Abstract
E2F is a family of transcription factors that regulate the expression of genes involved in a wide range of cellular processes, including cell-cycle progression, DNA replication, DNA repair, differentiation, and apoptosis. E2F1, the founding member of the family, undergoes posttranslational modifications in response to DNA damage, resulting in E2F1 stabilization. In some cases, E2F1 is important for DNA damage-induced apoptosis through the transcriptional activation of p73 and perhaps other proapoptotic target genes. However, in other contexts, E2F1 can stimulate DNA repair and promote survival in response to DNA damage. The E2F1 protein accumulates at sites of both DNA double-strand breaks and UV radiation-induced damage, indicating that E2F1 has a nontranscriptional function at sites of damage. This review summarizes recent progress made in understanding the role of E2F1 in the DNA damage response, including transcription-independent activities that facilitate DNA repair in the context of chromatin.
Introduction
The E2F family of transcription factors controls the expression of genes involved in cell proliferation, differentiation, and apoptosis. There are 8 mammalian E2F genes (E2F1–8) and 3 related DP genes (DP1, DP2/3, and DP4). E2F1 through E2F6 require heterodimerization with a DP protein to create a functional DNA-binding factor, whereas E2F7 and E2F8 bind DNA independently of DP factors. The E2Fs were originally classified as either activators or repressors of transcription on the basis of in vitro reporter assays and expression patterns during the cell cycle. However, it is now clear that several E2F family members can function as either activators or repressors of transcription, depending on the cellular context and target gene (1).
The activities of E2F1 through E2F5 are regulated by binding to the retinoblastoma (RB) tumor suppressor protein and the related pocket proteins p107 and p130. In general, pocket protein binding blocks the transcriptional activity of E2F-DP dimers by masking the transcriptional activation domain located in the carboxy terminus of E2F1 through E2F5. This association prevents E2F-DP heterodimers from recruiting transcriptional coactivators, such as histone acetyltransferases, to the promoters of target genes. RB family members can also convert E2Fs to transcriptional repressors by recruiting other chromatin-modifying enzymes that block access to the basal transcription machinery. Cyclin-dependent kinases (CDK), particularly cyclin D–and cyclin E–associated kinases, phosphorylate RB and the other pocket proteins on multiple sites during cell-cycle progression. These phosphorylation events regulate RB interactions with corepressors and E2F family members to modulate the transcription of various genes involved in cell proliferation and other processes.
A number of E2F family members are known to be responsive to DNA damage. More than 10 years ago, it was shown that E2F1 is induced in response to various DNA-damaging agents, including ionizing radiation, UV radiation, and a number of chemotherapeutic drugs (2–4). This response primarily involves an increase in E2F1 protein stability and, in at least some cases, is associated with the induction of apoptosis (3, 5, 6). On the other hand, E2F4 levels decrease in response to drugs that cause DNA damage, and this may also be important for drug-induced apoptosis (5, 6). Although previous studies did not observe a change in E2F3 levels following DNA damage, a recent study shows E2F3a induction following treatment with several chemotherapeutic drugs (7). Moreover, E2F3 was shown to transcriptionally upregulate E2F1 and contribute to the induction of apoptosis in drug-treated cells. E2F7 and E2F8 are also upregulated in response to DNA-damaging drugs, but in this case, E2F7 and E2F8 repress apoptosis (8). When induced, E2F7 and E2F8 bind to and repress the E2F1 gene promoter, which may limit E2F1-induced apoptosis in response to damage. DNA damage also induces the expression of DP2/3 and DP4 (9, 10). These heterodimeric partners positively (DP2/3) or negatively (DP4) regulate E2F-mediated apoptosis.
The mechanisms by which most of the E2F family members respond to DNA damage are poorly understood. Moreover, the role they play in the DNA damage response has not been extensively explored, and in many cases, research has focused on how these E2F family members regulate the apoptotic activity of E2F1. In contrast, much more is understood about how DNA damage signaling modifies E2F1 and the role these modifications play in regulating E2F1 functions. Thus, this review focuses on the progress made in understanding E2F1 regulation and function in response to DNA damage.
Posttranslational Modifications of E2F1 in Response to DNA Damage
Given the key role of the ataxia telangiectasia mutated (ATM) kinase in orchestrating many of the cellular responses to DNA damage, Nevins and colleagues explored the role of ATM in E2F1 stabilization (5). ATM was found to phosphorylate E2F1 on serine 31, a site not conserved in other E2F family members. E2F1 was not stabilized in response to DNA double-strand breaks in cells lacking ATM activity or when serine 31 was mutated to alanine (5). The ATM and Rad3 related (ATR) kinase was also shown to phosphorylate E2F1 on serine 31, consistent with the finding that ATR-activating agents, such as UV radiation, also induce E2F1 levels (2, 4, 5). The 14-3-3τ protein binds to E2F1 specifically when it is phosphorylated on serine 31, and this blocks E2F1 ubiquitination and degradation by the proteosome (11). In the absence of 14-3-3τ, E2F1 is not stabilized, proapoptotic target genes like p73 and Apaf1 are not induced, and apoptosis is inhibited in response to DNA damage.
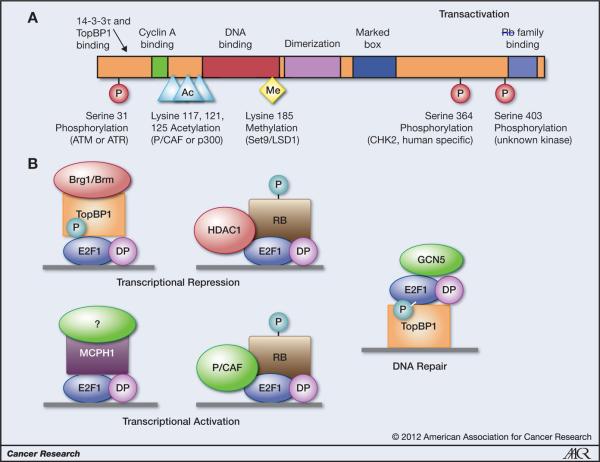
Several other DNA damage-inducible modifications also occur on E2F1 and may contribute to stabilization or altered activity (Fig. 1A). Checkpoint kinase 2 (CHK2), which is downstream of ATM in the DNA damage response, phosphorylates E2F1 on serine 364 (12). This phosphorylation site is not conserved in most other mammals but seems to contribute to the stabilization of human E2F1 through an unknown mechanism. It was recently reported that E2F1 is phosphorylated on serine 403 in response to doxorubicin treatment, which was associated with increased E2F1 transcriptional activity (13). Although serine 403 is conserved in mammals and some other species, the kinase responsible for this modification is unknown.
Figure 1.
A, DNA damage-inducible E2F1 posttranslational modifications. B, E2F1-containing complexes implicated in transcriptional regulation and DNA repair.
E2F1 is also acetylated in response to DNA damage on several lysine residues (K117, K120, and K125) near the DNA-binding domain by either P/CAF or p300 acetyltransferase (14–16). These conserved lysines are also found in E2F2 and E2F3, but it has not been determined whether these other E2F family members are also acetylated in response to DNA damage. Acetylation of E2F1 is stimulated by ATM-mediated phosphorylation at serine 31 but can also occur with reduced efficiency independent of serine 31. Acetylation contributes to E2F1 stabilization following DNA damage independently of serine 31 phosphorylation (15). Acetylation of E2F1 also enhances DNA-binding activity, particularly on the p73 gene promoter, and it is important for the induction of apoptosis in response to agents that cause double-stand breaks (17). Interestingly, E2F1 is not acetylated in response to UV radiation, so E2F1 does not transcriptionally activate p73 or promote apoptosis following UV irradiation (17). Consistent with the positive role that some acetyltransferases have on E2F1 activity, the SIRT1 deacetylase negatively regulates E2F1-mediated apoptosis. SIRT1 interacts with E2F1 and inhibits p73 induction and apoptosis in response to etoposide by directly deacetylating E2F1 and/or by deacetylating and inhibiting P/CAF (16, 18).
A recent report shows that E2F1 is also methylated on lysine 185 by the Set9 methyltransferase, which inhibits E2F1 transcriptional activity (19). In response to doxorubicin treatment, E2F1 is demethylated at this site by the LSD1 demethylase, resulting in increased E2F1 activity and the promotion of apoptosis. As might be expected, cross-talk seems to occur among these different modifications, as methylation of E2F1 inhibited E2F1 acetylation and phosphorylation on serine 364 (19).
Regulation of E2F1 Transcriptional Activity by Protein Partners in Response to DNA Damage
RB binds to E2F1 through 2 different mechanisms, one that is common to other E2F family members and occurs at the carboxy terminus and another specific to E2F1 that involves other regions of E2F1 (20). In response to DNA damage, the E2F1-specific interaction is reduced, whereas the carboxy-terminal interaction is enhanced by the phosphorylation of RB on serine 612 by the DNA-damage responsive CHK1 or CHK2 kinases (21). Surprisingly, E2F1–RB complexes formed following DNA damage not only participate in the transcriptional repression of cell-cycle genes like cyclin A2 but also in the activation of proapoptotic genes such as p73 and caspase-7 (22). Two different E2F1–RB complexes seem to form in response to DNA damage, a repressing complex containing histone deacetylase 1 (HDAC1) and an activating complex containing P/CAF (Fig. 1B; ref. 22).
The amino terminus of E2F1 interacts with 2 different proteins containing BRCA1 carboxy-terminal (BRCT) domains that have opposing activities in regulating E2F1-dependent transcription in response to DNA damage (Fig. 1B). Microcephalin (MCPH1/BRIT1) contains 3 BRCT domains; the second and third BRCT domains are required for binding to E2F1 (23). MCPH1 augments E2F1 transcriptional activity and can be found with E2F1 on target gene promoters that include p73, BRCA1, CHK1, and Caspase-7. The interaction between E2F1 and MCPH1 increases in response to DNA-damaging drugs, and MCPH1 is required for E2F1-mediated induction of p73 and apoptosis (23).
The TopBP1 protein also interacts with E2F1 in response to DNA damage, but in this case, E2F1 transcriptional activity is inhibited independently of RB (24, 25). TopBP1 is involved in the DNA damage response through its interaction with Rad9 and other DNA repair factors and as an activator of ATR. TopBP1 contains 9 BRCT domains (BRCT0–8), and the BRCT6 domain specifically binds E2F1 when it is phosphorylated on serine 31 (24). Other E2F family members that lack this phosphorylation site do not bind to and are not inhibited by TopBP1. This interaction with TopBP1 is important for suppressing E2F1-mediated cell-cycle progression and apoptosis following DNA damage. The mechanism by which TopBP1 represses E2F1 transcriptional activity relies upon the recruitment of Brg1 and hBrm, which are components of the SWI/SNF chromatin-remodeling complex, to E2F1 target gene promoters (25). E2F1 binding to TopBP1 is also regulated by Akt-mediated phosphorylation of TopBP1 (26). Phosphorylation of TopBP1 by Akt results in TopBP1 dimerization, which is required for E2F1 to bind TopBP1 (26).
TopBP1 and MCPH1 bind to the same region of E2F1, yet have opposite effects on E2F1 transcriptional activity, so it is likely that their binding is mutually exclusive. At present, it is unclear what determines whether E2F1 will bind to TopBP1 and be repressed or associate with MCHP1 to enhance transcriptional activity. One difference between these interactions is that TopBP1 binding is sensitive to E2F1 serine 31 phosphorylation, whereas MCPH1 binding seems to be independent of serine 31 phosphorylation (23). It is quite possible that RB interacts with E2F1 at the same time as TopBP1 or MCPH1, as RB interacts with the carboxy-terminus of E2F1, whereas TopBP1 and MCPH1 bind the amino terminus. Additional work will be required to determine the composition of different E2F1-containing complexes that form following DNA damage and their unique functions in the DNA damage response.
E2F1 Functions at Sites of DNA Damage
In addition to inhibiting E2F1 transcriptional activity, TopBP1 binding recruits E2F1 to sites of DNA double-strand breaks, to from foci that overlap with other DNA damage response proteins like BRCA1 (24). E2F1 foci formation at sites of double-strand breaks specifically requires the BRCT6 domain of TopBP1 and only the amino terminus of E2F1, including serine 31, but not the DNA-binding domain (24). E2F1 also rapidly accumulates at sites of UV-induced DNA damage, which is dependent on E2F1 serine 31 but independent of a functional DNA-binding domain (27). Like agents that cause double-strand breaks, UV radiation also stimulates the interaction between E2F1 and TopBP1 (27). Taken together, these findings suggest a model in which E2F1 is recruited to both DNA double-strand breaks and UV-induced damage through a phospho-specific interaction with TopBP1. Depending on the type of damage, either ATM or ATR is likely responsible for phosphorylating E2F1 on serine 31 to induce this interaction with TopBP1.
Only recently have we begun to understand the function of E2F1 at sites of DNA damage. Previous studies suggested that E2F1 activity promoted the repair of UV-induced DNA damage, but it was assumed that this was due to the transcriptional upregulation of nucleotide excision repair (NER) enzymes. However, more recently it was shown that E2F1 is required for the efficient recruitment of NER factors, including XPA and XPC, to sites of UV damage, independent of effects on the levels of these repair proteins (27). The ability of E2F1 to promote XPA recruitment and stimulate efficient NER required serine 31, the dimerization domain, and the Marked box domain of E2F1, but it did not require a functional DNA-binding domain or transcriptional activation domain (27). Thus, E2F1 protein domains important for transcriptional regulation, namely the DNA-binding and transactivation domains, are dispensable for E2F1 to stimulate NER. The ability of E2F1 to stimulate NER through this nontranscriptional mechanism provides a plausible explanation for the surprising finding that E2F1, which is usually thought of as a proapoptotic factor, suppresses apoptosis in response to UV irradiation (1).
E2F1 also promotes the recruitment or retention of repair factors to sites of DNA double-strand breaks. In cells knocked out or knocked down for E2F1, NBS1 foci formation following exposure to ionizing radiation or the radiomimetic drug neocarzinostatin is significantly impaired (28). As with the NER factors, NBS1 protein levels are not altered by E2F1 deficiency, although NBS1 phosphorylation in response to DNA damage is impaired (28). NBS1 is part of the MRN complex that also contains Mre11 and Rad50. The MRN complex is recruited to sites of double-strand breaks through several mechanisms, although the majority of MRN is thought to be recruited through a phospho-specific interaction with the mediator of DNA-damage checkpoint 1 (MDC1) protein, which in turn is recruited to large regions of DNA flanking a double-strand break through a specific interaction with phosphorylated H2AX (γH2AX). The absence of E2F1 does not inhibit the formation of γH2AX or 53BP1 foci, suggesting that E2F1 modulates a specific branch of the DNA damage response involving NBS1. Enhanced association between E2F1 and NBS1 following DNA damage was shown in coimmunoprecipitation experiments (28). Whether this association helps to stabilize the interaction between the MRN complex and MDC1 or whether E2F1 has another activity that promotes NBS1 foci formation remains to be determined.
The MRN complex is involved in DNA end processing to create single-stranded DNA that is required for homologous recombination and microhomology-mediated end joining. The replication protein A (RPA) complex associates with and stabilizes the single-stranded DNA flanking double-strand breaks. RPA is subsequently replaced by the Rad51 recombinase for homologous recombination. As with NBS1, the absence of E2F1 inhibits ionizing radiation–induced foci formation of RPA and Rad51, suggesting that E2F1 is important for end resection and the formation of single-stranded DNA at sites of double-strand breaks (28). Consistent with this idea, cells lacking E2F1 display genomic instability and inefficiently repair DNA breaks following ionizing radiation exposure (28).
It is possible that E2F1 indirectly enhances the recruitment of repair factors to sites of DNA damage by modifying chromatin structure. E2F1 was recently shown to recruit the GCN5 histone acetyltransferase to sites of UV-induced DNA damage (29). Moreover, UV radiation induces an increase in histone H3 lysine 9 (H3K9) acetylation, and this response requires both E2F1 and GCN5. As with E2F1, GCN5 deficiency inhibits the recruitment of NER factors to sites of UV damage and impairs DNA repair (29). Coimmunoprecipitation experiments show an association between E2F1 and GCN5 following UV damage, but the mechanism by which this association is regulated has not been determined. We also find an association between E2F1 and GCN5 in response to ionizing radiation, suggesting that GCN5 could also be involved in mediating E2F1-dependent functions at sites of DNA double-strand breaks (A.K. Biswas; unpublished data). Indeed, GCN5 was recently shown to acetylate H3 in nucleosomes containing γH2AX, which in turn promoted the recruitment of the SWI/SNF chromatin-remodeling complex (30). It is quite possible that E2F1, through its interaction with TopBP1, is responsible for GCN5 recruitment to sites of DNA double-strand breaks (Fig. 1B).
Conclusions and Future Directions
It is now clear that E2F1 has both transcriptional and nontranscriptional functions in the responses to various types of DNA damage. Damage-inducible posttranslational modifications play key roles in regulating E2F1 interactions with protein partners, such as TopBP1, as well as with specific sites in the genome, such as the p73 gene promoter. Much work remains to characterize the E2F1 complexes that form following DNA damage and their roles in regulating transcription and DNA repair. It will be of particular interest to determine whether E2F1 collaborates with the plethora of other proteins that associate with TopBP1 to regulate transcription, DNA repair, and checkpoint signaling.
The mechanism by which E2F1 promotes the recruitment of repair factors to sites of damage remains poorly understood. Although direct binding to some repair proteins could be involved, it is quite likely that modification of chromatin structure plays an important role in the ability of E2F1 to stimulate DNA repair. Indeed, E2F1-mediated recruitment of GCN5 and the stimulation of H3K9 acetylation may be important for both NER and double-strand break repair. How the interaction between E2F1 and GCN5, and perhaps other chromatin-modifying enzymes, is regulated in response to DNA damage is another area of research that needs to be addressed.
Finally, the physiologic relevance of E2F1 in the response to DNA damage remains to be established. E2F1 can behave as either an oncogene or a tumor suppressor gene in mouse models, depending on the experimental context. The ability of E2F1 to drive cell proliferation is likely responsible for its oncogenic activity, whereas the ability of E2F1 to induce apoptosis has been suggested to underlie its tumor suppressor activity. However, at present no experimental data support a role for apoptosis induction as the mechanism by which E2F1 inhibits cancer development. An alternative explanation is that E2F1 suppresses tumor development by enhancing DNA repair efficiency and maintaining genomic stability. Given that radiotherapy and many chemotherapeutic drugs function by causing DNA damage, it will also be of interest to determine how E2F1 modulates the response to cancer therapies by either enhancing repair and cancer cell survival or promoting apoptosis and tumor regression.
Acknowledgments
The authors thank Dr. Weei-Chin Lin for helpful comments and Hilary Graham, Christine Brown, and Becky Brooks for help in preparing the manuscript.
Grant Support Research from the Johnson laboratory described in this review was supported by the NIH (CA079648 to D.G. Johnson), through MD Anderson's Cancer Center Support Grant (CA016672), and by the National Institute of Environmental Health Sciences (P30ES007784).
Footnotes
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 2.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–13. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kato T, Yuan ZM. Role for E2F in DNA damage-induced entry of cells into S phase. Cancer Res. 1997;57:3640–3. [PubMed] [Google Scholar]
- 4.O'Connor DJ, Lu X. Stress signals induce transcriptionally inactive E2F-1 independently of p53 and Rb. Oncogene. 2000;19:2369–76. doi: 10.1038/sj.onc.1203540. [DOI] [PubMed] [Google Scholar]
- 5.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–44. [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Y, Freeman SN, Cress WD. E2F4 deficiency promotes drug-induced apoptosis. Cancer Biol Ther. 2004;3:1262–9. doi: 10.4161/cbt.3.12.1239. [DOI] [PubMed] [Google Scholar]
- 7.Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G, et al. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 2010;30:524–36. doi: 10.1128/MCB.00938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zalmas LP, Zhao X, Graham AL, Fisher R, Reilly C, Coutts AS, et al. DNA-damage response control of E2F7 and E2F8. EMBO Rep. 2008;9:252–9. doi: 10.1038/sj.embor.7401158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingram L, Munro S, Coutts AS, La Thangue NB. E2F-1 regulation by an unusual DNA damage-responsive DP partner subunit. Cell Death Differ. 2011;18:122–32. doi: 10.1038/cdd.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milton AH, Khaire N, Ingram L, O'Donnell AJ, La Thangue NB. 14-3-3 proteins integrate E2F activity with the DNA damage response. EMBO J. 2006;25:1046–57. doi: 10.1038/sj.emboj.7600999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang B, Liu K, Lin FT, Lin WC. A role for 14-3-3 tau in E2F1 stabilization and DNA damage-induced apoptosis. J Biol Chem. 2004;279:54140–52. doi: 10.1074/jbc.M410493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5:401–9. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- 13.Real S, Espada L, Espinet C, Santidrian AF, Tauler A. Study of the in vivo phosphorylation of E2F1 on Ser403. Biochim Biophys Acta. 2010;1803:912–8. doi: 10.1016/j.bbamcr.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Galbiati L, Mendoza-Maldonado R, Gutierrez MI, Giacca M. Regulation of E2F-1 after DNA damage by p300-mediated acetylation and ubiquitination. Cell Cycle. 2005;4:930–9. doi: 10.4161/cc.4.7.1784. [DOI] [PubMed] [Google Scholar]
- 15.Ianari A, Gallo R, Palma M, Alesse E, Gulino A. Specific role for p300/CREB-binding protein-associated factor activity in E2F1 stabilization in response to DNA damage. J Biol Chem. 2004;279:20830–30835. doi: 10.1074/jbc.M402403200. [DOI] [PubMed] [Google Scholar]
- 16.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol. 2003;5:552–8. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 17.Pediconi N, Guerrieri F, Vossio S, Bruno T, Belloni L, Schinzari V, et al. hSirT1-dependent regulation of the PCAF-E2F1-p73 apoptotic pathway in response to DNA damage. Mol Cell Biol. 2009;29:1989–98. doi: 10.1128/MCB.00552-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–31. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 19.Kontaki H, Talianidis I. Lysine methylation regulates E2F1-induced cell death. Mol Cell. 2010;39:152–60. doi: 10.1016/j.molcel.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003;12:639–49. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- 21.Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 2007;26:2083–93. doi: 10.1038/sj.emboj.7601652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ianari A, Natale T, Calo E, Ferretti E, Alesse E, Screpanti I, et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009;15:184–94. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SZ, Lin FT, Lin WC. MCPH1/BRIT1 cooperates with E2F1 in the activation of checkpoint, DNA repair and apoptosis. EMBO Rep. 2008;9:907–15. doi: 10.1038/embor.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol Cell Biol. 2003;23:3287–304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu K, Luo Y, Lin FT, Lin WC. TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 2004;18:673–86. doi: 10.1101/gad.1180204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu K, Paik JC, Wang B, Lin FT, Lin WC. Regulation of TopBP1 oligomerization by Akt/PKB for cell survival. EMBO J. 2006;25:4795–807. doi: 10.1038/sj.emboj.7601355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo R, Chen J, Zhu F, Biswas AK, Berton TR, Mitchell DL, et al. E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J Biol Chem. 2010;285:19308–15. doi: 10.1074/jbc.M110.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Zhu F, Weaks RL, Biswas AK, Guo R, Li Y, et al. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle. 2011;10:1287–94. doi: 10.4161/cc.10.8.15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo R, Chen J, Mitchell DL, Johnson DG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res. 2011;39:1390–7. doi: 10.1093/nar/gkq983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–45. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]