Abstract
Objective
We observed differences in atherosclerosis susceptibility in mouse inbred strains over the years as the health status of our animal rooms increased. Therefore, we investigated the effect of animal room health status on atherosclerosis susceptibility in different strains. As this data can also be used for genome-wide association mapping, we performed a mapping study and compared our results with previously found quantitative trait loci for atherosclerosis in mouse and human.
Methods and Results
Males and females from 48 inbred strains were housed in two animal rooms with different health status and given an atherogenic diet. We compared atherosclerosis susceptibility between animal rooms and between sexes and found that susceptibility is dependent on both health status and sex. Subsequently, the data were used for associations with loci on the mouse genome using 63,222 SNPs. Three loci in males and four loci in females were identified using the data from the low health status room. No significant associations were identified using the data from the high health status room.
Conclusion
Health status influences susceptibility to atherosclerosis and suggests that microbiological pressure plays an important role in the development of atherosclerosis in many strains. As we were only able to map susceptibility loci using the data from the lower health status room, we argue that susceptibility under these conditions is determined by a few key loci, while in the higher health status room different mechanisms might play a role in the differences in atherosclerosis susceptibility between strains and we did not have enough power to map the loci that are involved.
Keywords: atherosclerosis, inbred strains, QTL
Introduction
Cardiovascular disease (CVD) is the leading cause of death in the United States, and more than 81 million Americans suffer from some form of CVD. The pathological basis of CVD is atherosclerosis, a disease of the arterial blood vessels in which the walls of the blood vessels become thickened by the deposition of plaques composed of cholesterol, other lipids, inflammatory cells, and calcium deposits. Atherosclerosis susceptibility is complex and dependent on multiple environmental and genetic factors, and a number of genes have been identified and studied using both genotype- and phenotype-driven approaches. In a genotype-driven approach the function of a gene suspected to be involved in atherosclerosis is tested with a transgenic or knockout animal model 1-3, while in a phenotype-driven approach the atherosclerotic phenotype is linked to chromosome locations or quantitative trait loci (QTL) in a genome-wide scan 4,5.
QTL for atherosclerosis in mice have been found in various studies using mice that have either been induced with atherosclerosis with an atherogenic diet or in mice that are susceptible to atherosclerosis due to the lack of apolipoprotein E (APOE) or low-density lipoprotein receptor (LDLR). The APOE-deficient mouse develops atherosclerotic lesions even on a standard chow diet 6. The LDLR deficient mouse has elevated levels of LDL but no or very small lesions on the chow diet. When these mice are on a Western-type diet (containing 21% fat and 0.15% cholesterol)7, however, atherosclerotic lesions are detectable. Among the 43 QTL for atherosclerosis that have been identified to date, 12 were found in atherogenic diet models and 31 were found in mice deficient for APOE or LDLR 8.
Our lab has studied the genetics of atherosclerosis for many years using the differences in susceptibility between inbred strains. Over the years, our mouse facilities have become cleaner with respect to the microorganisms that are found in the mice. This resulted in a loss of the atherosclerosis phenotype using our standard protocol, which was an 8-week atherogenic diet starting at 8 weeks of age. Increased risk of atherosclerosis has been linked to a wide variety of infectious agents, including Chlamidia pneumoniae, Porphyromonas gingivalis, Helicobacter pylori, influenza A virus, hepatitus C virus, cytomegalovirus, and human immunodeficiency virus. Reports of risk, however, are mixed, depending on the pathogen. For example, in the case of H. pylori, one study reported that B6 female mice on a high-fat diet and infected with the bacteria had an equal prevalence of atherosclerosis but more foam cells in the plaques 9, while another study reported that both male B6 and male LDLR-deficient mice on a high-fat diet did not show any effect 10. Overall, it has been observed that C. pneumoniae and some of the periodontal organisms such as P. gingivalis contribute to the risk of atherosclerosis, but for all the other infectious agents, reports are conflicting 11.
To test atherosclerotic lesion development in mice kept in animal rooms of different health statuses, we conducted a study in both males and females in 48 inbred strains. These results were then used for genome-wide association mapping to identify the loci that cause differences in atherosclerosis susceptibility. Identification of new genes involved in the development of atherosclerosis in mice is a potentially powerful approach since these results can be used to find genes related to atherosclerosis in humans. Human orthologs of these novel genes can then be studied for variation and tested for correlation with atherosclerosis. This could be a significant step in the understanding of CVD and for the innovation of new therapies, but it will be imperative to put this in the context of such environmental factors as microbiological status in humans.
Materials and Methods
Animals and Housing
Ten males and 10 females of 48 mouse inbred strains (Table 1) were purchased from The Jackson Laboratory (Bar Harbor, ME). At the age of 6 weeks, animals were transferred from breeding rooms to one of two animal rooms designated as having either a low-health status (low-status) or a high-health status (high-status). Colonies were regularly monitored for viruses, bacterial species, ectoparasites and endoparasites (see Table 2 for a complete list). The only differences between rooms were the detection of Pneumocystis carinii, Pasturella pneumotropica, and Helicobacter spp. in the low-status room, which were not detected in the high-status room. All mouse rooms at The Jackson Laboratory are monitored in the same way by the Lab Animal Health Service using standard methods. All details of the JAX monitoring program are publically available at jaxmice/jax.org/genetichealth/index.html. Same-sex mice were housed four per pen in duplex polycarbonate cages equipped with pressurized individually ventilated mouse racks (Thoren Caging Systems Inc.) with HEPA-filtered air supply and exhaust. Water and food pellets containing 6% fat (Lab diet 5K52, PMI Nutritional International, Bentwood, Mo) were provided ad libitum. Mouse rooms were maintained at an ambient temperature of 21–23°C and a 12:12 hour light:dark cycle. All animals started on an atherogenic diet (18.5% dietary fat, 1.9% corn oil, 50% sucrose, 4.1% cellulose, 20% casein, 1% cholesterol, 0.5% cholic acid, 5% mineral mix, 1% vitamin mix, 0.3% DL-methione, 0.13% α-tocopherol, 1% choline chloride) at 9 weeks of age as previously described 5. Animals in the low-status room were kept on this diet for eight weeks and then sacrificed, while the animals in the high-status room were kept on the diet for 17 weeks and then sacrificed. All animal protocols were approved by The Jackson Laboratory Animal Care and Use Committee. Mouse handling and care complied with the Public Health Service animal welfare policy.
Table 1.
List of inbred strains used in this study
| Strains | |||
|---|---|---|---|
| 129S1/SvImJ | C57BL/6J | I/LnJ | PERA/EiJ |
| A/J | C57BLKS/J | JF1/Ms | PL/J |
| AKR/J | C57BR/cdJ | KK/HLJ | PWK/PhJ |
| BALB/cByJ | C57L/J | LP/J | RBF/DnJ |
| BALB/cJ | C58/J | MA/MyJ | RF/J |
| BPH/2J | CAST/EiJ | MOLF/EiJ | RIIIS/J |
| BPL/1J | CBA/J | MSM/Ms | SEA/GnJ |
| BPN/3J | CE/J | NOD/LtJ | SJL/J |
| BTBR T<+> tf/J | CZECHII/EiJ | NON/LtJ | SM/J |
| BUB/BnJ | DBA/1J | NZB/BlNJ | SPRET/EiJ |
| C3H/HeJ | DBA/2J | NZW/LacJ | SWR/J |
| C57BL/10J | FVB/NJ | P/J | WSB/EiJ |
Table 2.
List of agents monitored in the Jackson Laboratory's animal rooms.
| Viruses | |
|---|---|
| Ectromelia virus | Mouse hepatitus virus (MHV) |
| GDVII virus | Mouse parvovirus (MPV) |
| Hantaan virus | Mouse thymic virus (MTV) |
| K virus | Pneumonia virus of mice (PVM) |
| Lactic dehydrogenase elevating virus | Polyoma virus |
| Lymphocytic choriomeningitis (LCMV) | Reovirus 3 (REO 3) |
| Mouse minute virus (MMV) | Rotavirus |
| Mouse adenovirus (MAV) | Sendai virus |
| Mouse cytomegalovirus (MCMV) | |
| Bacteria & mycoplasma | |
| CAR bacillus | Mycoplasma pulmonis |
| Citrobacter rodentium | Salmonella spp. |
| Clostridium piliforme | Streptobacillus moniliformis |
| Parasites | |
| Encephalitozoon cuniculi | Pinworms |
| Fleas, fur mites, lice | Roundworms and other helminths |
| Follicle mites | Tapeworms |
| Opportunistic organisms | |
| Bordetella bronchiseptica | Pneumocystis carinii |
| Corynebacterium kutscheri | Pseudomonas spp. |
| Helicobacter spp. | Staphylococcus aureus |
| Klebsiella pneumoniae | Streptococcus spp. |
| Murine norovirus (MNV) | Opportunistic protozoa (eg, Giardia) |
| Pasteurella pneumotropica | |
Measurement of HDL Cholesterol Levels and Atherosclerotic Lesions
Blood was collected via retro-orbital sinus from animals that were food-deprived for 4 hours in the morning. Serum HDL cholesterol was analyzed on a Beckman Coulter Synchron CX 5 Delta autoanalyzer (Beckman Coulter, Inc., Brea, CA). The heart and aorta were washed in saline, immersed in a 4% formalin solution overnight, and transferred to a 10% formalin solution. Tissues were then processed as previously described 12. In brief, hearts were embedded in 25% gelatin and sectioned in a cryostat at –25°C. Fatty streak aortic lesion size was averaged from five aortic sections of 10 μm cut at 80-μm intervals. Staining was performed with Oil Red O (neutral lipids) and hematoxylin and counterstained with light green. All results are expressed as log(lesion size +1). Atherosclerosis and HDL datasets are deposited in the Mouse Phenome Database (phenome.jax.org) as Paigen1 and Paigen2.
Statistics
Statistical analyses were performed using JMP7 (SAS Institute). Strain data are presented as the mean ± standard deviation (SD). For the genome-wide association mapping, SNPs were selected from the following sources: JAX, Oxford, Merck, GNF and Perlegen (see www.jax.org/phenome for detailed information on the sources). The mouse genome was divided into non-overlapping 40-kb intervals. For each interval, one SNP that best met the following criteria was selected: highly polymorphic among 25 widely used classical laboratory strains, missing in a low number of genotypes, and evenly distributed across the genome. A total of 63,222 SNPs were selected. A hidden Markov model (HMM) was applied, fitting five states at each SNP, for the primary purpose of missing genotype imputation and for the secondary purpose of haplotype identification 13. A total of 580,781 missing genotypes (28.70%) were imputed for this particular dataset. All SNPs with imputed genotypes had a confidence score greater than 0.6, and the average filling accuracy of the imputed genotypes across the whole genome was 89.9%. At each SNP, a strain distribution pattern (SDP) was determined using the HMM smoothed haplotype states (HMMpath). We computed F-test statistics to measure the strength of association between genotype and phenotype. Its significance was estimated to detect haplotype groups with different mean phenotypes. The segregation of strains into haplotype groups varied widely over haplotype blocks; therefore, P-values of the F-test statistic were compared between haplotype blocks. Type I error rates for multiple testing due to genome-wide searching were controlled for using family-wise error rate control (FWER)14. The strain label in the phenotype data was shuffled to keep the genotype data intact. The minimum P-value was recorded on each permutation, and percentiles of their distribution were used to provide approximate multiple test-adjusted thresholds. The genome-wide type I error thresholds were estimated based on 1000 permutation tests. Peaks corresponding to P-value thresholds adjusted for global significance were defined as significant at α<0.05 level. Due to the genetic relatedness between genomes of inbred strains, HAM analysis has limited power to detect small genetic effects. Furthermore, FWER methods generally yield conservative results. To detect peaks with small genetic effects that were biologically relevant, the protection against type I error was relaxed and HAM peaks that exceed an α<0.20 were considered suggestive evidence of true genetic association. All analysis was done in the MATLAB computing environment (The Mathworks, http://www.mathworks.com), except the imputation of missing genotypes.
Results
Differences in Atherosclerosis Susceptibility Between Health Statuses
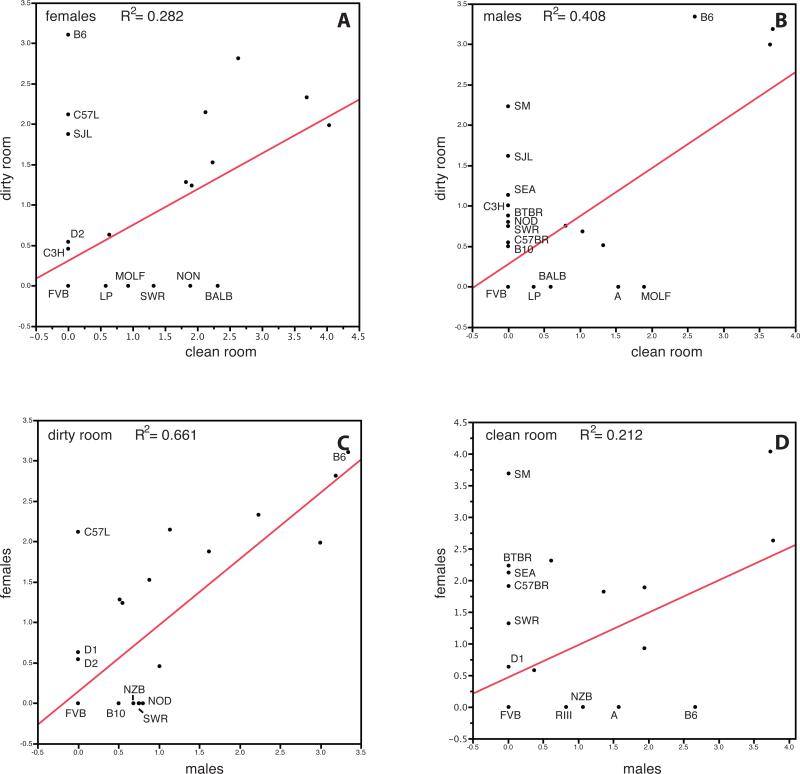
As previously shown, there is variation in atherosclerosis susceptibility among mouse inbred strains (Supplementary Figure 1). The differences between the low-status and high-status rooms were the detection of additional pathogens Pneumocystis carinii, Pasturella pneumotropica, and Helicobacter spp. in the low-status room. Strain distributions in the low-status versus the high-status rooms are plotted in Figure 1A and 1B. Although some strains seem to have similar susceptibility in both rooms (e.g., C57BR and BTBR females), many strains developed lesions only in the low-status room (males — SM, SJL, SEA, C3H, BTBR, NOD, SWR, C57BR, B10; females — B6, C57L, SJL, D2, C3H) but not in the high-status room even after longer exposure to the atherogenic diet.
Figure 1.
Differences in atherosclerosis susceptibility between mouse rooms. (A) strain distributions for females in the low-status and high-status rooms by log lesion area, (B) males in the low-status and high-status rooms by log lesion area, (C) males and females in the low-status room by log lesion area, (D) males and females in the high-status room by log lesion area.
Differences in Atherosclerosis Susceptibility Between Males and Females
In humans, few studies have been conducted on differences in the etiology and progression of CVD between males and females. The general perception is that CVD is predominantly a male disease, primarily because women develop CVD almost 10 years later. The reason for cardio-protection in females is still unclear, however, and remains under investigation. In this study, we extensively surveyed the formation of atherosclerotic lesions present in both female and male mice. The differences between females and males in the low-status and high-status rooms are shown in Figure 1C and D. In the low-status room we see that C57L females develop atherosclerotic lesions, while C57L males do not. In the high-status room we see more strains with differences between males and females: SM, BTBR, SEA, C57BR, and SWR females show lesions but males do not; RIII, NZB, and A males show lesions, but females do not. It is believed that elevated levels of plasma HDL cholesterol protects against cardiovascular disease in humans. We therefore measured HDL in all animals and looked for a correlation between HDL levels and atherosclerosis both at the individual level (Supplementary Figure 2) as at the strain level (data not shown). We did not observe any correlation in either males or females, in either room. This suggests that HDL cholesterol levels are not a major determining factor for the development of atherosclerosis in our inbred strains.
Genome-wide Association Analysis Reveals Differences in Genetic Loci
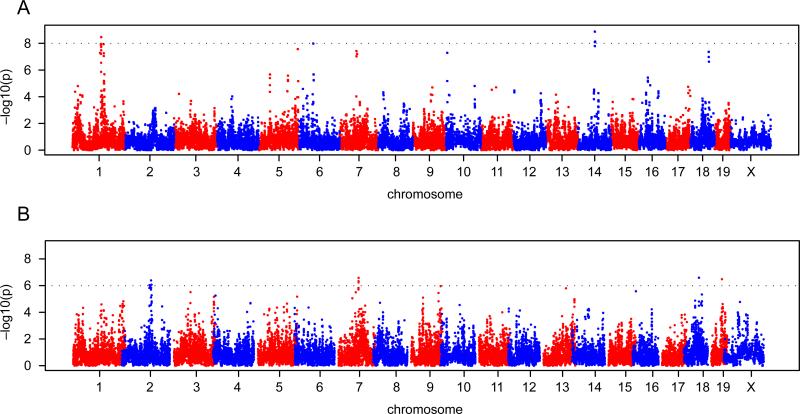
The datasets that we assembled for the above comparisons are suitable for genome-wide association mapping. We used the logarithm of the lesion size in males and females in both rooms. We did not observe any significant associations in either males or females in the high-status room, but we did observe significant loci in both males and females in the low-status room (Figure 2 shows the scan for females). We found 3 loci in males with a P-value less than 10-8 and 4 loci in females with a P-value less than 10-6 (Table 3). Interestingly, 5 of the 7 loci are within the confidence intervals of previously mapped QTL for atherosclerosis.
Figure 2.
Genome-wide haplotype association mapping in male mice (A) and female mice (B) from 42 inbred strains in the low-status room. The 0.63, 0.10, and 0.05 alpha thresholds, as determined by permutation testing, are indicated.
Table 3.
Results of the genome-wide analysis. Loci were only found using the mice with the lower health status.
| Sex | Chr | Locus start | Locus end | P-value | Genes in interval | QTL strains | QTLref |
|---|---|---|---|---|---|---|---|
| M | 1 | 108,137026 | 108,799,193 | 3.4×10-9 | Kdsr, Bcl2, Serpinb5, Phlppl, Vps4b, | B6 × 129 | 16 |
| 6 | 53,791,464 | 53,903,686 | 1.0×10-8 | Cpvl | |||
| 14 | 67,543894 | 67,957,095 | 3.4×10-9 | Bnip3l, Ppp2r2a, Ebf2 | B6 × FVB | 18 | |
| F | 2 | 109,951,892 | 110,501,333 | 4.0×10-7 | Ccdc34, Bbox1, Fibin, Slc5a12 | ||
| 7 | 85,652,862 | 85,927,942 | 2.6×10-7 | Ntrk3, Gm9885, Mrps11 | B6 × A | Ath326 | |
| 18 | 58,022,123 | 58,291,010 | 2.5×10-7 | Slc12a2, Fbn2 | |||
| 19 | 42,044,028 | 42,195,636 | 3.0×10-7 | Mms19, Ubtd1, Ankrd2, Hoga1, Mom4, Pi4k2a | B6 × FVB | 18 |
Discussion
In this study we completed a strain survey for atherosclerotic lesions in inbred mice that were on an atherogenic diet in animal facilities with different health statuses. We showed that (1) difference in health status alters the atherosclerosis susceptibility of different inbred strains, with some strains more susceptible in the low-status room and others more susceptible in the high-status room; and (2) susceptibility differs between males and females, with males more susceptible than females in some strains and females more susceptible in others.
Because the duration of the atherogenic diet was different between the low-status and high-status rooms, we are not able to draw any conclusions from the quantitative data, but as many of the groups showed a clear presence or absence of lesions, we are able to examine the differences between those groups (e.g., when a strain shows lesions at 8 weeks of diet in the low-status room and has no lesions after 17 weeks on the atherogenic diet in the high-status room). The presence of lesions in either males or females of 14 inbred strains in the low-status room but not in the high-status room demonstrates that atherosclerosis susceptibility is higher in the room with a lower health status even though mice were on the atherogenic diet for a shorter time span. We did see some strains (e.g., MOLF and BALB) that developed lesions in the high-status room but not in the low-status room. However, this might be an effect of the duration of the diet.
We also observed differences between males and females within the low-status and high-status rooms. In the low-status room, we saw lesions in C57L, DBA/1, and DBA/2 females but not in males. In the high-status room, we saw lesions in SM, BTBR, SEA, C57BR, SWR, and DBA/1 females but not in males. We made opposite observations for other strains for which only the males were susceptible; this difference depended on the room. SWR mice are interesting because only females developed lesions in the high-status room, while only males developed lesions in the low-status room. It is often reported that in mice, females are more susceptible to atherosclerosis than males, and that in humans, males are more susceptible. However, our data show that susceptibility depends on the individual inbred strains and their health status. Furthermore, as we did not find any correlation between HDL cholesterol levels and lesions, HDL cholesterol levels are not a major determining factor for the development of atherosclerosis in our inbred strains.
The data that we collected are suitable for genome-wide association mapping, a method similar to human genome-wide association. Performing the analysis for males and females, and for the low-status and high-status rooms separately, we conclude that mapping of loci was affected by the health status of the room. While we were able to map loci for males and females in the low-status room, we did not obtain any results from the high-status room. One way to explain this observation for genes that determine atherosclerosis susceptibility is that in the low-status room, the number of genes is small, but relative effects are large, while in the high-status room, the number of genes is larger, but relative effects are smaller. Our analysis would not have enough power to detect these small effect genes. If this explanation is true, then it would follow that atherosclerosis susceptibility in the different rooms is determined (in part) by different pathways.
The loci that were mapped in the low-status room differed between males and females, suggesting different pathways. Five of the 7 loci are within the confidence intervals of previously mapped QTL for atherosclerosis, while the loci on Chr 2 in females and Chr 6 in males were regions that have never been mapped in any previous studies. When comparing our new QTL with QTL from previously published studies, it would be interesting to consider the health status of each study, whether overlapping QTL were found under similar microbiological conditions, and whether differences among studies can be explained by differences in strains or health status or both. Unfortunately, these data are not available.
The variation in atherosclerosis susceptibility within the same strain and the differences in mapping of loci by genome-wide association analysis between the rooms can be attributed only to the difference in microbiological environment. Although Pneumocystis carinii, Pasturella pneumotropica, and Helicobacter spp. are the only differences that we detected, our microbiological screens are by no means exhaustive, and we cannot rule out the presence of other bacteria or viruses in one room and not the other (it is even possible that the high-status room contains a microorganism that is absent in the low-status room). Multiple investigations have shown that infectious agents affect cellular and molecular changes that could contribute to atherogenesis 15. The development of atherosclerosis is influenced by either (1) infection of the vascular cells or (2) the effects of cytokines or acute phase proteins produced due to infection at non-vascular sites 11. This causes an activation of the innate immune response and adds to the chronic inflammation that is already present in the atherosclerotic plaque. Helicobacter pylori is a pathogen that has been found in the human atherosclerotic plaque, but studies have reported both positive and negative roles in cardiovascular disease development in mice. In a study by Chen et al, it was shown that Helicobacter pylori infection enhances atherosclerosis in C57BL/6 mice on a high-cholesterol diet9. However, another in vivo study performed in C57BL/6 and LDLR-deficient mice demonstrated that Helicobacter pylori infection does not contribute to the development of atherosclerotic lesion formation 10. Overall, previous data provide conflicting results on the role of Helicobacter pylori in atherogenesis. Although the previous data from various studies are conflicting, the influence of Helicobacter spp. on the pathogenesis of atherosclerotic plaque formation in different inbred strains of mice in our study is certainly a possibility. Although less commonly considered, it is also possible that the immune response caused by these bacteria exert a protective effect in some strains but not others, thus contributing to the differences seen within the strains between the different rooms.
In conclusion, we surveyed atherosclerotic lesions in inbred strains of mice in 2 animal rooms with different health statuses. The intent of this large study was to investigate development of atherosclerotic lesions in different genetic backgrounds under different microbiological conditions and to subsequently identify loci that regulate predisposition to atherosclerosis. We observed that a small percentage of inbred strains developed lesions and that susceptibility was dependent on sex and the health status of the room in which the mice were kept. Genome-wide association mapping using the data from our strain surveys demonstrate that mapping results are not only dependent on sex, with different loci mapped in males and females, but also on health status of the animal room. Therefore, health status is an important factor in genetic studies for atherosclerosis and should be a consideration in the experimental design.
Supplementary Material
Table 4.
Summary of previously found QTL for atherosclerosis susceptibility.
| Chr | Peak (Mb) | LOD | 95% CI | Induct. | Sex | Cross | High allele | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 105 | 5.0 | 75-135 | Apoe-/- | M+F | (B6. Apoe-/-×129. Apoe-/-)F2 | 129 | 16 |
| 163 | 6.8 | 151-173 | Apoe-/- | M+F | (B6. Apoe-/-×129. Apoe-/-)F2 | 129 | 16 | |
| 163 | 161-164 | ath diet | F | B×H and C×B RI | B6 | 17 | ||
| 100 | ath diet | F | (B6×A)F2 | B6 | unpub | |||
| 180 | (2.3) | 160-194 | Apoe-/- | M+F | (B6. Apoe-/-×FVB, Apoe-/-)F2 | ? | 18 | |
| 2 | 149 | 2.8 | 131-162 | Ldlr-/- | M+F | (PERA×B6. Ldlr-/-)×B6 Ldlr-/- | B6 | 19 |
| 3 | 148 | 5.1 | Ldlr-/- | M | (B6. Ldlr-/-×FVB. Ldlr-/-)F2 | B6 | 20 | |
| 4 | 53 | (2.6) | 10-106 | Apoe-/- | F | (B6. Apoe-/-×C3H. Apoe-/-)F2 | B6 | 21 |
| 60 | 3.6 | 45-105 | ath diet | F | (SM×NZB)F2 | SM | 22 | |
| 76 | 4.6 | Apoe-/- | F | (A×B6. Apoe-/-)F2 | A | unpub | ||
| 130 | 6.2 | 83-134 | Ldlr-/- | F | (MOLF× B6. Ldlr-/-)× B6. Ldlr-/- | MOLF | 23 | |
| 6 | 134 | 6.7 | 93-145 | HF diet | M+F | (CAST×B6)F2 | B6 | 24 |
| 134 | 6.7 | 121-145 | Ldlf-/- | M+F | (MOLF× B6. Ldlr-/-)× B6. Ldlr-/- | B6 | 23 | |
| 7 | 57 | 3.7 | HF diet | F | (B6×D2)F2 | D2 | 25 | |
| 88 | ath diet | (A×B)F2 and B×A RI | B6 | 26 | ||||
| 8 | 22 | 2.8 | ath diet | F | (B6×A)F2 | A | unpub | |
| 61 | 3.4 | HF diet | F | (B6×D2)F2 | D2 | 25 | ||
| 60 | Apoe-/- | F | (A×B6. Apoe-/-)F2 | A | unpub | |||
| 9 | 25 | 4.1 | ath diet | F | (B6×A)F2 | B6 | unpub | |
| 42 | Apoe-/- | F | (A×B6. Apoe-/-)F2 | A | unpub | |||
| 44 | 5.0 | 34-70 | Apoe-/- | F | (B6. Apoe-/-×C3H. Apoe-/-)F2 | B6 | 21 | |
| 47 | ath diet | F | SWR×(SWR×SJL) | SJL | unpub | |||
| 61 | 6.9 | 47-71 | Apoe-/- | M+F | (B6. Apoe-/-×129. Apoe-/-)F2 | B6 | 16 | |
| 10 | 21 | 13.1 | Ldlr-/- | M+F | (B6. Ldlr-/-×FVB. Ldlr-/-)F2 | FVB | 20 | |
| 12 | 2.8 | Apoe-/- | F | (A×B6. Apoe-/-)F2 | A | unpub | ||
| 25 | 5.1 | 0-50 | Apoe-/- | M | (B6. Apoe-/-×FVB, Apoe-/-)F2 | FVB | 18 | |
| 20 | 3.9 | ath diet | F | (B6×A)F2 | A | unpub | ||
| 82 | 4.5 | HF diet | F | (B6×D2)F2 | D2 | 25 | ||
| 68 | 6.6 | 60-72 | ath diet | F | (B6×129S1)F2 | 129 | 27 | |
| 108 | ath diet | F | (B6×129S1)F2 | ? | 27 | |||
| 11 | 120 | ath diet | F | (B6×129S1)F2 | ? | 27 | ||
| 12 | 17 | 2.5 | 11-20 | ath diet | F | (B6.db/db × BKS)F2 | BKS | 28 |
| 25 | 6.0 | Ldlr-/- | M+F | (B6. Ldlr-/-×FVB. Ldlr-/-)F2 | B6 | 20 | ||
| 36 | 3.7 | ath diet | F | (B6×129S1)F2 | ? | 27 | ||
| 84 | ath diet | F | SWR×(SWR×SJL) | SWR | unpub | |||
| 100 | ath diet | F | (B6×129S1)F2 | ? | 27 | |||
| 14 | 48 | (2.5) | Apoe-/- | M | (B6. Apoe-/-×FVB, Apoe-/-)F2 | B6 | 18 | |
| 16 | 32 | (2.5) | Apoe-/- | M+F | (B6. Apoe-/-×FVB, Apoe-/-)F2 | FVB | 18 | |
| 17 | 36 | 4.3 | 33-53 | Apoe-/- | M | (D2. Apoe-/-×AKR. Apoe-/-) | AKR | 29 |
| 18 | 22 | 2.5 | Apoe-/- | F | (A×B6. Apoe-/-)F2 | B6 | unpub | |
| 19 | 41 | (3.8) | 35-47 | Apoe-/- | M | (B6. Apoe-/-×FVB, Apoe-/-)F2 | FVB | 18 |
Acknowledgments
We thank Cynthia McFarland for expert technical assistance.
Sources of Funding
This study was funded by U.S. National Institutes of Health grants HL077796, HL081162, and HL095668 and the National Cancer Institute Core grant CA034196.
Footnotes
Disclosures
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Agah R, Topol EJ. Genetic testing for coronary heart disease: the approaching frontier. Expert Rev. Mol. Diagn. 2002;2(5):448–460. doi: 10.1586/14737159.2.5.448. [DOI] [PubMed] [Google Scholar]
- 2.Humphries SE, Morgan L. Genetic risk factors for stroke and carotid atherosclerosis: insights into pathophysiology from candidate gene approaches. Lancet Neurol. 2004;3(4):227–235. doi: 10.1016/S1474-4422(04)00708-2. [DOI] [PubMed] [Google Scholar]
- 3.Lusis AJ, Mar R, Pajukanta P. Genetics of atherosclerosis. Annu Rev Genomics Hum Genet. 2004;5:189–218. doi: 10.1146/annurev.genom.5.061903.175930. [DOI] [PubMed] [Google Scholar]
- 4.Allayee H, Ghazalpour A, Lusis AJ. Using mice to dissect genetic factors in atherosclerosis. Arterioscler Thromb Vasc Biol. 2003;23(9):1501–1509. doi: 10.1161/01.ATV.0000090886.40027.DC. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Ishimori N, Korstanje R, Rollins J, Paigen B. Identifying novel genes for atherosclerosis through mouse-human comparative genetics. Am J Hum Genet. 2005;77(1):1–15. doi: 10.1086/431656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meir KS, Leitersdorf E. Atherosclerosis in the apolipoprotein-E-deficient mouse: a decade of progress. Arterioscler Thromb Vasc Biol. 2004;24(6):1006–1014. doi: 10.1161/01.ATV.0000128849.12617.f4. [DOI] [PubMed] [Google Scholar]
- 7.Jawie J, Nastałek P, Korbut R. Mouse models of experimental atherosclerosis. J. Physiol. Pharmacol. 2004;55(3):503–517. [PubMed] [Google Scholar]
- 8.Stylianou IM, Bauer RC, Reilly MP, Rader DJ. Genetic basis of atherosclerosis: insights from mice and humans. Circ Res. 2012;110(2):337–355. doi: 10.1161/CIRCRESAHA.110.230854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X-H, Wang J-B, Wang Y-S, Liu Z-M, Li Y. [Helicobacter pylori infection enhances atherosclerosis in high-cholesterol diet fed C57BL/6 mice]. Zhonghua Xin Xue Guan Bing Za Zhi. 2010;38(3):259–263. [PubMed] [Google Scholar]
- 10.Mach F, Sukhova GK, Michetti M, Libby P, Michetti P. Influence of Helicobacter pylori infection during atherogenesis in vivo in mice. Circ Res. 2002;90(1):E1–4. doi: 10.1161/hh0102.102270. [DOI] [PubMed] [Google Scholar]
- 11.Rosenfeld ME, Campbell LA. Pathogens and atherosclerosis: Update on the potential contribution of multiple infectious organisms to the pathogenesis of atherosclerosis. Thromb. Haemost. 2011;106(5):858–867. doi: 10.1160/TH11-06-0392. [DOI] [PubMed] [Google Scholar]
- 12.Paigen B, Morrow A, Holmes PA, Mitchell D, Williams RA. Quantitative assessment of atherosclerotic lesions in mice. Atherosclerosis. 1987;68(3):231–240. doi: 10.1016/0021-9150(87)90202-4. [DOI] [PubMed] [Google Scholar]
- 13.Szatkiewicz JP, Beane GL, Ding Y, Hutchins L, Pardo-Manuel de Villena F, Churchill GA. An imputed genotype resource for the laboratory mouse. Mamm Genome. 2008;19(3):199–208. doi: 10.1007/s00335-008-9098-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westfall PH, Young SS. Resampling-Based Multiple Testing: Examples and Methods for p-Value Adjustment (Wiley Series in Probability and Statistics) 1st ed. Wiley-Interscience; 1993. p. 360. [Google Scholar]
- 15.Liuba P, Pesonen E. Infection and early atherosclerosis: does the evidence support causation? Acta Paediatr. 2005;94(6):643–651. doi: 10.1111/j.1651-2227.2005.tb01958.x. [DOI] [PubMed] [Google Scholar]
- 16.Tomita H, Zhilicheva S, Kim S, Maeda N. Aortic arch curvature and atherosclerosis have overlapping quantitative trait loci in a cross between 129S6/SvEvTac and C57BL/6J apolipoprotein E-null mice. Circ Res. 2010;106(6):1052–1060. doi: 10.1161/CIRCRESAHA.109.207175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phelan SA, Beier DR, Higgins DC, Paigen B. Confirmation and high resolution mapping of an atherosclerosis susceptibility gene in mice on Chromosome 1. Mamm Genome. 2002;13(10):548–553. doi: 10.1007/s00335-002-2196-1. [DOI] [PubMed] [Google Scholar]
- 18.Dansky HM, Shu P, Donavan M, Montagno J, Nagle DL, Smutko JS, Roy N, Whiteing S, Barrios J, McBride TJ, Smith JD, Duyk G, Breslow JL, Moore KJ. A phenotype-sensitizing Apoe-deficient genetic background reveals novel atherosclerosis predisposition loci in the mouse. Genetics. 2002;160(4):1599–1608. doi: 10.1093/genetics/160.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seidelmann SB, De Luca C, Leibel RL, Breslow JL, Tall AR, Welch CL. Quantitative trait locus mapping of genetic modifiers of metabolic syndrome and atherosclerosis in low-density lipoprotein receptor-deficient mice: identification of a locus for metabolic syndrome and increased atherosclerosis on chromosome 4. Arterioscler Thromb Vasc Biol. 2005;25(1):204–210. doi: 10.1161/01.ATV.0000149146.32385.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teupser D, Tan M, Persky AD, Breslow JL. Atherosclerosis quantitative trait loci are sex and lineage-dependent in an intercross of C57BL/6 and FVB/N low-density lipoprotein receptor-/- mice. Proc Natl Acad Sci USA. 2006;103(1):123–128. doi: 10.1073/pnas.0509570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SS, Shi W, Wang X, Velky L, Greenlee S, Wang MT, Drake TA, Lusis AJ. Mapping, genetic isolation, and characterization of genetic loci that determine resistance to atherosclerosis in C3H mice. Arterioscler Thromb Vasc Biol. 2007;27(12):2671–2676. doi: 10.1161/ATVBAHA.107.148106. [DOI] [PubMed] [Google Scholar]
- 22.Korstanje R, Eriksson P, Samnegård A, Olsson PG, Forsman-Semb K, Sen S, Churchill GA, Rollins J, Harris S, Hamsten A, Paigen B. Locating Ath8, a locus for murine atherosclerosis susceptibility and testing several of its candidate genes in mice and humans. Atherosclerosis. 2004;177(2):443–450. doi: 10.1016/j.atherosclerosis.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Welch CL, Bretschger S, Latib N, Bezouevski M, Guo Y, Pleskac N, Liang CP, Barlow C, Dansky H, Breslow JL, Tall AR. Localization of atherosclerosis susceptibility loci to chromosomes 4 and 6 using the Ldlr knockout mouse model. Proc Natl Acad Sci USA. 2001;98(14):7946–7951. doi: 10.1073/pnas.141239098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehrabian M, Wong J, Wang X, Jiang Z, Shi W, Fogelman AM, Lusis AJ. Genetic locus in mice that blocks development of atherosclerosis despite extreme hyperlipidemia. Circ Res. 2001;89(2):125–130. doi: 10.1161/hh1401.093458. [DOI] [PubMed] [Google Scholar]
- 25.Colinayo VV, Qiao J-H, Wang X, Krass KL, Schadt E, Lusis AJ, Drake TA. Genetic loci for diet-induced atherosclerotic lesions and plasma lipids in mice. Mamm Genome. 2003;14(7):464–471. doi: 10.1007/s00335-002-2187-2. [DOI] [PubMed] [Google Scholar]
- 26.Stewart-Phillips JL, Lough J, Skamene E. ATH-3, a new gene for atherosclerosis in the mouse. Clin Invest Med. 1989;12(2):121–126. [PubMed] [Google Scholar]
- 27.Ishimori N, Li R, Kelmenson PM, Korstanje R, Walsh KA, Churchill GA, Forsman-Semb K, Paigen B. Quantitative trait loci analysis for plasma HDL-cholesterol concentrations and atherosclerosis susceptibility between inbred mouse strains C57BL/6J and 129S1/SvImJ. Arterioscler Thromb Vasc Biol. 2004;24(1):161–166. doi: 10.1161/01.ATV.0000104027.52895.D7. [DOI] [PubMed] [Google Scholar]
- 28.Mu JL, Naggert JK, Svenson KL, Collin GB, Kim JH, McFarland C, Nishina PM, Levine DM, Williams KJ, Paigen B. Quantitative trait loci analysis for the differences in susceptibility to atherosclerosis and diabetes between inbred mouse strains C57BL/6J and C57BLKS/J. J Lipid Res. 1999;40(7):1328–1335. [PubMed] [Google Scholar]
- 29.Smith JD, Bhasin JM, Baglione J, Settle M, Xu Y, Barnard J. Atherosclerosis susceptibility loci identified from a strain intercross of apolipoprotein E-deficient mice via a high-density genome scan. Arterioscler Thromb Vasc Biol. 2006;26(3):597–603. doi: 10.1161/01.ATV.0000201044.33220.5c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.