Abstract
We previously showed that reduced infectivity of HIV with incompletely processed capsid-spacer protein 1 (CA-SP1) is rescued by cellular activation or increased expression of HSP90AB1, a member of the cytosolic heat shock protein 90 family. Here we show that HSP90AB1 is present in HIV virions and that HSP90AB1, but not nonfunctional mutated HSP90AB1E42A+D88A, restores infectivity to HIV with mutations in CA that alter core stability. Further, the CA mutants were hypersensitive to pharmacological inhibition of HSP90AB1. In agreement with Roesch, et al. (PLoS Pathog, e1002792, 2012), we found that culturing HIV at 39.5 °C enhanced viral infectivity up to 30-fold in human peripheral blood mononuclear cells (p = 0.002) and rescued CA-mutant infectivity in nonactivated cells, concurrent with elevated expression of HSP90AB1 during hyperthermia. In sum, the transdominant effect of HSP90AB1 on CA-mutant HIV infectivity suggests a potential role for this class of cellular chaperones in HIV core stability and uncoating.
Keywords: HSP90AB1, Hyperthermia, HIV, capsid, postentry
Introduction
The highly ordered proteolysis of HIV Gag is required for condensation of capsid (CA) monomers into an intact and stable core lattice (Wright et al., 2007). This sequential maturation process prepares the HIV core for uncoating in the infected cell (Vogt, 1996). Although the precise mechanism and location of HIV uncoating are not currently known, the early postentry steps in HIV infection are presumed to be regulated by host proteins, including cyclophilin A and TRIM5α, in the infected cell (Goff, 2007; Luban et al., 1993; Stremlau et al., 2006).
Infectious HIV virions regularly display multiple and amorphously shaped cores (Carlson et al., 2008), and deviations from optimal core stability often result in nonproductive infection (von Schwedler et al., 2003). Through extensive mutational analysis of the HIV CA, Forshey and colleagues described several key residues that are crucial for intrinsic core stability and optimal virus infectivity (Forshey et al., 2002). Recently, the same group identified additional second-site suppressor mutations within HIV CA that restore infectivity (Yang et al., 2012) but that, surprisingly, do not alleviate the changes in core stability caused by the original mutations. In an in vitro uncoating assay, the P38A mutation rendered the HIV CA core unstable, but the E45A mutation resulted in a hyperstable core. Yang et al. found that introducing the T216I and R132T suppressor mutations in HIV CA restored infectivity to both the P38A and the E45A mutant viruses. Unexpectedly, the compensatory suppressor mutations did not rescue the altered stability of the CA-mutant cores, leading the authors to propose that while a stable core is a prerequisite for successful uncoating, alternate mutations can arise to preserve critical CA-host factor interactions.
In a previous report (Joshi and Stoddart, 2011), we observed that drug-resistance mutations in HIV protease (PR) resulted in virions harboring incompletely processed CA molecules with uncleaved CA-spacer peptide 1 (CA-SP1). This apparent defect greatly reduced PR-mutant HIV infectivity in nonactivated cells, and infectivity of the virus containing uncleaved CA-SP1 was arrested before proviral DNA synthesis. The PR-mutant virus displayed robust replication in activated cells, which suggested that the cellular status of the target cell dictated the infectivity of PR-mutant HIV. Through a functional genetic screen, we identified heat shock protein 90 kDa alpha (cytosolic) class B member 1 (HSP90AB1) as a host factor that alleviated this block to infectivity in activated cells. The influence of HSP90AB1 was not restricted to the defective CA-SP1 phenotype, as pharmacologic inhibition (IC50 = 0.17 ± 0.03 μM) of this cellular chaperone decreased wild-type (WT) HIV infectivity.
In this report, we show that HSP90AB1 is incorporated into virions but outside of the HIV core. The relevance of HSP90AB1 incorporation is currently under investigation, but here we extend our previous observations to demonstrate that expression of HSP90AB1 can rescue the impairment imposed by mutations in HIV CA. For this study, we selected mutations in CA that recapitulate the CA-SP1 PR-mutant phenotype in that the CA-mutant viruses are arrested after entry and before proviral DNA integration. Consistent with rescue of the CA-SP1 PR-mutant phenotype, the CA-mutants infected phytohemagglutinin (PHA)-activated human peripheral blood mononuclear cells (PBMC) and unactivated Jurkat T cells expressing native HSP90AB1. Further, the CA mutants were hypersensitive to pharmacological inhibition of HSP90AB1 compared to WT HIV. The expression of HSP90AB1 responds to changes in ambient temperature and consistent with a recent report by Roesch et al. (Roesch et al., 2012), we found that raising the incubation temperature to 39.5 °C enhanced HIV infectivity up to 30-fold, concurrent with an increase in HSP90AB1 expression. More importantly, CA-SP1 HIV and the panel of CA mutants were infectious in target cells incubated at 39.5 °C.
Results
Cellular HSP90AB1 is associated with HIV virions
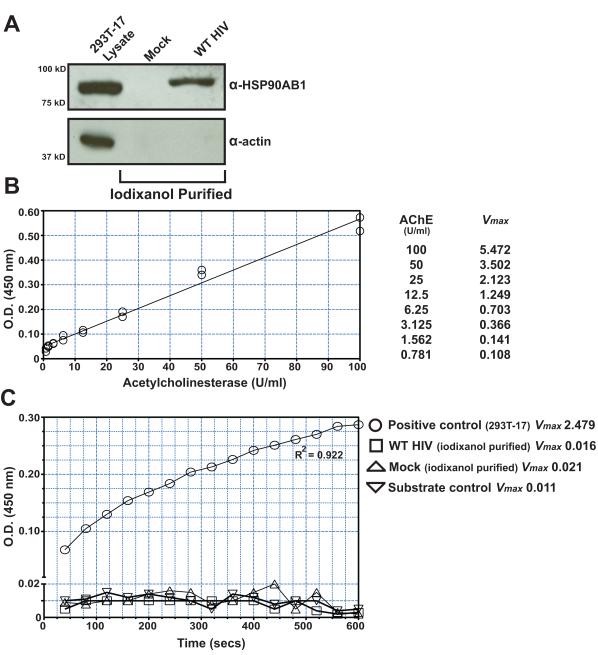
Mature HIV virions are known to incorporate cellular proteins, and recent studies have identified a vast array of virion-associated host proteins (Ott, 2008), so we therefore investigated whether HSP90AB1 was also associated with HIV virions. In general, cytoplasmic organelles, vesicles, and physiological nanoparticles are similar to enveloped retroviruses in terms of size and buoyant density (Cantin et al., 2008). Due to these similarities, even carefully purified retrovirus preparations are often contaminated with these nonviral cytoplasmic organelles and vesicles. To ensure that HSP90AB1 is a bona fide cellular protein associated with HIV virions, we tested highly purified virus preparations for the presence of this abundant cellular protein. Previous studies have successfully described the use of iodixanol velocity gradients to fractionate retrovirus preparations and have shown that in vitro-generated HIV can be exclusively concentrated from culture supernatant when overlaid on 8.4% iodixanol (Cantin et al., 2008; Dettenhofer and Yu, 1999). We first purified virions from the culture supernatant over a conventional 20% sucrose cushion. The virus pellet was resuspended in saline and subjected to an additional purification on 8.4% iodixanol, and the contents of the virus preparation were assayed by western blot (Fig. 1A). The purified virus preparation was also probed for alpha smooth muscle actin to rule out any trace contaminating cellular debris. We detected the presence of HSP90AB1 in this highly purified HIV preparation. To confirm the absence of contaminating cytoplasmic organelles and vesicles, we tested the virus preparation for acetylcholinesterase (AChE) activity in an enzyme assay (Ellman et al., 1961) with sensitivity extending below 1 U/ml, as determined by using serial dilutions of Electrophorus electricus AchE (Fig. 1B). The level of AChE activity in the iodixanol-purified pellets from WT HIV and mock-transfected culture supernatants was comparable to the substrate control (Fig. 1C). A previous study by Gurer et al. also investigated the presence of different heat shock proteins in virions and did not detect heat shock protein 90 (Gurer et al., 2002), but more recent studies have detected HSP90AB1 in purified virions using mass spectrometry (Chertova et al., 2006; Santos et al., 2012). Our results demonstrate that HSP90AB1 is indeed associated with HIV virions.
FIGURE 1.
HSP90AB1 is associated with HIV virions. (A) Western blot analysis of WT HIV virions purified from transfected 293T-17 cell culture supernatant on 8.4% iodixanol. The purified pellet from mock- and WT HIV-transfected samples were probed for HSP90AB1 and alpha smooth muscle actin, and the results are representative of three independently purified cell culture supernatant preparations. (B) The sensitivity of the AChE assay was determined by serial dilution of Electrophorus electricus AChE, and detection of 0.78 U/ml of AChE was reliably achieved with these assay conditions. (C) Pelleted virus and mock-transfected samples were assayed for contaminating cytoplasmic organelles and vesicles by assessing AChE activity. The positive control represents AChE activity in cytoplasmic organelles and vesicles sedimented from a comparable volume of 293T-17 culture supernatant. The mean AChE Vmax ± SEM for the positive control (293T-17 supernatant pellet) was 2.71 ± 0.41, and the graph represents one of three independent assays.
HSP90AB1 is incorporated into HIV virions but is not detected in the core
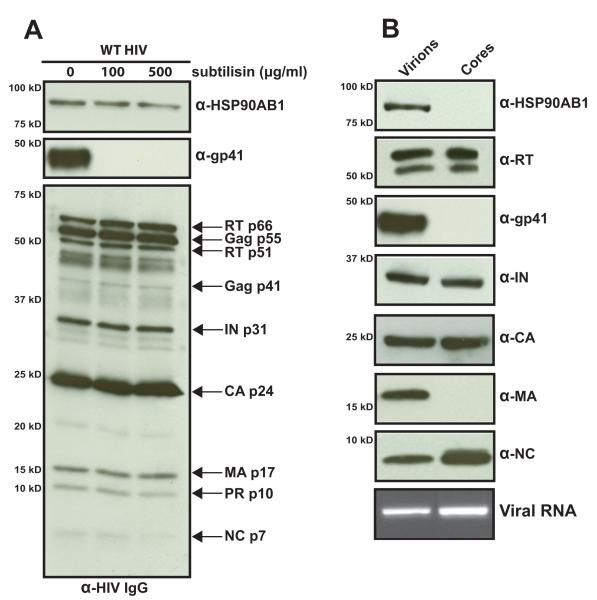
Due to the nature of retrovirus assembly and release from the plasma membrane, cellular proteins are associated with the surface or within different compartments of the virus. To determine the location of HSP90AB1 with respect to HIV virions, we exploited the nonspecific activity of the bacterial subtilisin protease (Fig. 2A). Subtilisin treatment strips the virions of surface proteins, including the extracellular domain of the gp41 transmembrane protein (Devroe et al., 2005; Ott, 2009). We incubated purified HIV with increasing subtilisin concentrations overnight, and virions were recovered by iodixanol purification. The absence of surface gp41 indicates the effective protease activity of subtilisin and the detection of HSP90AB1 in these virus preparations indicates that this cellular protein is found within virions.
FIGURE 2.
HSP90AB1 is incorporated into HIV virions. (A) Subtilisin digestion of purified HIV virions. Iodixanol-purified virions were incubated with the indicated concentration of subtilisin for 18 h at 37 °C. Virions were recovered by centrifugation, lysed in SDS-PAGE loading buffer, and individual virus proteins were detected with a human anti-HIV IgG antibody cocktail. An equal volume of subtilisin-treated virus lysate was probed independently for HSP90AB1 and gp41. (B) HSP90AB1 is incorporated within HIV virions but is not present in the core. Western blot analysis of purified intact virions and HIV cores recovered from Triton X-100-treated intact virus probed individually for HSP90AB1 and the indicated HIV proteins. HIV genomic RNA was detected by RT-PCR. The results are representative of three independent iodixanol-purified virus and core preparations.
In an attempt to further determine the location of HSP90AB1 in HIV virions, we subjected intact virus to brief detergent lysis (Welker et al., 2000). Purified HIV was treated with Triton X-100 to strip off the virus envelope, exposing the core. The extracted cores were extensively washed, and their protein composition was compared to that of intact virions (Fig. 2B). Characteristic of detergent-lysed virus (Welker et al., 2000), the extracted cores lacked the presence of transmembrane gp41 and matrix (MA) protein, and the integrity of the extracted cores was confirmed by the presence of their expected contents (i.e., reverse transcriptase (RT), integrase (IN), CA p24, nucleocapsid (NC), and viral genomic RNA). HSP90AB1 was not detected in the extracted core preparations, indicating that this cellular protein is packaged within the lumen of HIV virions between the virus membrane and the core. Due to the nature of this assay, it is not possible to determine whether HSP90AB1 is physically associated with the virus inner membrane, with MA, or with the HIV core. Affinity-binding studies between HSP90AB1 and HIV structural proteins are underway to determine if HSP90AB1 is actively recruited into assembling virions.
HSP90AB1 is present in virions produced from an HIV-inoculated T-cell line and overexpression of HSP90AB1 does not influence its incorporation
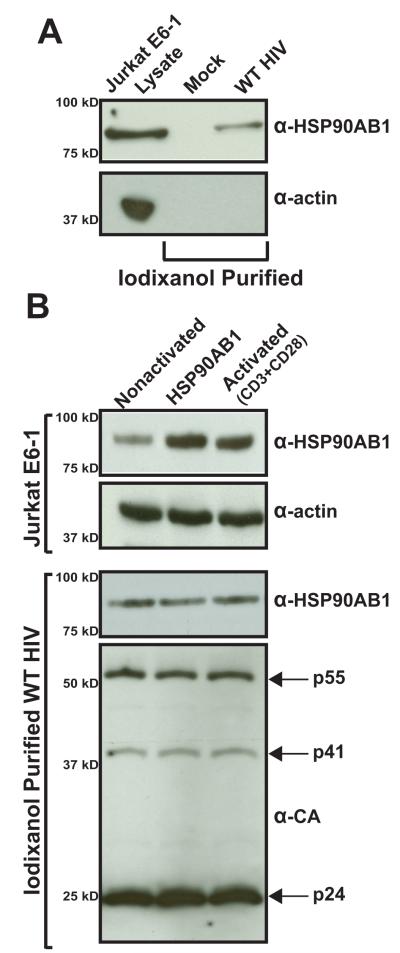
Host cellular proteins incorporated into the HIV virion vary between different virus-producing cell types and different procedures to generate infectious virus in the lab (Cantin et al., 1996; Giroud et al., 2011; Tremblay et al., 1998). The presence of HSP90AB1 was detected in virions produced from Jurkat E6-1 cells inoculated with WT HIV (Fig. 3A). The bona fide presence of HSP90AB1 in virions was confirmed by the absence of AChE activity in the sample. Also, the original association between HIV infection and HSP90AB1 was identified in activated cells and subsequently confirmed in cells overexpressing this cellular protein. To determine if overproduction of HSP90AB1 influences the extent of its incorporation, we tested virions purified from HIV-infected Jurkat E6-1 cells that were either nonactivated, transduced with a retroviral vector intended to overexpress exogenous native HSP90AB1, or activated with anti-CD3 and -CD28 antibodies (Fig. 3B). In comparison to normal Jurkat E6-1 cell lysate, we observed an increase in HSP90AB1 protein in Jurkat E6-1 cells expressing exogenous recombinant HSP90AB1 and in Jurkat E6-1 activated with anti-CD3 and -CD28 antibodies (top panel), but these elevated protein levels did not alter the amount of HSP90AB1 incorporated into virions (lower panel). In addition, virus infectivity was tested in the TZM-bl reporter cell line, and we did not observe any significant change in virus infectivity produced from activated cells or HSP90AB1-overexpressing Jurkat E6-1 cells (data not shown). We tried to knock down HSP90AB1 by siRNA to determine if lowering the host factor expression level would affect its incorporation or alter HIV infectivity. Given that the cytosolic HSP90 isoforms are constitutively expressed and account for 1-2% of all cellular proteins (Whitesell and Lindquist, 2005), we were unable to effectively reduce HSP90AB1 expression in both, 293T-17 and Jurkat E6-1 cell lines without causing permanent cell damage. Taken together, these results demonstrate that HSP90AB1 is detected in multiple HIV preparations and, under the present assay conditions its incorporation is not influenced by increased HSP90AB1 protein level in the cell.
FIGURE 3.
HSP90AB1 is incorporated into HIV virions collected from infected Jurkat E6-1 cells and is not altered by overexpression of recombinant HSP90AB1 or cellular activation. (A) Western blot analysis of virions purified from WT HIV-inoculated Jurkat E6-1 culture supernatant. The results are representative of three independently purified virus preparations. (B) Jurkat E6-1 cells were either transduced by a retroviral vector expressing recombinant HSP90AB1 or were CD3- and CD28-activated for 24 h, subsequently infected with WT HIV, and cultured for 4 days. The treated Jurkat E6-1 cells were lysed, and 1 μg of protein was probed for HSP90AB1. Probing for alpha smooth muscle actin served as a loading control. HIV virions were iodixanol-purified from the culture supernatant and probed for HSP90AB1 and CA. The results are representative of three independent assays.
CA-mutant HIV can infect activated human primary cells
Our initial study focused on mutations in HIV protease that prevent the enzyme from completely processing the CA precursor, resulting in virions with a substantial proportion of uncleaved CA-SP1 molecules. This defect alone greatly reduced PR-mutant infectivity; nevertheless, the PR-mutant virus was infectious in PHA-activated PBMC. In this study, we sought to determine if cellular activation could also rescue HIV with CA mutations that reduce virus infectivity. HIV CA has been subjected to extensive mutational analysis with the intent to determine structurally and functionally important residues and motifs responsible for optimal core formation (Dismuke and Aiken, 2006; Forshey et al., 2002; Ganser-Pornillos et al., 2004; Yamashita et al., 2007). CA mutations that resulted in either unstable or hyperstable HIV cores were selected and similar to the CA-SP1 phenotype, infectivity of the CA-mutant viruses was arrested after entry but before complete proviral DNA synthesis. The selected mutations did not affect virion formation, endogenous RT activity, genomic RNA packaging, or CA p24 production (Forshey et al., 2002).
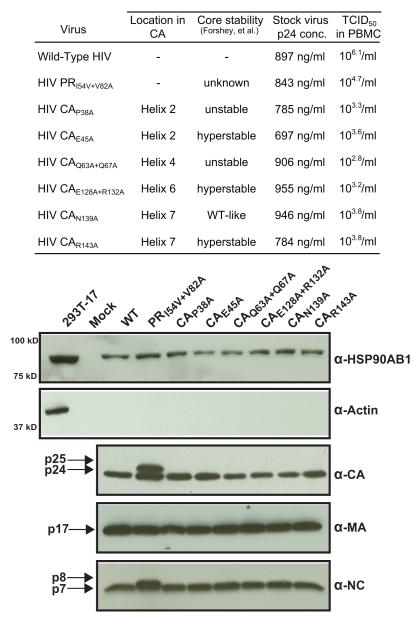
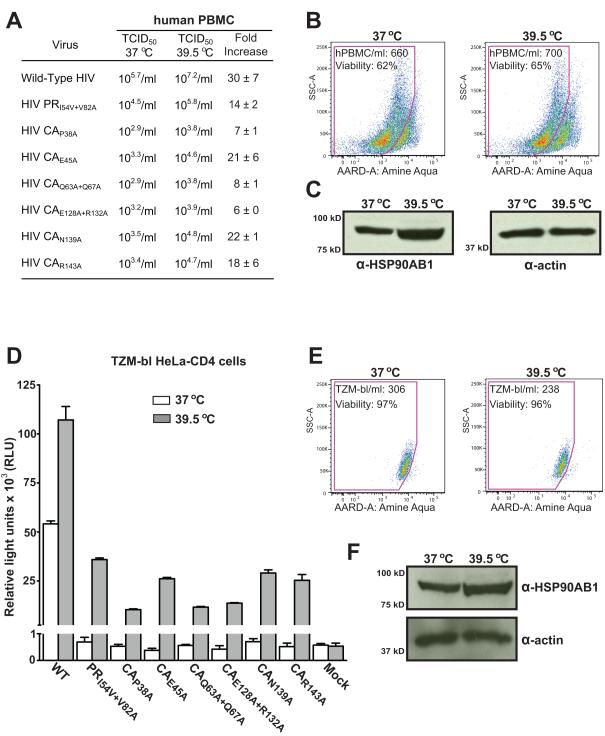
The selected CA mutants were previously shown to reduce HIV infectivity from 1.2- to 34-fold in a single-cycle infection assay (von Schwedler et al., 2003), although these authors observed variation in infectivity when the CA mutants were tested in a transformed T-cell line continuous culture assay. We first tested the ability of these CA mutants to infect PHA-activated PBMC and determined that the CA p24 concentrations of WT, PR-mutant, and CA-mutant virus stocks were comparable, confirming that these mutations did not affect virus yield (Fig. 4). To quantify the infectivity of CA mutants in PHA-activated PBMC, we opted for a highly sensitive 50% endpoint dilution assay in which the infectious titer is determined from serial half-log dilutions of the stock virus. The selected CA mutants infected activated primary human cells to the extent that an infectious dose could be accurately calculated for each virus stock (Fig. 4). As observed previously (von Schwedler et al., 2003), the variation in CA-mutant infectivity between a single-cycle infection assay and a spreading virus infection assay can be attributed to delayed replication kinetics of the mutant viruses. Nonetheless, these results suggest that CA-mutant HIV can establish productive infection in activated cells and is reminiscent of the significantly increased infectivity of CA-SP1 PR-mutant HIV in activated PBMC (Joshi and Stoddart, 2011).
FIGURE 4.
CA-mutant HIV can infect PHA-activated PBMC. (Table) The yield of WT and mutant virus was determined by p24 concentration for each stock virus. Infectivity of the CA mutants was determined by TCID50 values derived from serial half-log dilutions of each virus stock in quadruplicate wells of PHA-activated PBMC (1 × 105 cells/well) and calculated as a reciprocal of the dilution at which 50% of the wells contained detectable (≥50 pg/ml) HIV p24 measured by ELISA 7 days after inoculation. The results represent the mean value of three independent titrations for each virus stock with no significant deviation from the mean in each triplicate assay (p = 1). (Western blot) Mutations in CA do not preclude incorporation of HSP90AB1 into virions. Western blot analysis of WT and CA-mutant virions purified from transfected 293T-17 cell culture supernatant. The pelleted virus was probed for HSP90AB1, and 1 μg of 293T-17 protein lysate was used as positive control. In addition, virus preparations were probed for CA, MA, and nucleocapsid proteins. The results are representative of three independent iodixanol-purified virus preparations.
To determine if mutations in CA or the CA-SP1 phenotype affect incorporation of HSP90AB1 into virions, we assayed highly purified virions harboring the different mutations (Fig. 4). Neither the CA-SP1 phenotype nor the CA mutations appeared to alter incorporation of HSP90AB1 into virions. Consistent with previous reports, the CA mutations did not block Gag processing, as evidenced by detection of CA, MA, and NC (Forshey et al., 2002). The PR-mutant HIV phenotype displayed the characteristic presence of CA-SP1 (p25) and uncleaved NC-spacer peptide 2 (p8).
CA-mutant HIV is rescued by and hypersensitive to in vitro inhibition of HSP90AB1
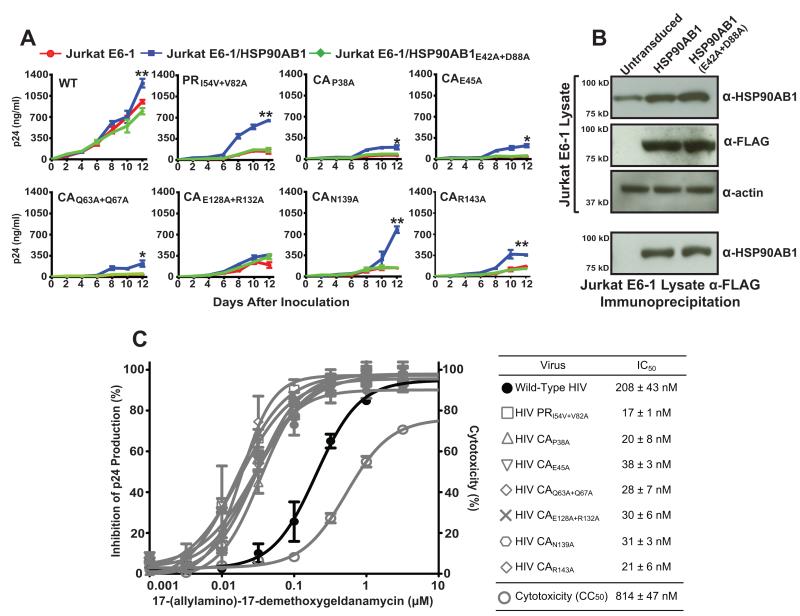
Consistent with previous reports (von Schwedler et al., 2003; Yamashita et al., 2007), the selected mutations in HIV CA reduced productive infection of Jurkat E6-1 cells, but cellular expression of HSP90AB1 compensated for the constraints imposed by these CA mutations (Fig. 5A). At the end of a 12-day continuous culture assay, CA-mutant infection of Jurkat E6-1 cells transduced with native HSP90AB1 increased by 1.7-fold for CAE128A+R132A to 5.4-fold for CAN139A over CA-mutant infection of nonactivated Jurkat E6-1 cells (Table 1). The P38A and Q63A+Q67A CA-mutant viruses are reported to have unstable cores, while the E45A and E128A+R132A mutations resulted in greater numbers of hyperstable cores per virion. A significant increase in infection was observed in HSP90AB1-transduced Jurkat E6-1 cells for three of the CA mutants irrespective of core stability. The N139A mutation in CA displays a normal WT-like stable core and normal core numbers per virion with no discernable defects in assembly, maturation, or viral RNA packaging, yet this mutation impairs HIV infectivity. N139A appeared to most benefit from cellular expression of HSP90AB1 and exhibited exponential growth with the potential to establish a spreading infection. The R143A mutation in CA reduces the number of cores per virion but paradoxically results in a hyperstable core. As with the other CA mutants, infectivity of the R143A mutant HIV increased in the presence of HSP90AB1. In parallel, we included the PR-mutant PRI54V+V82A HIV as a positive control to validate the rescue by overexpression of HSP90AB1.
FIGURE 5.
Reduced infectivity of CA-mutant HIV is rescued by HSP90AB1. (A) Jurkat E6-1 cells transduced with retroviral vectors expressing native and mutant HSP90AB1 were inoculated with WT, PR-mutant, and CA-mutant HIV at a MOI of 0.05. Results are expressed as mean supernatant HIV p24 levels ± SEM (error bars) and each experiment represents independent transduction of Jurkat E6-1 cells followed by inoculation repeated three separate times. CA-mutant p24 levels in Jurkat E6-1/HSP90AB1 were compared to Jurkat E6-1 (*p<0.05, **p<0.001, Student’s t test). (B) Western blot analysis of transduced Jurkat E6-1 cells to demonstrate comparable expression of native and mutant HSP90AB1. (top panel) 1 μg of protein from transduced Jurkat E6-1 cell lysate was probed for HSP90AB1, FLAG, and alpha smooth muscle actin. Comparable detection of alpha smooth muscle actin served as a loading control. (lower panel) Detection of HSP90AB1 in Jurkat E6-1 cell lysates does not discriminate between endogenous protein and the protein contributed by vector-mediated exogenous HSP90AB1 expression. Transduced Jurkat E6-1 cell lysates were immunoprecipitated with an anti-FLAG antibody and subsequently probed for HSP90AB1. The results are representative of three independent assays. (C) CA mutations render HIV hypersensitive to pharmacological inhibition of HSP90AB1 in vitro. PHA-activated PBMC inoculated at a MOI of 0.001 with each CA mutant were treated with serial half-log dilutions of 17-AAG, and 50% inhibitory concentrations (IC50) were calculated after 7 days. The 50% cytotoxic concentration (CC50) of the drug was determined by MTT assay in uninfected PBMC. The graph represents the mean IC50 ± SEM (error bars) of three independent assays.
TABLE 1.
Cellular expression of HSP90AB1 can rescue infection-impaired CA-mutant HIV.
| Virus | Relative Infectivity (%) to WT HIV |
|||
|---|---|---|---|---|
| Jurkat E6-1 | HSP90AB1 | Fold- Increase |
Hsp90AB1 E42A+D88A |
|
| Wild-Type HIV | 100 | 133 ± 11 | 1.3 | 83 ± 7 |
| HIV PRI54V+V82A | 15 ± 4 | 69 ± 3 | 4.6 | 17 ± 4 |
| HIV CAP38A | 6 ± 1 | 20 ± 4 | 3.4 | 8 ± 1 |
| HIV CAE45A | 5 ± 1 | 22 ± 2 | 4.4 | 6 ± 1 |
| HIV CAQ63A+Q67A | 5 ± 1 | 23 ± 6 | 4.6 | 5 ± 1 |
| HIV CAE128A+R132A | 22 ± 3 | 38 ± 3 | 1.7 | 34 ± 4 |
| HIV CAN139A | 15 ± 1 | 81 ± 8 | 5.4 | 15 ± 1 |
| HIV CAR143A | 17 ± 1 | 37 ± 3 | 2.1 | 13 ± 1 |
Relative infectivity of WT and mutant HIV in Jurkat E6-1/HSP90AB1 and Jurkat E6-1/HSP90AB1E42A+D88A cells compared to WT HIV infectivity in Jurkat E6-1 cells after 12 days of continuous culture. Results are expressed as mean infectivity ± S.E. for three independent transduction experiments each performed in triplicate.
We ruled out the possible influence of retroviral-mediated cellular upregulation in Jurkat E6-1 cells simultaneously transduced with the same retroviral vector expressing the catalytically defective HSP90AB1E42A+D88A mutant. Overexpression of the nonfunctional HSP90AB1E42A+D88A mutant protein failed to increase CA-mutant infectivity beyond the level observed in nonactivated Jurkat E6-1 cells. In addition, to validate rescue by native HSP90AB1, we tested the transduced Jurkat E6-1 cells for expression levels of both the native and mutant HSP90AB1 (Fig. 5B, top panel). First, equal amounts of Jurkat E6-1 lysate extracted from nonactivated HSP90AB1-transduced and mutant HSP90AB1E42A+D88A-transduced cells were probed for HSP90AB1 and the FLAG epitope, and equal protein loading was confirmed by comparable detection of actin. Second, because direct probing of Jurkat E6-1 cell lysates does not discriminate between endogenous HSP90AB1 and the amount of protein contributed by transduced exogenous expression of native or mutant HSP90AB1, we immunoprecipitated the exogenously expressed N-terminal FLAG-tagged native and mutant HSP90AB1 from the cell lysates using anti-FLAG affinity gel (Fig. 5B, lower panel). Further, to ensure the IP was not carried out under saturating conditions, the Jurkat E6-1 cell lysate from the first-round IP was subjected to additional IP using fresh anti-FLAG affinity gel and subsequently probed for HSP90AB1. The absence of HSP90AB1 in the second-round IP indicates that recombinant protein expression did not saturate the anti-FLAG affinity gel. Under these conditions, we found comparable protein expression between the native- and mutant HSP90AB1-transduced Jurkat E6-1 cells. Taken together, these results confirm the rescue of CA-mutant HIV by native HSP90AB1 and rule out the possible influence of cellular side effects resulting from retroviral transduction or protein overexpression. To determine if inhibition of HSP90AB1 in activated primary cells would adversely affect infectivity of the CA mutants variants, we determined the antiviral activity of 17-(allylamino)-17-demethoxy-geldanamycin (17-AAG), a pharmacological inhibitor of HSP90AB1 (Powers and Workman, 2007) (Fig. 5C). 17-AAG is an analog of the naturally occurring benzoquinone ansamycin class of antibiotics and specifically competes with cellular ATP for the N-terminal nucleotide binding site in HSP90AB1. Similar to HIV with the CA-SP1 phenotype, the CA mutants were also hypersensitive to HSP90AB1 inhibition, suggesting a functional association between this cellular protein and the HIV core.
Hyperthermia enhances HIV infection
Members of the heat shock protein family respond to variation in ambient temperature and, Roesch et al. (Roesch et al., 2012) recently reported that raising the culture temperature to 39.5 °C enhances HIV replication, without changing the surface expression of CD4 or other cell surface markers, or accelerating HIV fusion. In our hands, the infectious titer of WT HIV was increased 30-fold while PR-mutant and CA-mutant titers increased from 6-fold to 22-fold (Fig. 6 A). We inoculated PBMC at 37 °C in duplicate and then incubated the cells at 37 °C or 39.5 °C for the duration of the assay. Elevated culture temperatures may affect cell growth and viability, which in turn might influence the infectivity of WT and mutant HIV variants. At the end of a 7-day titration assay, the absolute cell number and viability of PBMC incubated at 39.5 °C were comparable to those observed at 37 °C (Fig. 6B). Incubation at 39.5 °C also resulted in a mean 3.6-fold increase in HSP90AB1 expression (Fig. 6C) and, taken together, these results provide evidence that hyperthermia enhances HIV infectivity.
FIGURE 6.
Hyperthermia enhances HIV infectivity. (A) Comparison of WT, PR-mutant, and CA-mutant infectivity assayed at 37 °C and 39.5 °C was determined by TCID50 values derived from serial half-log dilutions of each virus stock in quadruplicate wells of PHA-activated PBMC (1 × 105 cells/well) and calculated as a reciprocal of the dilution at which 50% of the wells contained detectable (≥50 pg/ml) HIV p24 measured by ELISA 7 days after inoculation. Results are represented as mean fold-increase in HIV infectivity at 39.5 °C ± SEM for three independent titration experiments. (B) Flow cytometry analysis of PBMC cultures incubated at 37 °C and 39.5 °C for 7 days using the Trucount assay for absolute PBMC cell number and amine aqua exclusion dye to determine cell viability. The flow cytometry plots are one of three independent assays, and the mean total cell number/ml ± SEM was 1171 ± 470 for PMBC incubated at 37 °C and 1190 ± 501 for PBMC incubated at 39.5° C. The mean percent cell viability ± SEM was 60 ± 3 for PMBC incubated at 37 °C and 63 ± 2 for PBMC incubated at 39.5 °C. (C) Hyperthermia increases cellular HSP90AB1 levels. Western blot analysis on 1 μg of protein from cell lysate extracted from PBMC incubated at 37 °C and 39.5 °C for 7 days and probed for HSP90AB1 and alpha smooth muscle actin. The mean fold-increase in HSP90AB1 expression ± SEM was 3.6 ± 0.14, and the western blot represents one of three independent assays. (D) Comparison of WT, PR-mutant, and CA-mutant infectivity assayed at 37 °C and 39.5 °C in a single-cycle infection assay. The TZM-bl luciferase reporter cell line was assayed for relative light units (RLU) 48 h after inoculation. The graph represents mean RLU ± SEM (error bars) from three independent inoculation assays. (E) Flow cytometry analysis of TZM-bl cultures incubated at 37 °C and 39.5 °C for 48 h using the Trucount assay for absolute cell number and amine aqua exclusion dye to determine cell viability. The flow cytometry plots are representative of three independent assays, and the mean total cell number/ml ± SEM was 223 ± 33 for TZM-bl cells incubated at 37 °C and 172 ± 43 for TZM-bl cells incubated at 39.5 °C. The mean percent cell viability ± SEM was 97 ± 0.6 for TZM-bl cells incubated at 37° C and 96 ± 0.8 for TZM-bl cells incubated at 39.5 °C. (F) Hyperthermia increases cellular HSP90AB1 levels. Western blot analysis of 1 μg of protein from lysate extracted from TZM-bl cells incubated at 37 °C and 39.5 °C for 48 h and probed for HSP90AB1 and actin. The mean fold-increase in HSP90AB1 expression ± SEM was 2.8 ± 0.21, and the western blot represents one of three independent assays.
We next assayed the CA-mutant panel in a single-cycle infection assay to determine if the elevated temperature enhances HIV infectivity in nonactivated cells. The TZM-bl luciferase reporter cell line was inoculated with WT, PR-mutant, and CA-mutant HIV at 37 °C, and duplicate cultures were subsequently incubated at 37 °C and 39.5 °C for the duration of the assay. The infectivity of WT HIV increased 2-fold at 39.5 °C and the PR-mutant and CA-mutant HIV, although noninfectious at 37 °C, were able to infect the heat-shocked reporter cell line (Fig. 6D). We confirmed that incubating the TZM-bl cell line at 39.5 °C did not affect viability or absolute cell number (Fig. 6E). As observed for PBMC, incubating the TZM-bl cell line at 39.5 °C resulted in a mean 2.8-fold increase in HSP90AB1 expression (Fig. 6F). Our observation that hyperthermia enhances WT HIV infectivity up to 30-fold suggests a functional role for the heat-shock response in HIV infection. The observation that reduced infectivity of the PR-mutant and CA-mutant viruses was rescued under hyperthermic conditions implies that hyperthermia may influence the early postentry stage of the HIV life cycle.
Discussion
HSP90AB1 is one of two cytosolic isoforms of the heat shock protein 90 family of cellular chaperones (Pearl et al., 2008). Heat shock protein 90 family members are some of the most conserved proteins in the kingdom of life and with the exception of the Archaea, all prokaryotes and eukaryotes express one or more genes encoding heat shock 90 proteins (Taipale et al., 2010). Heat shock protein 90 members are at the hub of protein homeostasis, executing conformation-dependent protein maturation and regulating the deleterious effects of misfolded proteins during embryogenesis and development (Richter and Buchner, 2011). The heat shock protein 90 machinery operates as a multisubunit complex on a wide array of “client” proteins to achieve their active conformation (Li et al., 2012). Further, heat shock protein 90 members appear to influence the genetic variation of organisms in response to environmental stress (Jarosz and Lindquist, 2010).
We have previously shown that HSP90AB1 rescues the loss in HIV infectivity caused by incompletely processed CA-SP1 and in this report, we extend our initial findings to evaluate the relevance of HSP90AB1 in WT and CA-mutant infectivity. Analysis of highly purified HIV revealed that HSP90AB1 is incorporated within virions and is located within the virion but outside of the virus core. The nature and content of cellular proteins incorporated into HIV virions vary greatly between different susceptible cell types and also depending on the method of virus production. To rule out a bias for HSP90AB1 incorporation into HIV virions produced by transient transfection of the human embryonic kidney-derived 293T-17 transformed cell line, HSP90AB1 was also detected in purified virions generated by infecting the human leukemic T-cell-derived Jurkat E6-1 immortalized cell line. In addition, Jurkat E6-1 cells were either activated or transduced to overexpress recombinant HSP90AB1 before inoculation with WT HIV. Overexpression did not influence the virion concentration of HSP90AB1, and biochemical assays are underway to determine the relevance of the presence of HSP90AB1 within the HIV virion.
Core stability determines the fate of HIV infection and mutations in CA often lead to nonproductive infection. The PR and CA mutations did not affect the yield of virus production and, when tested in a serial-dilution titration assay, the CA mutants infected activated human primary cells. In addition, the PR and CA mutations did not prevent incorporation of HSP90AB1 or affect virion formation. The selected CA mutations result in unstable cores that rapidly dissociate, hyperstable cores that resist disassembly, or cores with WT-like characteristics (Forshey et al., 2002). Here, we show that CA-mutant HIV with unstable, hyperstable, or WT-like cores can productively infect target cells that express HSP90AB1. These observations are in agreement with the second-site suppressor CA mutation for which the T261I and R132T compensatory mutations restored infectivity to CA-mutants with both unstable cores and hyperstable cores (Yang et al., 2012). Further, the CA-mutant viruses were hypersensitive to HSP90AB1 inhibition irrespective of core stability, suggesting a functional role for this class of cellular chaperone in early postentry HIV infection.
Heat shock proteins respond to changes in ambient temperature (Nakai and Ishikawa, 2001; Wandinger et al., 2008), and a recent report (Roesch et al., 2012) demonstrated that hyperthermia enhances HIV infectivity. In accordance with this report, we find that elevating the incubation temperature to 39.5 °C enhanced WT HIV infectivity, though we observed a more pronounced 30-fold increase in virus infection. More importantly, the elevated incubation temperature restored infectivity to a panel of viruses with mutant CA. This rescue effect was observed in a continuous-culture assay and in a single-cycle infection assay with a concurrent increase in cellular HSP90AB1 levels.
Of the six CA mutants studied, the N139A mutant virus appeared to most benefit from expression of HSP90AB1in the target cell as well as from increased temperature. The proposed structural model for monomeric CA places the N139 residue within helix 7, a flexible linker region that connects the large N-terminal domain to the smaller C-terminal domain of CA. The N139A mutation does not affect core stability or morphology but, because maturation of the HIV core occurs in preparation for uncoating, mutations at this position might prevent access to essential CA-interacting host factors in the infected cell. This is in line with the view by Vogt (Vogt, 1996) in that “Teleologically, one can think of proteolytic maturation of Gag and Gag-Pol proteins as readying the virion for a new round of infection. In this view maturation destabilizes the virus core, facilitating its dissociation in the newly infected cell and perhaps facilitating reverse transcription of the genome.” The transdominant effects of cell activation, HSP90AB1 expression, and increased temperature independently rescued the CA-mutant viruses, and further analysis of the N139A mutant will determine if structural motifs in or around this residue regulate HIV core uncoating in an infected cell.
In conclusion, altering the protein composition of the HIV core most likely precludes essential host factors from facilitating the postentry events of infection but despite that, nonlethal mutations in virus structural proteins routinely occur in response to immune pressure, evolutionary constraints, and drug therapy. Detection of HSP90AB1 in HIV virions and the observation that hyperthermia enhances virus infectivity suggest a functional role for this cellular chaperone in the HIV life cycle. Further analysis will reveal if HSP90AB1 directly interacts with the HIV core or if expression of this cellular chaperone in turn induces an environment conducive suboptimally assembled virions to uncoat in an infected cell.
Material and methods
Cells, virus, and expression constructs
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Jurkat clone E6-1 from Dr. Arthur Weiss (Weiss et al., 1984), the TZM-bl cell line from Dr. John C. Kappes, Dr. Xiaoyun Wu and Tranzyme Inc. (Derdeyn et al., 2000; Wei et al., 2002), and the pNL4-3 infectious molecular proviral DNA clone from Dr. Malcolm Martin (Adachi et al., 1986). The 293T-17 cell line was obtained from the ATCC (Manassas, VA). pNL4-3 was transfected into 293T-17 cells using Lipofectamine2000 (Invitrogen) to generate WT, PR-mutant, and CA-mutant HIV stocks. The 50% tissue culture infectious dose (TCID50) of WT, PR-mutant, and CA-mutant stock viruses was determined with serial half-log dilutions of each virus stock in quadruplicate wells of PHA-activated PBMC (1 × 105 cells per well). The TCID50 was calculated as a reciprocal of the dilution at which 50% of the wells contained detectable (≥ 50 pg/ml) HIV p24 measured by ELISA (Perkin Elmer) 7 days after inoculation. CA mutations were introduced individually into pNL4-3 by standard site-directed mutagenesis (Agilent Technologies). The I54V and V82A PR-mutant HIV clone has been described earlier (Joshi and Stoddart, 2011). Native HSP90AB1 and catalytically inactive mutant HSP90AB1E42A+D88A were cloned into a murine leukemia virus (MLV)-based retrovirus expression vector, as described earlier (Joshi and Stoddart, 2011).
Purification of HIV virions
A subconfluent culture of 293T-17 cells was transfected with the respective WT and mutant pNL4-3 clones for 4 h using Lipofectamine2000 followed by a change of culture medium. The supernatant was harvested 48 h later, clarified by low-speed centrifugation, and filtered through a 0.22-μm membrane, and virions were sedimented through a 20% sucrose cushion at 100,000 × g for 2 h at 4 °C. The virus pellet was resuspended in phosphate-buffered saline (PBS), treated with DNase (RNase-free, Roche) and overlaid on 2 ml of 8.4% iodixanol (OptiPrep, Sigma). HIV virions were sedimented at 100,000 × g for 1.5 h, resuspended in PBS, and assayed for p24 content (Perkin Elmer). The pelleted WT, PR- and CA-mutant samples were diluted to 10 μg p24 per ml in PBS. Culture supernatant from mocktransfected 293T-17 cells was treated identically, and the resulting “pellet” was resuspended in the same volume of PBS as the virus samples.
Acetylcholinesterase activity assay
AChE activity was measured following a procedure described previously by Ellman et al. (Ellman et al., 1961). Briefly, 50 μl of the iodixanol-purified HIV pellet was diluted to 100 μl in PBS and mixed with 100 μl of 1.25 mM acetylthiocholine (pH 8) and 0.1 mM 5,5-dithio-bis(2-nitrobenzoic acid) (pH 7) in PBS. The incubation was performed at room temperature, but the test samples and the substrate solution were prewarmed to 37 °C for 10 min before reading the optical density. Changes in absorption were monitored at 450 nm for 10 min with a plate reader spectrophotometer. The sensitivity of the assay was determined using serial dilutions of Electrophorus electricus AChE (Sigma). As a positive control for AChE activity, cytoplasmic organelles and vesicles from a comparable volume of 293T-17 culture supernatant were sedimented at 100,00 × g for 1.5 h, and AChE activity was determined in 50 μl of the resuspended pellet. The enzyme kinetics (Vmax) of AChE in the iodixanol-purified HIV sample, mock transfected sample, and positive control were calculated using the Michaelis-Menten protocol.
Western blot analysis
The equivalent of 100 ng p24 from purified WT, PR-, and CA-mutant preparations was resuspended in Laemmli SDS-PAGE loading buffer (Sigma). Sample volume from the mock transfection was normalized to the virus preparations by loading the same volume size as the purified WT HIV preparation. Cellular protein from Jurkat E6-1 and 293T-17 cell lines was extracted using the CelLytic M solution (Sigma), and protein concentration was estimated using bicinchoninic acid (Sigma). High-molecular-weight HIV proteins were separated on 8–20% linear gradient polyacrylamide gels, and smaller HIV proteins were resolved on 15% gels. Cellular and HIV protein samples for the detection of HSP90AB1 and alpha smooth muscle actin were resolved on a stacked 12% polyacrylamide gel. The proteins were electroblotted onto Immobilon PVDF membranes (Millipore) and reacted individually with specific primary antibodies. The following primary antibodies were obtained from the AIDS Reagent Program; CA (cat. #6521), MA (cat. #4811), RT (cat. #11338), IN (cat. #7374), gp41 (cat. #7623), and HIV-immunoglobulin (cat. #3957). The NC primary antibody was obtained from the National Cancer Institute AIDS Vaccine Program. Rabbit anti-HSP90AB1, mouse anti-alpha smooth muscle actin primary antibodies, and peroxidase-conjugated secondary antibodies were purchased from Abcam, and immune complexes were visualized by enhanced chemiluminescence (Pierce).
Subtilisin treatment of purified HIV
Virions from WT HIV-transfected 293T-17 cells were purified as described above. The pelleted virus was resuspended in 500 μl of 10 mM Tris pH 8.0, 100 mM NaCl and mixed with an equal volume of 2x digestion buffer (40 mM Tris pH 8.0, 2 mM CaCl2) containing the bacterial subtilisin protease (Sigma), as previously described (Ott, 2009). Samples were incubated with a final concentration of 100 μg/ml or 500 μg/ml subtilisin at 37 °C for 18 h and reactions stopped by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma) for 20 min at room temperature. The samples were diluted in PBS, and the virus was purified on 8.4% iodixanol as described above. Pelleted virions were resuspended in SDS-PAGE buffer containing 1 mM PMSF.
Isolation of HIV cores
Intact WT HIV virions were purified as described above, and the cores were purified using a standard protocol (Welker et al., 2000). Briefly, the pelleted virus was resuspended in 100 μl of PBS and mixed with an equal volume of 200 mM NaCl, 100 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0). Intact virions were lysed by addition of Triton X-100 to a final concentration of 0.5%, and the HIV cores were recovered by centrifugation in a microcentrifuge (16,000 × g) for 8 min at 4 °C. The pelleted cores were washed three times with 100 mM NaCl, 50 mM MOPS (pH 7.0), resuspended in SDS-PAGE buffer, and subjected to immunoblotting. Prior to resuspending the extracted cores in SDS-PAGE buffer, a small fraction of the sample was subjected to RT-PCR to detect the gag gene sequence by standard methods.
Detection of HSP90AB1 in virions harvested from inoculated Jurkat E6-1 cells
A subconfluent culture of Jurkat E6-1 cells was inoculated with WT HIV at a multiplicity of infection (MOI) of 0.5 and cultured for 4 days. Culture supernatant from HIV- and mock-inoculated Jurkat E6-1 cells was sedimented on iodixanol and the sample pellets were tested for AChE activity as described above.
Overexpression of HSP90AB1 in Jurkat E6-1 cells
The design and construction of a retroviral-based gene transfer vector intended to express human HSP90AB1 preceded by a short FLAG epitope has been described earlier (Joshi and Stoddart, 2011). Jurkat E6-1 cells were transduced with 0.1 MOI of the retroviral vector carrying the native HSP90AB1 gene for 24 h. The transduced cells were resuspended in fresh culture medium, inoculated at a MOI of 0.5 with WT HIV, and cultured for an additional 4 days. Simultaneously, Jurkat E6-1 cells were activated in the presence of anti-CD3 and anti-CD28 antibody-coated magnetic beads as recommended by the manufacturer (Invitrogen) for 24 h. Activated cells were treated similar to the HSP90AB1 transduced cells and four days after inoculation, the Jurkat E6-1 cells were washed in PBS and lysed in CelLytic M solution (Sigma). Simultaneously, virions were purified from the cell-free culture supernatant as described above. Cellular and HIV proteins were resuspended in SDS-PAGE buffer and subjected to immunoblotting.
CA-mutant inoculation of HSP90AB1-expressing cells
Jurkat E6-1 cells were transduced at a MOI of 0.1 with retroviral vectors carrying the native HSP90AB1 gene and a catalytically inactive mutant of HSP90AB1E42A+D88A, as described previously (Joshi and Stoddart, 2011). The transduced Jurkat E6-1 cells were inoculated at a MOI of 0.05 with WT HIV and CA-mutant HIV, and the PR-mutant HIV PRI54V+V82A served as a positive control for rescue of reduced infectivity of the mutant viruses. Productive infection and HIV growth kinetics were observed over 12 days by measuring HIV p24 levels in the supernatant by standard ELISA (Perkin Elmer). Overexpression of both native HSP90AB1 and the catalytically inactive mutant HSP90AB1E42A+D88A was compared on transduced Jurkat E6-1 cell lysates by immunoblotting. Additionally, the cell lysates were subjected to immunoprecipitation (IP) using the anti-FLAG M2 affinity gel (Sigma) for 18 h at 4 °C. To ensure the IP was not performed under saturating conditions, the cell lysate was subjected to an additional IP with the anti-FLAG M2 affinity gel. The IP protein was immunoblotted and subsequently probed with anti-HSP90AB1.
In vitro antiviral activity assay
Antiviral activity of 17-AAG (Sigma) was assayed in PHA-activated PBMC as previously described (Joshi and Stoddart, 2011). Briefly, PBMC were inoculated in bulk at a MOI of 0.001 of each virus for 2 h, cells were washed, and 1 × 105 cells in 100 μl were seeded in triplicate wells of a 96-well plate. Wells were treated with 100 μl of serial half-log dilutions of 17-AAG or with medium alone, and supernatants were assayed at day 7 for p24 by ELISA. Parallel cellular toxicity of the drug was determined on uninfected cells by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay (Sigma) on day 7. The IC50 (50% inhibition of virus replication) and CC50 (50% reduction in cell viability) were calculated as the mean of three independent assays.
PBMC hyperthermia conditions and assays
The difference in infectivity of WT, PR-mutant, and CA-mutant stock viruses in PHA-activated PBMC cultured at 39.5 °C compared to 37 °C was determined from TCID50 values, as described above. To ensure the elevated temperature did not influence HIV adsorption or entry, the 2-h inoculation period was carried out at 37 °C, after which duplicate plates were incubated at 37 °C and 39.5 °C for the duration of the assay. Simultaneously, uninoculated PBMC were cultured at 37 °C and 39.5 °C for 7 days and assayed for cell viability and HSP90AB1 protein content by western blot as described above. The absolute number of PBMC was determined by flow cytometry with Trucount tubes (BD Biosciences), and the viability of the 7-day culture was estimated by adding amine aqua (Invitrogen) exclusion dye. The fold-increase in HSP90AB1 expression was calculated using the ImageJ software (http://imagej.nih.gov/ij/, 1997-2012).
TZM-bl hyperthermia conditions and assays
The difference in infectivity of WT, PR-mutant, and CA-mutants was determined in a single-cycle infection assay using the TZM-bl HeLa CD4 cell line, as previously described (Wei et al., 2002). Briefly, 20,000 TZM-bl cells were seeded into 12-well plates in duplicate and inoculated with 0.01 MOI of the WT, PR-mutant, and CA-mutant stock viruses. To ensure the elevated temperature did not influence HIV adsorption or entry, the 4-h inoculation period was carried out at 37 °C, after which duplicate plates were incubated at 37 °C and 39.5 °C for the duration of the assay. After 48 h, the cultured cells were washed with PBS, and the Bright-Glo luciferase assay substrate and buffer (Promega) were added directly to each well to lyse the cells. Following a 2-min incubation, the cell lysate was transferred to a black 96-well plate, and luminescence was measured on a SpectraMax M2 plate reader (Molecular Devices). Simultaneously, uninoculated TZM-bl cells were incubated at 37 °C and 39.5 °C for 48 h and assayed for HSP90AB1 protein content and cell viability, as described above.
Highlights.
We previously identified HSP90AB1 as a host factor essential for HIV replication.
HSP90AB1 expression rescued infectivity of HIV with incomplete CA-SP1 cleavage.
Here we show that HSP90AB1 can also rescue HIV with defective cores.
HSP90AB1 expression increases with hyperthermia.
Hyperthermia increased HIV infectivity up to 30-fold and rescued CA-mutant virus.
Acknowledgments
We thank Mike McCune for a critical reading of the manuscript. This project has been funded in part with Federal funds from NIAID, NIH, under contract no. HHSN266200700002C/N01-AI-70002, the California HIV/AIDS Research Program (CHRP) Grant No. ID09-SF-051 to C. S., by the UCSF AIDS Research Institute and the Harvey V. Berneking Living Trust, and P01 GM083658 and P30 AI036214 to B. E. T.
The abbreviations used are
- PR
Protease
- CA-SP1
capsid-spacer peptide 1
- AChE
Acetylcholinesterase
- HSP90AB1
heat shock protein 90 kDa alpha (cytosolic), class B member 1
- 17-AAG
17-(allylamino)-17-demethoxygeldanamycin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59(2):284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantin R, Diou J, Belanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J Immunol Methods. 2008;338(1-2):21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Cantin R, Fortin JF, Tremblay M. The amount of host HLA-DR proteins acquired by HIV-1 is virus strain- and cell type-specific. Virology. 1996;218(2):372–381. doi: 10.1006/viro.1996.0206. [DOI] [PubMed] [Google Scholar]
- Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Muller B, Grunewald K, Krausslich HG. Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis. Cell Host Microbe. 2008;4(6):592–599. doi: 10.1016/j.chom.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr., Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74(18):8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettenhofer M, Yu XF. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J Virol. 1999;73(2):1460–1467. doi: 10.1128/jvi.73.2.1460-1467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroe E, Silver PA, Engelman A. HIV-1 incorporates and proteolytically processes human NDR1 and NDR2 serine-threonine kinases. Virology. 2005;331(1):181–189. doi: 10.1016/j.virol.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80(8):3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres V, Jr., Feather-Stone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Forshey BM, von Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol. 2002;76(11):5667–5677. doi: 10.1128/JVI.76.11.5667-5677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J Virol. 2004;78(5):2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroud C, Chazal N, Briant L. Cellular kinases incorporated into HIV-1 particles: passive or active passengers? Retrovirology. 2011;8:71. doi: 10.1186/1742-4690-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5(4):253–263. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Gurer C, Cimarelli A, Luban J. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J Virol. 2002;76(9):4666–4670. doi: 10.1128/JVI.76.9.4666-4670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330(6012):1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Stoddart CA. Impaired infectivity of ritonavir-resistant HIV is rescued by heat shock protein 90AB1. J Biol Chem. 2011;286(28):24581–24592. doi: 10.1074/jbc.M111.248021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 2012;1823(3):624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell. 1993;73(6):1067–1078. doi: 10.1016/0092-8674(93)90637-6. [DOI] [PubMed] [Google Scholar]
- Nakai A, Ishikawa T. Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. Embo J. 2001;20(11):2885–2895. doi: 10.1093/emboj/20.11.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE. Cellular proteins detected in HIV-1. Rev Med Virol. 2008;18(3):159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- Ott DE. Purification of HIV-1 virions by subtilisin digestion or CD45 immunoaffinity depletion for biochemical studies. Methods Mol Biol. 2009;485:15–25. doi: 10.1007/978-1-59745-170-3_2. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem J. 2008;410(3):439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007;581(19):3758–3769. doi: 10.1016/j.febslet.2007.05.040. [DOI] [PubMed] [Google Scholar]
- Richter K, Buchner J. Closing in on the Hsp90 chaperone-client relationship. Structure. 2011;19(4):445–446. doi: 10.1016/j.str.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Roesch F, Meziane O, Kula A, Nisole S, Porrot F, Anderson I, Mammano F, Fassati A, Marcello A, Benkirane M, Schwartz O. Hyperthermia Stimulates HIV-1 Replication. PLoS Pathog. 2012;8(7):e1002792. doi: 10.1371/journal.ppat.1002792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos S, Obukhov Y, Nekhai S, Bukrinsky M, Iordanskiy S. Virus-producing cells determine the host protein profiles of HIV-1 virion cores. Retrovirology. 2012;9(1):65. doi: 10.1186/1742-4690-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Tremblay MJ, Fortin JF, Cantin R. The acquisition of host-encoded proteins by nascent HIV-1. Immunol Today. 1998;19(8):346–351. doi: 10.1016/s0167-5699(98)01286-9. [DOI] [PubMed] [Google Scholar]
- Vogt VM. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol. 2003;77(9):5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283(27):18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46(6):1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Wiskocil RL, Stobo JD. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pretranslational level. J Immunol. 1984;133(1):123–128. [PubMed] [Google Scholar]
- Welker R, Hohenberg H, Tessmer U, Huckhagel C, Krausslich HG. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J Virol. 2000;74(3):1168–1177. doi: 10.1128/jvi.74.3.1168-1177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. Embo J. 2007;26(8):2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3(10):1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Shi J, Byeon IJ, Ahn J, Sheehan JH, Meiler J, Gronenborn AM, Aiken C. Second-site suppressors of HIV-1 capsid mutations: restoration of intracellular activities without correction of intrinsic capsid stability defects. Retrovirology. 2012;9(1):30. doi: 10.1186/1742-4690-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]