, two inter-subunit salt bridges are observed between the amine nitrogen (Nz) atom and Nε atom of Arg8/Arg123 of one subunit and the Oε1 and Oε2 atoms of Glu40/Glu129 in the neighboring subunits. These ionic interactions may explain the salt dependency of ring formation observed in solution (Roy et al., 2011).
The structure of a single S-terminase protomer can be divided into four distinct domains (Figure 1C,D): (1) an N-terminal α-helical core formed by 6 α-helices (α1–α6), which builds most of the gear-like ring. (2) a long β-hairpin (β1–β2), extending from the loop connecting helices α2–α3 (residues 41–69), which exposes at its tip a Tyr/Gln (YQ) motif. (3) a porin-like β-strand (residues 126–139) that forms with its neighbors the 9-stranded β-barrel. This is the first observed β-barrel in a soluble protein with more than 8 strands (Galdiero et al., 2007). Finally, (4) residues 140–162 are not present in the crystal form used for high-resolution refinement due to proteolytic cleavage in the crystallization drop (Roy et al., 2011). The same moiety has no discernable electron density in crystals of fl-S-terminase grown in the presence of PMSF (Figure S1C).
Structural diversity of viral S-terminases
At least one S-terminase structure is available for each of the three families of tailed bacteriophages, which include Podoviridae, Siphoviridae and Myoviridae (Ackermann, 2003). This provides a unique opportunity to identify structural similarities even in the absence of significant sequence conservation. For instance, despite only 15% sequence identity, the structure of P22 S-terminase presented in this paper shares an overall similar organization as the octameric S-terminase of bacteriophage Sf6 (Zhao et al., 2010). Podoviridae P22 (Figure 2A,B) and Sf6 (Figure 2C,D) S-terminases have equal height (~66 Å) and comparable diameter (~99 versus 95 Å), but differ in oligomerization state. Moreover, in P22 the internal channel is large enough throughout its length to accommodate hydrated dsDNA, while in Sf6 the channel diameter is ~27 Å in the helical core and narrows at the C-terminal end of the protein to ~17 Å, which, in the conformation observed crystallographically, is too narrow to accommodate dsDNA (Zhao et al., 2010). Superimposition of P22 and Sf6 protomers (Figure S2A) reveals structural similarity in five (helices α2–α6) of the six helices forming the α-helical core, while helix α1 is not visible in the Sf6 model. The most noticeable difference between these two structures is that the long β-hairpin forming the β-stranded dome (residues 44–63) in P22 is missing in the Sf6 S-terminase (Figure S2A). Furthermore, the C-terminal domain of both terminases folds into a porin-like barrel, but while in Sf6 a second β-sheet follows strand β3, in P22 residues 140–162 downstream of strand β3 are unstructured.
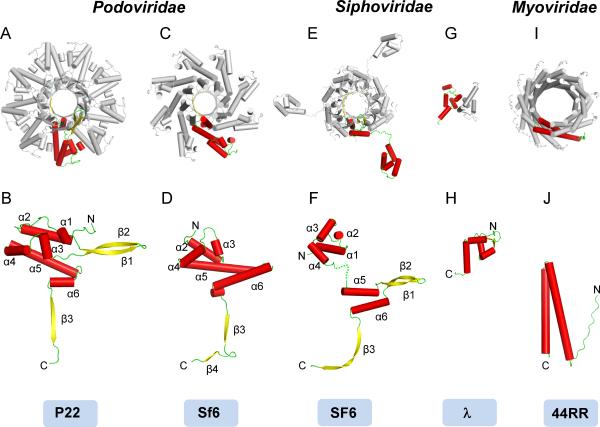
Figure 2. Conservation of S-terminase in tailed bacteriophages.
Oligomer and protomer structure of S-terminase subunits in Podoviridae P22 (A, B) and Sf6 (C,D) (pdb 3HEF); Siphoviridae SF6 (E,F) (pdb 3ZQQ) and λ (G,H) (pdb 1J9I); Myoviridae T4-like phage 44RR (I,J) (pdb 3TXQ). In all cases, the S-terminase is displayed from the top with α-helices represented as cylinders; only one protomer per oligomer is colored by secondary structure elements, while the other subunits are in gray. Numbering of secondary structure elements in panels (B, D, F, H, J) is relative to P22 S-terminase. See also Figure S2.
Partial structures of S-terminase subunits are also available for the SPP1-like Bacillus phage SF6 and λ, two members of the Siphoviridae family. SF6 (Figure 2E,F) (Buttner et al., 2012) and P22 (Figure 2A,B) S-terminases share a nonameric quaternary structure and a similar organization of the β-stranded dome and 9-stranded porin-like channel (Figure 1A), which are superimposable (r.m.s.d. ~2.2 Ǻ). However, the β-hairpin β1–β2 (forming the β-dome) originates at a topologically different position in the two terminases. In P22, it extends the loop between α-helices α2–α3 in the α-helical core (Figure 2B), while in SF6, it projects from helices α5–α6 that form the oligomerization core (Figure 2F). Furthermore, there is a significant difference in the way the N-terminal helical domain connects to the oligomerization core. In P22, the connection between these two domains is rigid (Figure 2B), while in a 4 Å structure of the SF6 fl-S-terminase (Buttner et al., 2012) no continuity is observed between N-termini and the oligomerization core (Figure 2E,F), suggesting a flexible hinge. A dimeric fragment of phage λ S-terminase (gpNu1) that comprises the DNA-binding domain (DBD) is also available (de Beer et al., 2002) (Figure 2G,H). This fragment (residues 1–68 of a 181-residue protein) assembles into a dimer in solution and possesses a pair of winged helix-turn-helix DNA-binding motifs. λ-DBD can be tentatively superimposed onto the P22 helical core with an r.m.s.d. ~3.6 Ǻ (Figure S2B,C). This structural alignment is poor, however, and only 32 of the 68 residues of λ-DBD superimpose onto P22 helices α1–α3. In addition, there is a topological inversion between the two polypeptide chains that point in opposite directions and P22 S-terminase lacks a `wing', essential for DNA binding in winged helix-turn-helix DNA-binding motifs (Wintjens and Rooman, 1996). Finally, most of the residues in λ-DBD directly involved in DNA-binding (Lys5 in helix αA and Arg17, Thr18, Gln20, Asn21, Gln23 in helix αB) (de Beer et al., 2002) do not have a direct counterpart in P22. Finally, the structure of the T4-like phage 44RR S-terminase (gp16) (Figure 2I,J) also shows dramatic structural difference as compared to P22, to the point that structural superimposition was not attempted. Phage 44RR S-terminase is built by a simple α-helical hairpin and presents a significantly larger central channel (>35 Å in diameter depending on oligomeric state, 11-mer or 12-mers) (Sun et al., 2012). Thus, viral S-terminase subunits differ dramatically in structure and oligomeric state, even within closely related phages that infect similar hosts. The lack of a winged helix-turn-helix DNA-binding motif in the N-terminus of P22 S-terminase suggests that this domain may not be directly involved in DNA-binding.
S-terminase C-terminus is partially structured
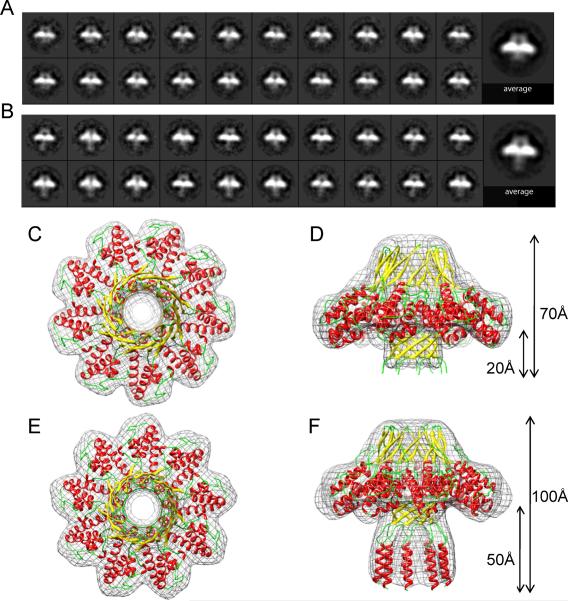
Both in solution and in crystal, the C-terminus of P22 S-terminase is highly susceptible to proteolysis (Roy et al., 2011). The amino acid sequence of residues 140–162 contains some of the signature features of intrinsically disordered proteins (Dyson and Wright, 2005), such as low sequence complexity and high amino-acid compositional bias (e.g. few bulky hydrophobic amino acids, high proportion of polar and charged amino acids like Arg, Glu, Lys, etc.) (Figure S3A). However, unlike natively disordered proteins (Dyson and Wright, 2005), the residue sequence 142–156 (`DRDKRRSRIKELFNR') has a high propensity to fold into a basic α-helix (Figure S3B,E). These observations led us to hypothesize that the C-terminus of S-terminase is structurally heterogeneous in solution, existing as an unstructured polypeptide chain in equilibrium with a folded α-helix. To test this hypothesis, we analyzed particles of the fl-S-terminase subunit by negative-stain electron microscopy (EM) single particle analysis (Figure S3F). Reference-free alignment and classification of 22,665 automatically selected particles revealed the existence of two populations of S-terminase that appeared identical in top views, but had distinct features in side views. To perform a more detailed assessment of their differences, a subset of 8,219 side-view particles from both populations underwent an additional round of alignment and classification. Whereas ~80% of the side-view particles exhibited a `short barrel' (Figure 3A), 20% of the particles presented a ~30 Å density extending outward from the 9-stranded β-barrel channel (referred to as `extended barrel') (Figure 3B). The two differing populations of particles were separated into two datasets and, together with top views, were used to compute a 3D-reconstruction of S-terminase in the two conformational states (Figure 3C–F). The reconstruction of `short-barrel' S-terminase at 18 Å resolution, (estimated by Fourier shell correlation at 0.5 criteria (Harauz and van Heel, 1986) (Figure S3G)) fits well with the crystal structure of S-terminase lacking the DNA-binding domain (Figure 3C,D). Since the sample used for EM-analysis was minimally degraded on SDS-PAGE, it is unlikely that this reconstruction corresponds exclusively to particles lacking C-terminal residues 140–162. Instead, the population of S-terminase with a `short barrel' represents particles with an unstructured C-terminus, which by negative stained EM-analysis is indistinguishable from cl-S-terminase. In contrast, the population of S-terminase with an `extended barrel' (Figure 3E,F), also solved at 18 Å resolution (Figure S3G), fits well with a model of the fl-S-terminase that contains a `structured' C-terminal domain, with residues 142–156 possibly folded into a straight α-helix (Figure S3C–E). In this hypothetical model, nine α-helices assemble together to extend S-terminase barrel by ~30 Å (Figure 3F). Although at this resolution we cannot determine if the putative α-helices lining the central channel are straight or bent with respect to the channel central axis, the fit between negative stain reconstruction and putative model of fl-S-terminase is remarkably good (Figure 3F). Interestingly, this model predicts that at least three basic side chains (Lys146, Arg150, Arg157) (Figure S3D–E) project into the central channel of each S-terminase, which would generate a highly basic inner core. Thus, single particle analysis of S-terminase particles supports the hypothesis of a plastic conformation of P22 S-terminase C-terminus.
Figure 3. Single particle analysis of negatively stained fl-S-terminase reveals two structurally distinct populations.
Class averages of fl-S-terminase reveal particles characterized by a `short-' (A) and an `extended-barrel' (B). Particles in (B) exhibit an extension of the DNA-channel by ~30Å as compared to particles in (A). A 3D-negative stain reconstruction (in gray) of S-terminase with a `short barrel' is overlaid to a ribbon model of the crystallographic structure of cl-S-terminase spanning residues 1–139, shown in top (C) and side (D) views. Top (E) and side (F) views of a 3D-negative stain reconstruction of S-terminase particles with an `extended barrel' are overlaid to a model of the fl-S-terminase that includes the crystallographic structure and an hypothetical model of C-terminal residues 140–158 produced by Modeller (Sali and Blundell, 1993) (Figure S3 F,G).
The C-terminus of P22 S-terminase mediates DNA-binding and formation of an assembly complex with L-terminase
We used an electrophoretic mobility shift assay (EMSA) on agarose gel to investigate S-terminase DNA-binding activity. We PCR amplified P22 gene 3 (referred to as gp3-DNA), which contains a pac site between nucleotides 265–286 (Casjens et al., 1987; Casjens and King, 1974; Wu et al., 2002). Incubation of fl-S-terminase with gp3-DNA (~500 bps) efficiently slowed down its electrophoretic mobility (Figure 4A). Quantification of this band-shift suggested a cooperative DNA-protein interaction (Figure 4C). Since the C-terminus of P22 S-terminase is highly basic (Figure S3A), we tested its putative involvement in DNA-binding. Strikingly, a deletion construct of S-terminase lacking residues 140–162, ΔC140-S-terminase, failed to bind gp3-DNA under identical experimental conditions (Figure 4B,C). To rule out the possibility that the C-terminal deletion had changed the oligomeric state of P22 S-terminase (as observed for bacteriophage SF6 (Buttner et al., 2012) and R44 (Sun et al., 2012) S-terminases) and its biochemical activity, we repeated the band-shift assay using cl-S-terminase, obtained by limited proteolysis of fl-S-terminase with chymotrypsin, which readily cleaves the last 23 residues (Roy et al., 2011). Proteolyzed S-terminase also showed complete loss of DNA binding (data not shown), confirming the results obtained with ΔC140-S-terminase.
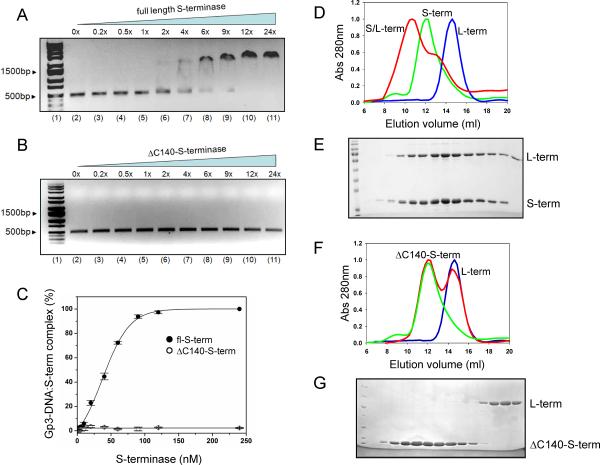
Figure 4. P22 S-terminase DNA-binding activity.
EMSA of fl-S-terminase (A) or ΔC140-S-terminase (B) binding to gp3-DNA. In both panels, lanes 2–11 show a titration of 0- to 24-fold equivalents of fl- and ΔC140-S-terminase incubated with the gp3-DNA and separated on a 1.5% agarose gel followed by ethidium bromide staining. (C) Quantification of EMSAs in (A,B) based on four independent repeats. The binding of fl-S-terminase (D) or ΔC140-S-terminase (F) to L-terminase was characterized by SEC on a Superose 12 column; elution peaks for S-terminase, L-terminase and the S/L-terminase complex are shown in green, blue and red, respectively. (E) and (G) show SDS-analysis of fractions eluted from gel filtration in panel (D) and (F), respectively.
To test if the nonameric S-terminase of bacteriophage P22 is competent for assembly to L-terminase, we performed binding studies using purified terminase subunits. By analytical size exclusion chromatography (SEC), S- and L-terminase migrated with elution volumes (Evol) of ~12.1 and 15.1 ml (Figure 4D), corresponding to globular species of ~180 KDa (a nonamer) and 60 KDa (a monomer) (Nemecek et al., 2007), respectively. Incubating a 2-fold molar excess of L-terminase with S-terminase resulted in a larger species migrating with Evol ~ 10.1 ml, corresponding to ~ 300 KDa (Figure 4D). SDS-page analysis (Figure 4E) confirmed this species had both S- and L-terminase in approximate 3:2 molar ratio. Interestingly, ΔC140-S-terminase (or proteolyzed cl-S-terminase) incubated with L-terminase under identical experimental conditions failed to assemble into a complex (Figure 4F,G), yielding two distinct species migrating separately by SEC. Thus, the C-terminus of P22 S-terminase is essential for DNA-binding and assembly to L-terminase; deletion of C-terminal residues 140–162 (without altering the nonameric quaternary structure) results in complete loss of S-terminase function.
S-terminase is a DNA-dependent ATPase-activating protein
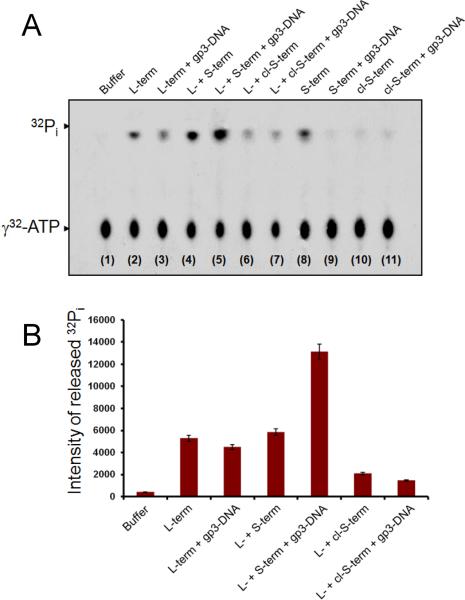
To determine how S-terminase modulates the ATPase activity of L-terminase, we carried out an ATPase assay with radioactive 32γ-ATP, using S- and L-terminase subunits, followed by thin layer chromatography on polyethyleneimine cellulose (PEI-TLC) (Figure 5A). Given the sensitivity of this radioactive assay, all factors used in the assay were >99% pure to avoid co-purification of contaminating ATPases from the expression host. At 37° C, in the absence of cold ATP (single-turnover ATPase assay), the intrinsic ATPase activity of L-terminase resulted in distinct release of 32Pi that was ~18-fold greater than that caused by spontaneous hydrolysis of 32γ-ATP in aqueous buffer (Figure 5A, lane 2 and 1, respectively). The basal ATPase activity of L-terminase was reduced by ~10% in the presence of gp3-DNA (Figure 5A, lane 3). This small but reproducible drop in activity (Figure 5B) is likely due to the nuclease activity of L-terminase that generates free dNTPs, potentially competing with 32γ-ATP for binding to L-terminase. Addition of nonameric S-terminase did not increase the ATPase activity of L-terminase in a statistically significant manner (Figure 5B, lane 4). While the intensity of released 32Pi in Figure 5A, lane 4 appears greater than that in lane 2 (free L-terminase), this value has to be corrected for the 32Pi released by free S-terminase in lane 8. As previously reported for at least three other S-terminases (gpNu1 in λ, gp16 in T4 and gp1 in SPP1 (Rao and Feiss, 2008)), ATPase activity was in fact observed for the fl-S-terminase of P22 (Figure 5A, lane 8), which lacks a classical Walker A- or Walker B-type ATP binding motif (Rao and Feiss, 2008). This fl-S-terminase ATPase activity was completely abolished by either adding gp3-DNA (Figure 5A, lane 9) or by cleaving off the DNA- and L-terminase binding domain (Fig 5A, lane 10). These data strongly suggest that P22 S-terminase is not a bona fide ATPase; the putative ATPase activity observed in vitro (Figure 5A, lane 8) likely results from the non-specific binding of 32γ-ATP to basic residues in the DNA-binding domain, which may facilitate its hydrolysis. Furthermore, since addition of gp3-DNA dramatically reduced S-terminase ATPase activity (Figure 5A, lane 9), this rules out that the observed ATPase activity seen in our assay is due to contaminating ATPases, which, if present, would not be inhibited by DNA. In contrast, the ATPase activity of L-terminase was significantly stimulated by S-terminase in the presence of gp3-DNA (Figure 5A, lane 5). The synergistic action of L-, S-terminase and gp3-DNA enhanced 32Pi release by ~42-fold as compared to spontaneous hydrolysis (Figure 5B). The ATPase activity of L-terminase in the presence of S-terminase and gp3-DNA was ~2.5-fold greater as compared to free L-terminase and this effect was specifically and directly dependent on the presence of gp3-DNA (Figure 5B). Deletion of S-terminase DNA- and L-terminase binding domain reduced the ATPase activity of L-terminase by more than 10-fold (Figure 5B) both in the presence and absence of gp3-DNA (Figure 5A, lane 7 and 6, respectively). Thus, P22 S-terminase bound to its own DNA functions as a specific ATPase-activating factor for L-terminase.
Figure 5. S-terminase activates the ATPase activity of L-terminase in the presence of gp3-DNA.
(A) ATPase assay resolved on PEI-TLC in the presence of different reactants and γ32-ATP. The position of γ32-ATP and 32Pi is indicated by arrows (B). Quantification of 32Pi released during the ATPase assay. The intensity of 32Pi released by L-terminase in the presence of S-terminase and gp3-DNA (lanes 4–5 in Figure 5A) was corrected by subtracting the intensity of 32Pi released in control reactions containing only S-terminase, with and without gp3-DNA (lanes 8–9 in Figure 5A). Error bars are calculated from averaging the intensity of 32Pi released over five independent experiments carried out under identical conditions. The average standard deviation is usually less than 3%.
DISCUSSION
An open question in virology is how dsDNA bacteriophages and herpesviruses efficiently package their genomes into empty procapsids in the presence of a large excess (>100 fold) of host DNA. In a general transducing phage like P22, each round of infection results in only 2% of newly replicated particles that carry host DNA instead of the viral chromosome. Despite the large excess of host DNA present during packaging, the packaging reaction is robust and 98% of virions in vivo contain phage DNA (Ebel-Tsipis et al., 1972). This specificity implies the existence of a fine mechanism that allows P22 to discriminate, bind and package its genome with greater affinity than host DNA. Likewise, to avoid wasteful hydrolysis of ATP when non-specific DNA encounters the packaging-motor, viruses must have developed fine mechanisms to regulate genome-packaging ATPase activity before and during the packaging reaction. To account for this regulation, both specific binding to packaging initiation sites and specific stimulation of genome-packaging ATPase activity have been ascribed to the S-terminase subunit that synergizes with L-terminase to sustain genome-packaging (Rao and Feiss, 2008).
The high resolution crystal structure of nonameric P22 S-terminase and the biochemical analysis presented in this study shed light on several unexpected properties governing the biology of viral S-terminases. First, there is a fundamental structural diversity among tailed bacteriophage S-terminase subunits, both at the level of protomer conformation and oligomer quaternary structure (Fig. 2). P22 protomer is superimposable only to the closely related Podoviridae Sf6, albeit the latter forms octamers instead of nonamers. Intriguingly, the β-hairpin (β1–β2) that forms the β-dome in P22, is absent in Sf6, but conserved in the SPP1-like phage SF6 (a Siphoviridae). This suggests that a complex shuffling of structural blocks must have occurred throughout evolution of tailed bacteriophages (Casjens and Thuman-Commike, 2011). The structural divergence of viral S-terminases is extreme between P22 and the T4-like phage 44RR (a Myoviridae), whose S-terminase subunit is completely different than in P22, both in fold (a helical hairpin) and oligomerization state (11/12-mer versus 9-mer). Together with the structure, the functional mechanisms underlining DNA-recognition are likely to have evolved and diverged throughout evolution, to the point that a universal mechanism for DNA binding is unlikely to exist. For instance, both P22 S-terminase and λ gpNu1 bind efficiently and cooperatively to DNA (de Beer et al., 2002). However, the N-terminal domain of P22 S-terminase does not contain a winged helix-turn-helix DNA-binding motif, as seen in λ gpNu1 (Fig. S2B,C), and DNA-recognition in P22 is fully dependent on C-terminal residues 140–162 (Figure 4). Interestingly, this moiety of P22 S-terminase is highly flexible and poorly structured both in solution (Roy et al., 2011) and in crystal, which suggests the DNA-binding domain may become preferentially stabilized upon binding to DNA as opposed to being constitutively folded, as in λ. Deletion of C-terminal residues 140–162 completely abolishes both DNA- and L-terminase binding activity of P22 S-terminase (Figure 4). Since the structures of the fl- and cl-S-terminase are identical, it is unlikely that the C-terminal deletion used in this study alters the 3D-conformation and/or oligomeric state and hence binding activity towards DNA and L-terminase. The location of a C-terminal DNA-binding domain in P22 S-terminase is consistent with the observation that deletion of residues 143–162 eliminates nonspecific DNA-binding activity (Nemecek et al., 2008). Finally, our finding of a C-terminal L-terminase-binding domain in P22 S-terminase agrees well with the recently proposed hypothesis by Casjens and Thuman-Commike that, based on evolutionary consideration, the C-terminus of S-terminase mediates binding to L-terminase (Casjens and Thuman-Commike, 2011).
Eleven missense mutations that increase the frequency of phage P22 generalized transduction (HT-mutations) map to the gene encoding the S-terminase (Casjens et al., 1992). Intriguingly, only one out of eleven of these substitutions (Glu→Lys)152 lies in the C-terminus of S-terminase, which in P22 contains the DNA-binding domain. All other mutations mainly cluster in the center of the protein, within residues 80 and 112. This is unexpected, as, intuitively, one would expect mutations that increase efficiency of transduction to cluster in the putative DNA-binding domain. However, the high frequency of transduction phenotype is very complex and could be the resultant of completely indirect effects. For instance, the point mutation at position (Ala→Val)112 has been thought to increase the frequency of transduction by affecting the DNA-binding properties of S-terminase (Casjens et al., 1992). However, a similar mutation at the same position, (Ala→Thr)112 changes the oligomeric state of P22 S-terminase from nonamer to decamer (Nemecek et al., 2007) suggesting that the DNA-binding activity of S-terminase can be affected indirectly by point mutations that cause structural rearrangements in the oligomer conformation. Likewise, four of the eleven HT-mutations (Leu→Phe)80, (Glu→Lys)81, (Asp→Asn)91, (Val→Ile)95 are clustered at positions in close proximity to the pac site itself, which spans the region between codons 88 and 97 in gene 3 (Wu et al., 2002). This suggests that certain HT-missense mutations could affect the DNA recognition site more than the ability of S-terminase protein to recognize it.
Understanding the DNA- and L-terminase binding activity of P22 S-terminase led us to discover an unexpected property of this molecule. Upon binding to gp3-DNA, S-terminase becomes a specific activator of L-terminase ATPase activity. Both a construct of S-terminase lacking the DNA/L-terminase-binding domain (res. 140–162) and fl-S-terminase in the absence of gp3-DNA were unable to stimulate the ATPase activity of L-terminase (Figure 5), thus confirming that specific binding to DNA is required to stimulate genome-packaging. The role of DNA in this interaction may involve wide conformational stabilization of S-terminase that folds a C-terminal domain, thus `igniting' the packaging-engine to `fire' ATP hydrolysis. This, in turn, could lower the entropic barrier of inserting the linear end of the P22 genome into the procapsid, thus triggering the packaging reaction in the same manner as a spark ignites a combustion-engine.
How is viral DNA recognized by S-terminase? It was previously proposed that S-terminases may recognize viral DNA using a nucleosome-like recognition characterized by wrapping dsDNA around the outer rim of the S-terminase ring formed by its N-terminus domains (Buttner et al., 2012; de Beer et al., 2002; Nemecek et al., 2008; Sun et al., 2012; Zhao et al., 2010). This model has quickly gained popularity in the literature, although it lacks experimental validation. The structural and biochemical results presented in this work do not support this model. In the absence of a strong, biochemically detectable DNA-binding activity in the N-terminal domain, it would be energetically too costly to bend dsDNA around this domain. In addition, the nucleosome-like model does not assign a function to the central ~23 Ǻ diameter channel in P22 S-terminase that is large enough to accommodate hydrated dsDNA (Vlieghe et al., 1999). Our discovery of a C-terminal DNA-binding site makes it plausible to speculate that DNA could be threaded through the internal channel as in a portal protein (Olia et al., 2011) as opposed to being `wrapped' on the outside of S-terminase like a `gear', although other assembly models are certainly possible and cannot be ruled out a priori without experimental validation.
In conclusion, the structural diversity of viral S-terminase subunits, the lack of a unique quaternary structure oligomeric state, and the fundamentally different way by which viruses package DNA lend support to the idea that S-terminases recognize viral genomes differently, and a universal mechanism for DNA-binding is not likely to exist. Whatever mechanism P22 S-terminase uses to recruit gp3-DNA, this work demonstrates that DNA- and L-terminase binding activities require the protein C-terminus and DNA-binding is coupled to stimulation of L-terminase ATPase activity, which triggers ATP-dependent genome-packaging.
EXPERIMENTAL PROCEDURES
Molecular biology techniques
The genes encoding small (gp3) and large (gp2) terminase subunits were PCR amplified from P22 genomic DNA (Pedulla et al., 2003) and ligated between BamHI and HindIII restriction sites of expression vectors pMal-2cE (New England Biolabs) and pET-28a (Novagen), respectively. The mutant ΔC140-S-terminase was constructed by introducing a stop codon at position 140 of S-terminase. All plasmids were sequenced to confirm the fidelity of the DNA sequence. Expression and purification of fl- and ΔC140-S-terminase and limited proteolysis of fl-S-terminase with chymotrypsin to obtain cl-S-terminase (res 1–139) was carried out as described (Roy et al., 2011). L-terminase was also expressed in E.coli strain BL21 (DE3/pLysS) by induction at 16° C for 16 hours with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside. Cell pellets expressing L-terminase were lysed by sonication in Lysis Buffer (20 mM Tris-HCl, pH 7.5, 300 mM NaCl, 0.1% (v/v) CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate), 5 mM β-Mercaptoethanol, 1 mM PMSF). L-terminase was purified by Ni2+ affinity chromatography followed by size-exclusion chromatography on a Superdex 200 column (GE Life Sciences) equilibrated in 20 mM Tris-HCL, 150 mM Nacl, 5 mM β-Mercaptoethanol, 5% (v/v) glycerol, pH 7.5.
Structure determination
Crystallization of cl-S-terminase was previously described (Roy et al., 2011). The best crystals were obtained at pH 7.0 using 20% (w/v) PEG 3350 in the presence of 0.2 M potassium thiocynate. Expression of semet-derivatized S-terminase was carried out as described by Olia et al. (Olia et al., 2011). Purification of semet-derivatized S-terminase was identical to that of the wild type protein (Roy et al., 2011). The structure was solved by SAD (Single-wavelength Anomalous Dispersion) using a single semet-derivatized crystal at the National Synchrotron Light Source (NSLS) beam-line X6A at a wavelength of 0.976 Å (Table 1). The positions of 27 selenium sites (3 per subunit) were located using Phenix (Adams et al., 2002). Initial phases to 2.5 Ǻ resolution were improved by 9-fold ncs-averaging and extended to the resolution of the native data (1.75 Å). A partial model was built by Autobuild, in Phenix (Adams et al., 2002) and completed manually in Coot (Emsley and Cowtan, 2004). The structure was then subjected to seven cycles of positional and anisotropic B-factor refinement in Phenix (Adams et al., 2002). To lower the Rfree below 29%, ncs-restraints were not used during refinement. Peaks above 3σ in a Fo – Fc difference electron density map were modeled as water molecules. The structure of P22 S-terminase has been refined to a Rwork / Rfree of 17.73 / 21.65%, at 1.75 Å resolution. The final S-terminase model contains residues 1–139 for chain A; 5–139 for chain D; 6–139 for chain F; 7–139 for chains C and G; 8–139 for chains B, E, H, and I; respectively. The final model also contains 1,397 water molecules. The model has excellent geometry (Table 1), with 100% residues in the most favored regions of the Ramachandran plot, and a root mean square deviation (rmsd) of bond lengths and angles of 0.006 Å and 0.918°, respectively. All Ribbon models were generated using PyMol (DeLano, 2002) or Chimera (Pettersen et al., 2004). Crystallization and structure determination of fl-S-terminase are described in Supplementary Information.
Negative stain electron microscopy and single particle analysis
Full length nonameric S-terminase at a concentration of ~2 μM were applied to glow discharged carbon coated copper grids and negatively stained with 2% phosphotungstic acid, as previously described (Nemecek et al., 2007). Electron micrographs were automatically acquired on a Tecnai F20 transmission electron microscope operating at 120 keV using Leginon data collection software (Suloway et al., 2005). Micrographs were collected on a Tietz F415 CCD camera at a nominal magnification of 50,000× (1.63 Å/pixel) with an underfocus range of 0.7 to 1.5 microns. All processing was performed within the Appion software package (Lander et al., 2009). From an initial dataset of 88,147 automatically selected particles, a subset of 22,665 properly centered particles that did not have closely neighboring densities were extracted for image processing. The particles were aligned and classified to generate 2D class averages in a reference-independent method using the IMAGIC image processing software (van Heel et al., 1996). Side-view particles were separated from top-view particles, and the side-view particles were further classified into short- and extended-barrel datasets. The initial references for the 3D-reconstructions were generated by angular reconstitution of the class averages while imposing c9 symmetry. The 3D models were improved by refinement of the particles against the model over the course of 20 iterations using the EMAN2 software package (Tang et al., 2007). The reported resolutions of the reconstructions according to Fourier shell correlation at 0.5 (Harauz and van Heel, 1986) are 18.1 and 18.2 for the short- and extended-barrel structures, respectively (Figure S3). The negative stain reconstructions were visualized using the program Chimera (Pettersen et al., 2004). X-ray structures were fit in the reconstructions manually and then the fit was improved using the command “Fit Models in Maps” in Chimera (Pettersen et al., 2004).
EMSA, SEC and ATPase assay
DNA fragments corresponding to the coding region of P22 gene 3 (486 bp) were amplified by PCR from a P22 genomic DNA and purified by gel extraction. EMSAs were carried out by adding 0 to 24 molar equivalents of nonameric S-terminase (corresponding to 0 – 240 nM) to gp3-DNA (at a fixed concentration of 10 nM) in a 50 μL reaction volume, containing 50 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl pH 7.5. Samples were incubated at room temperature for 30 min and then electrophoresed in a 1.5% (w/v) agarose gel with 1 × TAE (Tris-Acetate-EDTA) buffer at 100 V for approximately 45 min, followed by ethidium bromide staining. Bands corresponding to the S-terminase:DNA complex were quantified using ImageJ (Abramoff et al., 2004), and the percentage of complex was plotted against the known concentration of S-terminase using the program Origin. Analytical SEC analysis was carried out on a Superose 12 column (G.E. Healthcare) pre-equilibrated in 10 mM Tris, pH 8.0, 150 mM NaCl and 5 mM β-Mercaptoethanol as described (Pumroy et al., 2012) (Supplementary Information). The ATPase-stimulating activity of S-terminase was measured using a radioactive ATPase assay as described by Leffers and Rao (Leffers and Rao, 2000). All factors used in this assay were >99% pure as judged by SDS-PAGE and Matrix-assisted laser desorption/ionization (MALDI) mass spectrometry analysis. Purified L-terminase (1 μM) was incubated with 5 μM purified S-terminase (e.g. fl-Sterminase or cl-S-terminase) in the presence of 75 nM [γ32P]ATP (specific activity, 3000 Ci/mmol; PerkinElmer, Inc) in ATPase buffer (50 mM Tris-HCl, pH 7.5, 0.1 M NaCl, 5 mM MgCl2). To test if DNA enhances S-terminase ATPase-activating activity, 20 nM gp3-DNA (corresponding to 1 copy of gp3-DNA per ~20 equivalents of nonameric S-terminase) or non-specific DNA (empty pUC19 vector) was added to the reaction mixture. Control experiments included free S-terminase with or without gp3-DNA. The total reaction volume was set to 20 μl and samples were incubated for 20 minutes at 37 °C. Reactions were terminated by adding EDTA to 50 mM final concentration and 1 μl reaction mixture was spotted on a TLC-PEI plate (Sigma-Aldrich). TLC plates were developed (Leffers and Rao, 2000), air-dried, autoradiographed and the intensity of each spot quantified using ImageJ (Abramoff et al., 2004). The intensity of 32Pi released by L-terminase in the presence of S-terminase was determined by subtracting the amount of 32Pi released by free S-terminase incubated with [γ32P]ATP under identical conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Vivian Stojanoff at the NSLS and to the macCHESS staff for beam time and assistance in data collection. We thank David Prince at the University of Iowa for useful suggestions on the ATPase assay. Electron microscopic imaging and reconstruction were conducted at the National Resource for Automated Molecular Microscopy which is supported by the NIH through a P41 program grant (RR17573) from the National Center for Research Resources. This work was supported by NIH grants 1R56 AI095974-01 to G.C. The coordinates and structure factors for the high resolution structure of S-terminase have been deposited in the protein Data Bank (accession code 3P9A).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Ackermann HW. Bacteriophage observations and evolution. Res Microbiol. 2003;154:245–251. doi: 10.1016/S0923-2508(03)00067-6. [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Baumann RG, Black LW. Isolation and characterization of T4 bacteriophage gp17 terminase, a large subunit multimer with enhanced ATPase activity. J Biol Chem. 2003;278:4618–4627. doi: 10.1074/jbc.M208574200. [DOI] [PubMed] [Google Scholar]
- Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, Chechik V, Jeschke G, Dykeman E, Benini S, Robinson CV, et al. Structural basis for DNA recognition and loading into a viral packaging motor. Proc Natl Acad Sci U S A. 2012;109:811–816. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278:23251–23259. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- Casjens S, Huang WM, Hayden M, Parr R. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J Mol Biol. 1987;194:411–422. doi: 10.1016/0022-2836(87)90671-1. [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Casjens S, Sampson L, Randall S, Eppler K, Wu H, Petri JB, Schmieger H. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J Mol Biol. 1992;227:1086–1099. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Catalano CE. Viral Genome Packaging Machines: Genetics, Structure and Mechanism. Landes. 2005:1–4. Bioscience/ Eurekah.com. [Google Scholar]
- Cingolani G, Moore SD, Prevelige PE, Jr., Johnson JE. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J Struct Biol. 2002;139:46–54. doi: 10.1016/s1047-8477(02)00512-9. [DOI] [PubMed] [Google Scholar]
- de Beer T, Fang J, Ortega M, Yang Q, Maes L, Duffy C, Berton N, Sippy J, Overduin M, Feiss M, Catalano CE. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–991. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- DeLano WL. 2002 www.pymol.org.
- Duffy C, Feiss M. The large subunit of bacteriophage lambda's terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–561. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ebel-Tsipis J, Botstein D, Fox MS. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972;71:433–448. doi: 10.1016/0022-2836(72)90361-0. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc Natl Acad Sci U S A. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero S, Galdiero M, Pedone C. beta-Barrel membrane bacterial proteins: structure, function, assembly and interaction with lipids. Curr Protein Pept Sci. 2007;8:63–82. doi: 10.2174/138920307779941541. [DOI] [PubMed] [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32:W500–502. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EN, Jackson DA, Deans RJ. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978;118:365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage t4 terminase. J Biol Chem. 2004;279:40795–40801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffers G, Rao VB. Biochemical characterization of an ATPase activity associated with the large packaging subunit gp17 from bacteriophage T4. J Biol Chem. 2000;275:37127–37136. doi: 10.1074/jbc.M003357200. [DOI] [PubMed] [Google Scholar]
- Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272:3495–3501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- Lorenzen K, Olia AS, Uetrecht C, Cingolani G, Heck AJ. Determination of stoichiometry and conformational changes in the first step of the P22 tail assembly. J Mol Biol. 2008;379:385–396. doi: 10.1016/j.jmb.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluf NK, Gaussier H, Bogner E, Feiss M, Catalano CE. Assembly of bacteriophage lambda terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–15268. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- Nadal M, Mas PJ, Blanco AG, Arnan C, Sola M, Hart DJ, Coll M. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. Proc Natl Acad Sci U S A. 2010;107:16078–16083. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Gilcrease EB, Kang S, Prevelige PE, Jr., Casjens S, Thomas GJ., Jr. Subunit conformations and assembly states of a DNA-translocating motor: the terminase of bacteriophage P22. J Mol Biol. 2007;374:817–836. doi: 10.1016/j.jmb.2007.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemecek D, Lander GC, Johnson JE, Casjens SR, Thomas GJ., Jr. Assembly architecture and DNA binding of the bacteriophage P22 terminase small subunit. J Mol Biol. 2008;383:494–501. doi: 10.1016/j.jmb.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olia AS, Prevelige PE, Jr., Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597–603. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Karthikeyan T, Houtz JM, Hendrix RW, Hatfull GF, Poteete AR, Gilcrease EB, Winn-Stapley DA, Casjens SR. Corrected sequence of the bacteriophage p22 genome. J Bacteriol. 2003;185:1475–1477. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pumroy RA, Nardozzi JD, Hart DJ, Root MJ, Cingolani G. Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin alpha5. J Biol Chem. 2012;287:2022–2031. doi: 10.1074/jbc.M111.315838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Roy A, Bhardwaj A, Cingolani G. Crystallization of the Nonameric Small Terminase Subunit of Bacteriophage P22. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2011;F67:104–110. doi: 10.1107/S174430911004697X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Cingolani G. Structure of P22 headful packaging nuclease. J Biol Chem. 2012 doi: 10.1074/jbc.M112.349894. under revisions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, Alonso JC, Antson AA. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10:592–598. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. Proc Natl Acad Sci U S A. 2012;109:817–822. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Gentz PM, Rossmann MG, Rao VB. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Sun S, Rao VB, Rossmann MG. Genome packaging in viruses. Curr Opin Struct Biol. 2010;20:114–120. doi: 10.1016/j.sbi.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Vlieghe D, Turkenburg JP, Van Meervelt L. B-DNA at atomic resolution reveals extended hydration patterns. Acta Crystallogr D Biol Crystallogr. 1999;55:1495–1502. doi: 10.1107/s0907444999007933. [DOI] [PubMed] [Google Scholar]
- White JH, Richardson CC. Gene 18 protein of bacteriophage T7. Overproduction, purification, and characterization. J Biol Chem. 1987;262:8845–8850. [PubMed] [Google Scholar]
- Wintjens R, Rooman M. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J Mol Biol. 1996;262:294–313. doi: 10.1006/jmbi.1996.0514. [DOI] [PubMed] [Google Scholar]
- Wu H, Sampson L, Parr R, Casjens S. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol Microbiol. 2002;45:1631–1646. doi: 10.1046/j.1365-2958.2002.03114.x. [DOI] [PubMed] [Google Scholar]
- Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, Casjens SR, Tang L. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. Proc Natl Acad Sci U S A. 2010;107:1971–1976. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.