Abstract
Surface tissues of the body such as the skin and intestinal tract are in direct contact with the external environment and are thus continuously exposed to large numbers of microorganisms. To cope with the substantial microbial exposure, epithelial surfaces produce a diverse arsenal of antimicrobial proteins that directly kill or inhibit the growth of microorganisms. In this Review, we highlight new advances in our understanding of how epithelial antimicrobial proteins protect against pathogens and contribute to microbiota–host homeostasis at the skin and gut mucosae. Further, we discuss recent insights into the regulatory mechanisms that control antimicrobial protein expression. Finally, we consider how impaired antimicrobial protein expression and function can contribute to disease.
The surfaces of the mammalian intestine, skin, respiratory tract and reproductive tract interface directly with the external environment. As a result, these epithelial tissues continuously encounter bacteria, fungi, viruses and parasites that could act as pathogens. In addition, each of these tissues is associated with indigenous communities of commensal microorganisms that comprise complex microbial ecosystems. The epithelium separating these microorganisms from mammalian internal tissues is generally only one or a few cell layers thick and represents an enormous surface area. In humans, the intestinal epithelium encompasses ~200 m2 of surface area1, with the skin contributing an additional ~2 m2 surface2. Thus, surface tissues are faced with the enormous challenge of defending a large surface area to maintain homeostasis with abundant communities of commensal microorganisms and to prevent pathogen invasion.
Epithelial antimicrobial proteins (AMPs) have an essential role in allowing epithelial surfaces to cope with these microbial challenges. These natural antibiotics are evolutionarily ancient innate immune effectors that are produced by almost all plants and animals3. Mammalian AMPs are members of a diverse array of protein families, all of which function to rapidly kill or inactivate microorganisms4. The epithelial cells lining the gut, skin and respiratory tract produce a rich arsenal of AMPs, probably reflecting the complexity of the microbial challenges faced by these tissues and the continuous threat of microbial invasion at these sites.
In this Review, we summarize recent advances in our understanding of how AMPs function to protect mammalian body surfaces. We analyse recent insights into the regulatory networks that control AMP expression and function at these sites. Further, we discuss how AMPs function to limit pathogen colonization, shape the composition of indigenous microbial communities, and promote the physical segregation of microbiota and host. Finally, we explore how impaired antimicrobial defences can contribute to disease. Although we focus this Review on the AMPs produced by the mammalian intestine and the keratinized areas of the skin (hereafter referred to as ‘skin’, but excluding mucosal skin epithelia), we aim to highlight general principles that are applicable to the understanding of AMPs of other surface tissues such as the respiratory and reproductive tracts.
Antimicrobial proteins
The AMPs of the gut and skin encompass representatives of several distinct protein families. These include defensins, cathelicidins, C-type lectins (such as the regenerating islet-derived protein (REG) family), ribonucleases (RNases, such as angiogenin 4 (ANG4)) and S100 proteins (such as calprotectin (also known as S100A8–S100A9) and psoriasin (also known as S100A7)). We do not discuss each of these exhaustively here, as this topic has been well covered in previous reviews4,5. Further, we have summarized the major characteristics of some of the main AMP families in gut and skin in TABLE 1. Other specialized epithelial surfaces, such as the oral and nasal mucosae, eye, lung and reproductive tract, are not discussed, but it is important to recognize that each interface has many characteristic AMPs that are uniquely necessary for that environment. Here, we briefly introduce a few key AMPs (mainly defensins, C-type lectins and cathelicidins) that are particularly relevant to our discussion below and serve to illustrate the general principle that the epithelial interface is the first line of defence of the immune system. As the antimicrobial action of cathelicidins and defensins has been widely confirmed (reviewed in REFS 3,5), and the physiological relevance of these large families of AMPs has been validated in several animal models (reviewed in REFS 3,5), our discussion of cutaneous AMPs will focus on the cathelicidins and β-defensins. Other AMPs, such as psoriasin and dermicidin, have a more limited spectrum of antimicrobial potency under in vivo conditions6,7, and so they are not emphasized here.
Table 1.
Major antimicrobial protein families in skin and intestine
| Family | Representative proteins | Mechanism of action | Tissue sites of expression | Cellular sites of expression | Target organisms | Refs |
|---|---|---|---|---|---|---|
| α-defensins (cryptdins in mice) | DEFA5, DEFA6 (in humans); cryptdins 1–16 (in mice) | Membrane disruption | Small intestine | Paneth cells, neutrophils, macrophages | Gram-positive bacteria, Gram-negative bacteria, fungi, viruses, protozoa | 8 |
| β-defensins | BD1, BD2, BD3 | Membrane disruption; lipid II binding (BD3) | Large intestine, skin, respiratory tract | Enterocytes, keratinocytes, respiratory tract epithelial cells | Gram-positive bacteria, Gram-negative bacteria, fungi, viruses, protozoa | 86,133 |
| Calprotectin (S100A8–S100A9) | NA | Metal chelation | Abscesses | Neutrophils | Staphylococcus aureus | 39 |
| Cathelicidins | LL37 (in humans); CRAMP (in mice) | Membrane disruption | Large intestine, skin, lung, urinary tract | Neutrophils, mast cells, epithelial cells | Gram-positive bacteria, Gram-negative bacteria, viruses, fungi | 36 |
| C-type lectins | HIP/PAP (in humans); REG3γ (in mice) | Peptidoglycan recognition; killing mechanism unknown | Small intestine | Epithelial cells (Paneth cells, enterocytes) | Gram-positive bacteria | 11,12,23 |
| Galectins | GAL4, GAL8 | Unknown | Intestine | Broad expression, including by epithelial cells | Bacteria bearing blood group antigens | 134 |
| Lipocalin | Lipocalin 2 | Sequestration of iron-laden siderophores | Broad expression, including intestine and lung | Macrophages, epithelial cells | Escherichia coli | 135,136 |
| Lysozyme | NA | Enzymatic attack on bacterial cell wall peptidoglycan | Intestine, eye, and more; secretions, including tears, saliva | Intestinal Paneth cells | Gram-positive bacteria; some activity against Gram-negative bacteria | 137 |
| Peptidoglycan recognition proteins | PGLYRP1–4 in mammals | Activation of bacterial two-component systems; PGLYRP2 is an amidase that targets peptidoglycan | Liver, intestine, skin, mammary gland | Epithelial cells | Gram-positive and Gram-negative bacteria | 138 |
| Phospholipase A2 | NA | Hydrolysis of bacterial membrane phospholipids | Intestine; secretions, including tears, inflammatory fluids | Paneth cells, macrophages | Gram-positive bacteria | 139 |
| Psoriasin (S100A7) | NA | Unknown | Skin | Keratinocytes | Escherichia coli | 12 |
| RNases | ANG4, RNase7 | Unknown | Skin, intestine | Epithelial cells | Gram-positive and Gram-negative bacteria | 26,38 |
ANG4, angiogenin 4; BD, β-defensin; CRAMP, cathelin-related antimicrobial peptide; DEFA, α-defensin; GAL, galectin; HIP/PAP, hepatointestinal pancreatic/pancreatitis-associated protein; NA, not applicable; PGLYRP, peptidoglycan recognition protein; REG3γ, regenerating islet-derived protein 3γ; RNase, ribonuclease.
The mammalian gut epithelium produces a diverse collection of AMPs, reflecting the complex microbial challenges confronted by the intestine. Among the most abundant, diverse and highly expressed AMP families in the gut are the α-defensins. These include α-defensin 5 (DEFA5; also known as HD5) and DEFA6 (also known as HD6) in humans, and cryptdins 1–16 (also known as DEFA1–16) in mice. The α-defensins are small peptides (~2–3 kDa) with a conserved three-dimensional structure that is characterized by an amphipathic arrangement of cationic and hydrophobic residues3, resulting in a positively charged surface that is spatially separated from a neighbouring hydrophobic region. This unique arrangement promotes attraction of α-defensins to the negatively charged cell surface and insertion into the lipid-rich membrane. In general, α-defensins have a broad spectrum of activity against both Gram-positive and Gram-negative bacteria, and in some cases are active against fungi, viruses and protozoa; however, particular defensin species have marked differences in their activity spectrum and expression patterns8.
Another key AMP family of the mammalian small intestine is the REG family of C-type lectins. REG3 lectins (such as REG3γ in mice and hepatointestinal pancreatic/pancreatitis-associated protein (HIP/PAP; also known as REG3α) in humans) are expressed in the small intestines of mice and humans9–11, and are produced in the large intestine during pathogen infections or inflammatory conditions10. These ~15 kDa proteins have canonical C-type lectin domains that bind to the glycan chains of peptidoglycan, which is an essential component of the bacterial cell wall11,12. In contrast to defensins, the bactericidal activities of REG3γ and HIP/PAP are selective for Gram-positive bacteria11. This selective activity is consistent with the accessibility of peptidoglycan on the cell surface of Gram-positive bacteria but not Gram-negative bacteria. The closely related lectin REG3β is usually co-expressed with REG3γ in mice. Although REG3β has been shown to bind peptidoglycan12, a direct demonstration of antimicrobial activity for REG3β is still lacking.
The mammalian epidermis also deploys a diverse and potent arsenal of AMPs. Key AMPs of the skin's repertoire are the cathelicidins13 and β-defensins14. The single cathelicidin gene (cathelicidin antimicrobial peptide (CAMP) in humans) encodes a precursor protein (CAP18; also known as CAMP)15. This protein can be alternatively cleaved to generate several active AMPs, including the 37-amino-acid peptide LL37 (REF. 16) and the murine peptide cathelin-related antimicrobial peptide (CRAMP)17. Cathelicidin precursor protein and mature peptide are abundantly expressed by resident mast cells of normal skin18 and expressed at lower levels in sweat and by resting keratinocytes19. Inflamed skin greatly increases the abundance of cathelicidin through increased expression of CAP18 by keratinocytes and increased local deposition by recruited neutrophils20,21. The β-defensins differ from the α-defensins in terms of the connectivity of the intramolecular disulphide bonds that establish the β-sheet structure of these peptides. In contrast to cathelicidin, approximately 90 β-defensin genes have been identified in mice and humans, with differences in activity and expression. Both cathelicidins and β-defensins are antimicrobial against a diverse range of skin pathogens, including Gram-negative and Gram-positive bacteria, fungi, viruses and parasites5.
Sites of expression
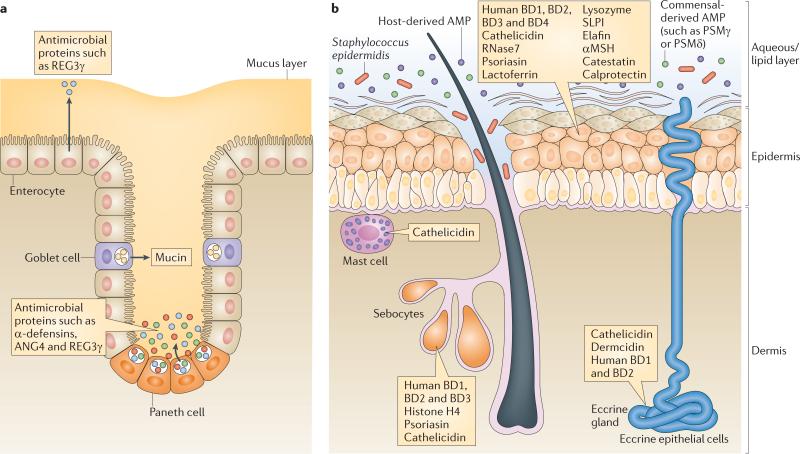
Epithelial cells produce the majority of AMPs in body surface tissues under steady-state conditions, although infiltrating immune cells can also contribute to AMP production during inflammation. Several distinct epithelial cell lineages comprise the intestinal epithelial surface, each of which expresses a distinct group of AMPs (FIG. 1a). The enterocyte is the most abundant epithelial cell lineage of both the small intestine and large intestine (colon). In the small intestine, enterocytes produce REG3γ and REG3β22,23, whereas colon enterocytes express β-defensins and cathelicidins24,25. Paneth cells are located at the base of crypts of Lieberkühn and are unique to the small intestine. Many AMPs are expressed abundantly by Paneth cells, including α-defensins8 and ANG4, an RNase26. Goblet cells constitute a third epithelial cell lineage that is present in both the small and large intestine. A major function of goblet cells is to secrete mucin glyco proteins that assemble to form a thick gel-like mucus layer that overlies the epithelium27 and functions in part to concentrate secreted AMPs at or near the epithelial surface28.
Figure 1. Epithelial barriers of the intestine and skin.
a The intestinal epithelium comprises several cell lineages. Enterocytes constitute the most abundant epithelial cell type, and secrete several antimicrobial proteins (AMPs) such as regenerating islet-derived protein 3γ (REG3γ). Paneth cells are unique to the small intestine and secrete abundant quantities of AMPs, such as α-defensins. Finally, goblet cells secrete mucin glycoproteins that assemble to form a thick mucus layer overlying the epithelium. The mucus layer seems to have a crucial role in concentrating secreted AMPs near the epithelial surface. b The epithelial barrier of the skin includes keratinocytes at the surface and the hair unit, and specialized secretory organs such as sebocytes and eccrine glands. Many diverse AMPs, including cathelicidins and defensins, are produced by these cells under steady-state and/or inflammatory conditions. The aqueous and lipid components of the skin surface combine with AMPs produced by microorganisms to enhance the barrier/protective function. The aqueous/lipid layer may serve a function that is similar to that of intestinal mucus by trapping AMPs at the epithelial surface. Resident bone marrow-derived cells in the dermis, such as mast cells, provide essential additional AMPs after skin injury or in early stages of infection. αMSH, α-melanocyte-stimulating hormone; ANG4, angiogenin 4; BD, β-defensin; PSM, phenol-soluble modulin; RNase7, ribonuclease 7; SLPI, secretory leukocyte protease inhibitor (also known as ALP).
In the skin, the keratinocyte is the most abundant cell type in the epidermis, and several specialized keratinocyte cell types populate skin appendages such as hair follicles (FIG. 1b). Follicular keratinocytes constitutively produce cathelicidin20,21,29–31 and β-defensins at a higher level than other keratinocytes, but all keratinocytes produce various AMPs to defend the skin barrier. Augmenting the action of keratinocytes, secretory cells of the skin, such as those of the sweat, apocrine and sebaceous glands, each contribute additional AMPs and antimicrobial lipids to the skin surface19. Below the surface, and adding to the complexity of the cutaneous barrier, bone marrow-derived resident mast cells in the dermis also have an important role in cutaneous defence. Mast cells normally occupy positions around blood vessels and store large amounts of cathelicidin in preformed granules. This location places the AMPs derived from mast cells in an ideal position to resist infections after skin injury and inoculation with either bacterial or viral pathogens32.
Mechanisms of action
Many AMPs target essential cell wall or cell membrane structures, which limits the ability of microorganisms to evolve resistance (BOX 1). Several AMPs are enzymes that kill bacteria by carrying out an enzymatic attack on cell wall structures. Lysozyme and phospholipase A2 (PLA2), which are both highly expressed by Paneth cells, function through such a mechanism. Lysozyme hydrolyses the glycosidic linkages between the N-acetylglucosamine and N-acetyl-muramic acid that constitute the carbohydrate backbone of cell wall peptidoglycan33, whereas secretory PLA2 (sPLA2) kills bacteria by hydrolysing bacterial membrane phospholipids34.
Many AMPs kill bacteria through non-enzymatic disruption of the bacterial membrane. Defensins comprise a major family of membrane-disrupting peptides in vertebrates. The clusters of cationic residues on most defensins interact with the bacterial membrane surface through electrostatic interactions with negatively charged phospholipid groups. This interaction is followed by the formation of defensin pores in the bacterial membrane that disrupt membrane integrity and promote lysis of the targeted microorganism35. Cathelicidins also typically kill microorganisms through membrane disruption. Like the defensins, cathelicidins are usually cationic, α-helical peptides that bind to bacterial membranes through electrostatic interactions, followed by membrane insertion and disruption36. Interestingly, in some mammalian species such as pig and cow, multiple cathelicidin genes are present, and some of these encode AMPs that function through mechanisms that do not involve direct membrane disruption37.
The antibacterial mechanisms of several AMPs remain a mystery. ANG4 is a bactericidal RNase that is secreted by Paneth cells26. Although ANG4 has weak RNase activity26, there is no evidence that this is required for its bactericidal function, and its mechanism of action is currently unknown. The mechanism of action of the REG C-type lectins also remains unclear. It is known that REG3γ recognizes its Gram-positive bacterial targets by binding to peptidoglycan11,12 and that the bactericidal action of REG3γ is accompanied by disruption of cell wall integrity11. It remains to be established whether REG3γ functions through membrane disruption, enzymatic attack or another mechanism. In the skin, there are several proteins with reported antimicrobial activity that might not function through membrane disruption. These include RNase7 (REF. 38), calprotectin39, psoriasin6 and dermcidin7. Calprotectin functions by metal chelation, thus regulating the availability of essential trace elements such as Zn2+ and Mn2+, and augments other host defence mechanisms for microbial killing40. In general, the other proteins have very low direct antimicrobial potency but are abundantly expressed in epithelia, thus arguing for a substantial role in host defence. Taken together, these findings suggest that different AMP families use distinct molecular mechanisms to kill microorganisms. The use of diverse killing strategies is probably important for limiting the evolution of microbial resistance to multiple AMPs.
Immunomodulation by antimicrobial proteins
Increasing evidence indicates that some AMPs can protect tissue surfaces by alternative mechanisms that are unrelated to their capacity to directly kill microorganisms. Thus, the capacity of microorganisms to develop antimicrobial resistance (BOX 1) might be irrelevant to the overall physiological effectiveness of several types of AMP. Cathelicidin and defensin peptides are major examples of this alternative activity. These peptides can function as potent immune regulators by signalling through chemokine receptors and by inhibiting or enhancing Toll-like receptor (TLR) signalling.
Chemotactic activity
The cathelicidins and α- and β-defensins have each been shown to have chemotactic activity and the capacity to recruit leukocytes by direct or indirect mechanisms. These actions can directly modify the inflammatory response. For example, the human cathelicidin peptide LL37 attracts neutrophils, monocytes and T cells, and in some experimental systems, this chemotactic activity is mediated by the G protein-coupled formyl peptide receptor-like 1 (FPRL1; also known as formyl peptide receptor 2)41. The mouse cathelicidin CRAMP, despite having a very different primary amino acid sequence to LL37, is also chemotactic for human cells and mouse cells in a manner dependent on FPRL1 or FPRL2 (also known as FPR3)42. Human α-defensins (DEFA1 (also known as neutrophil defensin 1) and neutrophil defensin 2) and β-defensins (BD3 (also known as DEFB103) and BD4 (also known as DEFB104)) have been reported to be chemotactic for monocytes and macrophages43, and BD2 (also known as DEFB4A) and LL37 attract mast cells44. The chemotactic activities of the different groups of AMPs are distinct from each other. For example, human α-defensins selectively induce the migration of human naive CD4+CD45RA+ and CD8+ T cells, but not CD4+CD45RO+ memory T cells45. By contrast, β-defensins are chemotactic for immature dendritic cells and CD4+CD45RO+ memory T cells. The chemotactic effect of human defensins is inhibited by antibodies specific for CC-chemokine receptor 6 (CCR6)46. An additional mode of action for the chemokine-like function of AMPs is through trans-activation of cell surface growth factor receptors. LL37 has been shown to activate the epidermal growth factor receptor on keratinocytes and induce migration by this mechanism47. Studies in Cramp–/– mice have confirmed the physiological relevance of the chemotactic effects of cathelicidins despite their low potency in vitro48.
Modulation of TLR responsiveness
An exciting new aspect of our understanding of AMP function has come from studies of their interactions with TLRs. TLRs are membrane-bound pattern recognition receptors (PRRs) that are activated by conserved microbial molecular patterns, such as lipopolysaccharide (LPS) or flagellin, triggering signalling cascades that activate nuclear factor-κB (NF-κB), which in turn drives transcription of pro-inflammatory cytokines. The consequences of AMP interactions with TLRs are complex, with potential for either inhibiting or activating inflammatory events. Cathelicidins have been extensively studied in this regard and seem to directly interact with TLR ligands such as endotoxin or nucleic acids, including self-DNA. This modifies cellular responsiveness to these ligands by altering membrane microdomain function or the internalization of the TLR ligand49,50. For example, the presence of active cathelicidin peptide (LL37) influences T helper 17 (TH17) cell maturation by interacting with DNA to activate TLR9, which in turn leads to increased production of type I interferons50. The consequences of these interactions imply that AMP expression during infection could kill pathogens and neutralize excessive inflammatory responses at the same time. However, inappropriate expression of some AMPs could contribute to dysregulation of inflammation and the development of autoimmune disorders, in part by modifying responsiveness to self-DNA. The pathophysiologies of the human diseases rosacea and psoriasis have each been associated with the interaction of AMPs with PRRs and are discussed later in this Review.
Together, these examples underscore the fact that AMP function is complex, extending beyond direct antibacterial activity to encompass general effects on downstream immune responses.
Antimicrobial protein regulation
The expression, secretion and activity of most AMPs are tightly controlled. This is necessitated in part by the toxic effects of many of these proteins on mammalian cell membranes. In addition, the chemokine-like activities and immune-modulating effects of many AMPs probably require tight control of expression in order to avoid triggering unnecessary inflammatory responses. In the following sections, we discuss the multifaceted ways in which AMP expression and activity are regulated at body surfaces.
Transcriptional regulation by bacteria
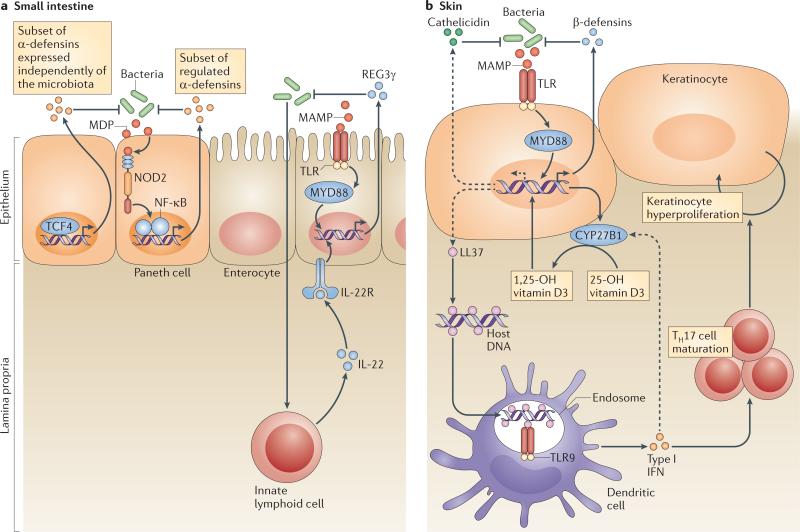
Germ-free animals are an indispensable experimental tool for determining which host functions are genetically encoded and which require interactions with microorganisms51. Studies of germ-free mice have revealed that some intestinal AMPs are expressed independently of the microbiota, whereas others require bacterial signals for their expression (FIG. 2a). For example, expression of most intestinal α-defensins requires the WNT pathway transcription factor TCF4 (REF. 52) but is independent of the microbiota53. Similarly, expression of lysozyme, sPLA2 and certain members of the β-defensin family does not require microbial signals24,26,54.
Figure 2. Regulatory mechanisms that shape antimicrobial protein expression and function.
a In the small intestine, the transcriptional control of α-defensin expression depends crucially on transcription factor 4 (TCF4)52. The pattern recognition receptor nucleotide oligomerization domain 2 (NOD2) also controls the expression and/or secretion of antimicrobial activities in the small intestinal crypt65. Regenerating islet-derived protein 3γ (REG3γ) mRNA expression in enterocytes and Paneth cells is controlled by microorganism-associated molecular patterns through Toll-like receptors (TLRs) and is dependent on the TLR signalling adaptor myeloid differentiation primary response protein 88 (MYD88)22,23. REG3γ mRNA expression also requires interleukin-22 (IL-22)-mediated signals from innate lymphoid cells60. b In the skin, TLR2 activation can directly induce β-defensin mRNA expression132 but indirectly influences cathelicidin expression. The activation of MYD88 by TLR2 results in a direct transcriptional increase in cytochrome p450, 27B1 (CYP27B1) and β-defensin expression. The increased expression of CYP27B1 hydroxylates 25-OH vitamin D3 to 1,25-OH vitamin D3, and this then induces cathelicidin mRNA expression76,77. β-defensins and cathelicidin released at the skin surface can function against microorganisms. Below the surface, the presence of cathelicidin peptide in the form of LL37 interacts directly with host DNA and can activate TLR9. TLR9-mediated activation of dendritic cells results in production of type I interferons (IFNα/β) and influences T helper 17 (TH17) cell maturation and the production of IL-17, which can drive keratinocyte hyperproliferation50. IL-22R, IL-22 receptor; MAMP, microbe-associated molecular pattern; MDP, muramyl dipeptide; NF-κB, nuclear factor-κB.
Other intestinal AMPs require microbial cues for their full expression. Members of the cryptdin-related sequence family of peptides are related to the α-defensins and have increased levels of expression in conventionally raised mice compared with germ-free mice53. Similarly, key members of the human β-defensin family, including BD2, are expressed under the control of bacterial signals24. For example, bacterial flagellin has been shown to be highly relevant for the induction of BD2 expression by keratinocytes55. Finally, the expression of both ANG4 and REG3γ is essentially absent in germ-free mice and is upregulated on colonization with a conventional microbiota11,26.
Host PRRs control the expression of some of these bacterially regulated AMPs. For example, stimulation of TLRs is required for REG3γ mRNA expression by intestinal epithelial cells. Studies in mice lacking myeloid differentiation primary response protein 88 (MYD88), an adaptor molecule that is common to several TLRs, have revealed that REG3γ and REG3β are expressed under the control of TLRs in vivo22,23,56,57. Further, the MYD88 dependence is intrinsic to epithelial cells, suggesting that epithelial cells, including enterocytes and Paneth cells, directly sense bacteria through TLRs and upregulate expression of REG3γ and REG3β in response22,23,56. Expression of these AMPs seems to be triggered by any of several TLRs, as mice deficient in individual TLRs do not have defects in REG3γ or REG3β expression22. This is consistent with the fact that both LPS22,56 and flagellin58 (which bind TLR4 and TLR5, respectively) are sufficient to trigger REG3γ expression. Microbial cues can also induce AMPs in a non-PRR-dependent manner; for example, production of the short-chain fatty acid butyrate in the colon through microbial fermentation of dietary fibres has been shown to be an important regulator of AMP production59.
Intestinal epithelial cell expression of REG3γ also requires signals from at least one immune cell lineage. Innate lymphoid cells (ILCs) reside in the lamina propria and phenotypically resemble natural killer cells60. ILCs produce large quantities of the cytokine interleukin-22 (IL-22), which binds to IL-22 receptors on epithelial cells to modulate epithelial cell function61. ILCs from germ-free mice produce low levels of IL-22 (REF. 62), suggesting that intestinal bacteria drive IL-22 expression by these cells. Interestingly, ILC-derived IL-22 seems to be required for epithelial cell expression of REG3γ mRNA62. Thus, REG3γ expression is dependent on both epithelial cell-intrinsic TLR signalling through MYD88 and IL-22 produced by ILCs. It seems possible to reconcile these disparate observations by proposing that IL-22 functions as an environmental cue that allows epithelial cells to become competent to express REG3γ. Epithelial cells must then receive an additional direct bacterial signal through TLRs in order to express REG3γ. Further studies will be required to unravel this complex regulatory network.
The expression of other intestinal AMPs is regulated by nucleotide oligomerization domain 2 (NOD2) (FIG. 2a). NOD2 is an intracellular PRR that is expressed by Paneth cells and macrophages in the small intestine63. NOD2 functions in the intracellular recognition of muramyl dipeptide (MDP) — a peptidoglycan motif that is common to Gram-positive and Gram-negative bacteria — triggering signalling cascades that activate the transcription factor NF-κB64. MDP has been shown to stimulate the bactericidal activity of intestinal crypts harbouring Paneth cells65. Also, Nod2–/– mice have pronounced alterations in the composition of their small intestinal microbiota65, as well as increased susceptibility to oral challenge with the gastrointestinal pathogen Listeria monocytogenes66. These results indicate that NOD2-stimulated antimicrobial defences are important for shaping the microbiota and protecting the epithelial barrier from pathogenic invasion.
Developmental regulation
All mammals are sterile during prenatal life and acquire indigenous microbial communities immediately after birth. Profound shifts in the composition of intestinal microbial communities occur during early postnatal life, particularly during weaning when young mammals transition from milk to solid food. Over time, the gut ecosystem stabilizes and by adulthood consists of an established climax community dominated by obligate anaerobes67. Several intestinal AMPs have developmentally regulated expression during neonatal and early postnatal life, suggesting that they might be crucial for maintaining immune homeostasis during key developmental transitions.
Both ANG4 and REG3γ are strongly induced in the small intestine during early postnatal life. In conventionally raised mice, the level of ANG4 expression increases approximately 20-fold during weaning (approximately day 17–21 in mice) and remains at adult levels thereafter26. The level of expression of REG3γ in mice increases by a remarkable 3,000-fold during the same period11. These findings suggest that ANG4 and REG3γ function in part to maintain mucosal homeostasis during weaning in the face of the associated change in microbial ecology and the withdrawal of the passive immune protection afforded by the mother's milk. By contrast, cathelicidin expression undergoes an inverse switch, as it is abundantly expressed during early neonatal life but begins declining just before weaning68. By adulthood, cathelicidin expression is extinguished in the small intestine but remains high in the colon. The expression of cathelicidin in the colon provides resistance to bacterial pathogens in mice69, but the physiological relevance of decreased cathelicidin expression with maturation of the small intestine is unclear.
Regulation during epithelial damage
A major function of AMPs is to respond rapidly to epithelial dis ruption and establish a temporary protective shield against infection. Early observations of inducible AMP expression with injury or infection were made in insects and amphibians70,71, and it was later recognized that a similar process takes place early in mammalian wound repair or infection13,21. The physical barrier of the epidermis and the antimicrobial barrier are intimately linked, with each influencing the function of the other72. Human BD3 can be induced by growth factors that are also important for wound repair. For example, after skin wounding, keratinocyte expression of this AMP is increased by activation of the epidermal growth factor receptor73. A particularly surprising observation came with the recognition that the human cathelicidin gene CAMP is under transcriptional control of a vitamin D response element (VDRE)74,75. Following skin injury or infection, 25-OH vitamin D3 is hydroxylated by the enzyme cytochrome p450, 27B1 (CYP27B1) to 1,25-OH vitamin D3, and this is stimulated locally by activation of TLR2 or local cytokines such as tumour necrosis factor (TNF) or type I interferons76,77 (FIG. 2b). This local enzymatic event enables rapid induction of CAMP expression through binding of 1,25-OH vitamin D3 to the VDRE. These observations suggest that AMP expression could be influenced by dietary vitamin D78 or vitamin D generated by exposure of the skin to sunlight79. This implies that the nutritional environment is probably a source of important signals that control AMP expression.
Post-translational regulation
A major problem faced by host cells that secrete membrane-toxic AMPs is the potential detrimental effects of these proteins on mammalian cell membranes80. As a result, the activities of many AMPs must be tightly regulated during storage in membrane-bound secretory granules. α-defensins are stored in Paneth cell granules as inactive pro-peptides. Mouse α-defensins are processed at their amino termini by matrix metalloproteinase 7 (MMP7) to produce mature bactericidally active peptides81. In humans, trypsin cleaves α-defensins to their mature forms82. REG3γ also requires N-terminal proteolytic processing to yield a bactericidally active protein. In both mice and humans, this processing is carried out by trypsin following secretion into the gut lumen83. The requirement for an inhibitory N-terminal peptide during secretory granule storage indicates that REG3γ might have membrane-disrupting activity, providing a potential clue about the mechanism of its bactericidal activity.
Different processing mechanisms generate the active forms of other AMPs. Similarly to defensins and REG3γ, cathelicidins are synthesized as pro-peptides. However, serine proteases are responsible for the processing of cathelicidins. In neutrophils, the pro-peptide is removed by protease 3 (REF. 84), whereas processing is carried out by kallikreins in keratinocytes85. Similarly, β-defensins are expressed as pro-peptides, but the processing mechanism remains to be established86.
The distinctive reducing environment of the intestinal lumen might provide another mechanism of post-translational regulation that protects host cells during storage of AMPs. Under oxidizing conditions, such as those present inside host cells, the human AMP BD1 has weak antimicrobial activity. However, under reducing conditions that mimic those encountered in the intestinal lumen, human BD1 undergoes marked structural changes, including reduction of its disulphide bridges and alterations in its secondary structure, that result in the unmasking of a potent antimicrobial activity87.
Regulated secretion
The secretion of AMPs into the gut lumen is also controlled by bacterial signals. As noted above, Paneth cells produce most of the AMPs in the small intestine, including α-defensins, ANG4 and lysozyme. Paneth cells secrete their granule contents in response to exposure to live bacteria or to bacterial molecules such as LPS88. Thus, AMP release is precisely regulated in response to potential bacterial threats, although the mechanisms of bacterial sensing that control AMP secretion by Paneth cells are not yet clear. In skin, the eccrine, apocrine and sebaceous cells carry out a similar role by releasing AMPs onto the skin surface. The sensors that recognize bacteria and mediate this regulated secretion also remain unknown.
Together, these findings show that a complex network of developmental, microbial and nutritional signals controls AMP expression and secretion. It will be important to sustain efforts to unravel these complex regulatory networks, which may allow the development of new therapeutic strategies aimed at enhancing endogenous AMP production.
Antimicrobial protein function in vivo
It has long been accepted that the diverse AMP arsenals of the intestine and skin evolved to manage the large numbers of microorganisms that are encountered at these body surfaces. Recent findings in genetically engineered mouse models have offered new insights into how epithelial AMPs function in vivo, showing that these proteins not only protect against pathogen colonization but also determine microbiota composition and limit access of the microbiota to host tissues (FIG. 3).
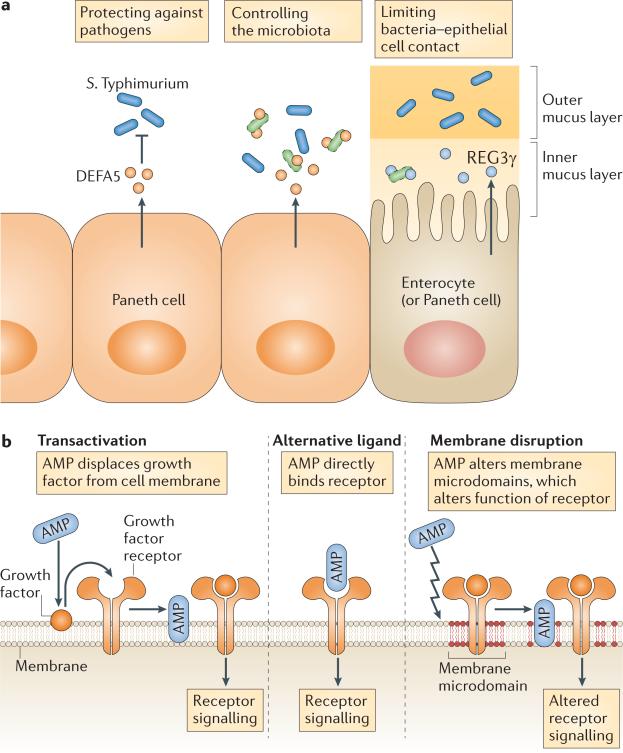
Figure 3. Functions of antimicrobial proteins in the defence of body surfaces.
a Forced expression of a human α-defensin 5 (DEFA5) transgene in Paneth cells limits colonization by Salmonella enterica subsp. enterica serovar Typhimurium89 and controls the composition of the microbiota in the small intestine96. Regenerating islet-derived protein 3γ (REG3γ) confines bacteria to the outer mucus layer, thus limiting bacterial contact with the small intestinal epithelial surface23. b Cathelicidins carry out additional immunomodulatory functions in skin defence by acting on cell membrane receptors. Depending on the cell type and condition, evidence for three potential mechanisms of action exists: transactivation by antimicrobial proteins (AMPs) releases membrane-bound growth factors; AMPs bind and directly activate receptors; or AMPs activate or inactivate receptors by disrupting membrane microdomains.
Protection against pathogens
One of the first studies to reveal the importance of intestinal AMPs in pathogen protection in vivo involved the analysis of mice deficient in MMP7, which is required for the generation of bactericidally active α-defensins in mice81. In vivo studies of Mmp7–/– mice showed increased susceptibility to oral challenge with the intestinal pathogen Salmonella enterica subsp. enterica serovar Typhimurium, revealing a crucial role of MMP7 in regulating immunity to enteric bacteria81. A more direct demonstration of the in vivo function of α-defensins was achieved with the development of a transgenic mouse overexpressing human DEFA5 in Paneth cells. DEFA5-expressing mice had greater resistance to oral challenge with S. Typhimurium than wild-type mice, pointing to an essential role for α-defensins in limiting colonization by gastrointestinal pathogens89. The development of a cathelicidin-deficient mouse was the first direct demonstration of the importance of AMPs in defending against skin infections, and this has now been confirmed with several diverse organisms, including group A streptococcus, vaccinia virus and Leishmania spp.90–92. The importance of cathelicidin for defending against infection has also been established in other organ systems, such as urinary tract infection by Escherichia coli93 and Pseudomonas aeruginosa keratitis of the eye94. Finally, antibody-mediated inactivation of REG3γ showed that this antimicrobial lectin is important for controlling colonization of the intestinal tract by Gram-positive opportunistic pathogens, such as Listeria monocytogenes56 and vancomycin-resistant Enterococcus faecalis95.
Controlling the microbiota
A recent elegant study96 showed that α-defensins regulate the composition of the intestinal bacterial community (BOX 2). The study used 16S ribosomal RNA gene sequencing to characterize the intestinal bacterial communities in Mmp7–/– and DEFA5-transgenic mice. Although no differences in the total numbers of colonizing bacteria were observed, there were substantial α-defensin-dependent changes in the composition of the microbial communities compared with wild-type mice. Further, the defensin-deficient Mmp7–/– mice and the defensin-complemented DEFA5-transgenic mice had reciprocal differences in community composition. These changes included alterations in the proportions of Firmicutes, the major Gram-positive phylum, and Bacteroidetes, the major Gram-negative phylum, of the mouse intestine. The DEFA5-transgenic mice also had a loss of segmented filamentous bacteria (SFB) relative to wild-type mice. Consistent with the fact that SFB increase the frequencies of intestinal TH17 cells97, the DEFA5-transgenic mice had lower frequencies of lamina propria TH17 cells compared with wild-type mice. This indicates that α-defensins have an important role in shaping intestinal microbiota composition and thus controlling the level of immune stimulation.
Limiting bacterial–epithelial cell contact
A key mechanism by which the mammalian intestine maintains homeostasis with its associated bacterial communities is to minimize contact between the bacteria and the intestinal epithelium. This is accomplished in part by enhancing the physical barrier through the production and assembly of a thick mucus layer at the epithelial surface. Visualization of spatial relationships between bacteria and the intestinal surface shows that the inner mucus layer remains relatively free of bacteria, whereas the outer mucus layer retains large numbers of bacteria23,27. Biochemical analysis of AMP localization has shown that most AMP activity is confined to the mucus layer and is essentially absent from the luminal content28. Thus, it seems likely that, in addition to functioning as a physical barrier, the mucus layer limits bacterial penetration by forming a diffusion barrier that concentrates AMPs near the epithelial cell surface.
Recent studies have shown that REG3γ interacts with the mucus layer to help maintain a relatively bacteria-free zone adjacent to the intestinal epithelial cell surface. Consistent with the fact that the bactericidal activity of REG3γ specifically targets Gram-positive bacteria, Reg3g–/– mice had increased colonization of the small intestinal epithelial surface by Gram-positive bacterial groups, including SFB23. Interestingly, these differences did not extend to the luminal bacterial communities, which were similar between Reg3g–/– mice and their wild-type littermates. This suggests that the antibacterial effects of REG3γ are confined to the inner mucus layer and do not extend into the lumen of the intestine. The mucosal niche-specific activity of REG3γ could be owing to restricted diffusion of REG3γ through the mucus barrier, the result of specific binding interactions between REG3γ and mucus glycoproteins, or because of a requirement for environmental conditions for REG3γ activity that are specific to the mucosal surface niche.
Antimicrobial proteins of the microbiota
An important component of the surface antimicrobial shield of the skin is produced by the resident commensal microorganisms themselves. Gram-positive bacteria such as Lactococcus, Streptococcus and Streptomyces spp. produce factors, known as bacteriocins, that inhibit the growth of other bacterial strains and species that could compete for nutrients and other resources. Staphylococcus epidermidis, the dominant commensal bacterium found in the skin microflora, produces several AMPs. For example, epidermin, pep5 and epilancin K7 are structurally unique as they contain the thioether amino acids lanthionine and/or methyllanthionine, and as such are also known as lantibiotics. S. epidermidis also produces peptides known as phenol-soluble modulin-γ (PSMγ) and PSMδ that function as AMPs98. These peptides cause membrane leakage in target bacteria, which indicates that they function in a manner similar to that of host-derived AMPs such as defensins and cathelicidins. PSMs are functional in vivo, as nanomolar concentrations decreased the survival of group A streptococcus on normal human skin but did not affect the survival of S. epidermidis from which the peptide was derived98. Another important example of the protective action of S. epidermidis in vivo was observed on the surface of the nasal cavity. In this study, nasal colonization by Staphylococcus aureus was inhibited in individuals who were colonized with specific strains of S. epidermidis that produced a serine protease with the capacity to destroy biofilms formed by S. aureus99. In addition, a thiolactone-containing peptide produced by S. epidermidis blocks the S. aureus agr quorum sensing system that controls the production of various virulence factors100. Thus, the selective activity of AMPs produced by commensal organisms might be an important part of a normal host defence strategy against pathogen colonization, and microbe-derived antimicrobials probably function together with host-derived proteins to establish the composition of the surface microbiota.
Antimicrobial proteins and disease
Given the crucial contributions of AMPs to maintaining homeostasis with microorganisms at body surfaces, it is not surprising that dysregulation of AMP production and function can be associated with disease. Abnormal AMP production is associated with both intestinal inflammatory diseases and common disorders of the human skin.
Intestinal disease
Inflammatory bowel disease (IBD) is a chronic inflammation of the intestine that affects 0.1% of the population in North America and northern Europe. Although the fundamental causes of IBD remain poorly understood, it is clear that dysregulated control of host–microbial interactions is involved in initiating and perpetuating IBD101. This idea is supported by the fact that patients with IBD frequently have increased numbers of epithelial surface-associated bacteria102, suggesting a breakdown in the immune mechanisms that normally limit direct contact between the microbiota and the intestinal mucosal surface.
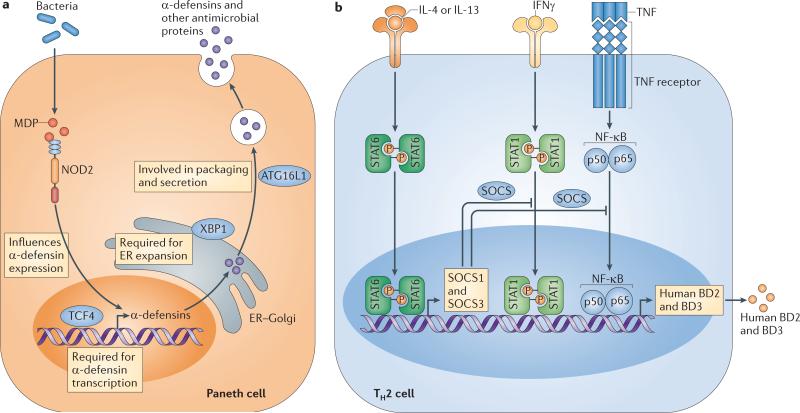
In support of this, several IBD risk alleles are associated with altered intestinal epithelial AMP production (FIG. 4a). For example, genetic variants in the promoter region of the WNT pathway transcription factor gene TCF4, which controls α-defensin expression52, are associated with ileal Crohn's disease103. The importance of the WNT pathway in maintaining α-defensin expression is further supported by a recent study that linked early onset ileal Crohn's disease to polymorphisms in low density lipoprotein receptor-related protein 6 (LRP6), an essential component of the WNT signalling pathway104. Polymorphisms in the NOD2 gene are also associated with ileal Crohn's disease105,106. In these patients, severe intestinal inflammation is coincident with decreased α-defensin expression107. Thus, mutations in TCF4, LRP6 and NOD2 could lead to decreased α-defensin production with consequent inflammation. A fourth Crohn's disease risk allele is autophagy related 16-like 1 (ATG16L1), which encodes a key component of the autophagy pathway that functions in part to regulate cellular membrane recycling and homeostasis108. Mice with a hypomorphic Atg16l1 allele have impaired exocytosis of Paneth cell secretory granules, resulting in decreased AMP release108. This suggests that the human ATG16L1 risk allele might impair the ability of Paneth cells to secrete AMPs, thereby increasing the likelihood of bacterial penetration and intestinal inflammation. Finally, hypomorphic alleles of the gene encoding the transcription factor X-box-binding protein 1 (XBP1) are linked to IBD in humans109. XBP1 is essential for the development of secretory cells, and thus mice lacking XBP1 have severe Paneth cell and goblet cell dysfunction that is coincident with spontaneous intestinal inflammation109. Thus, Paneth and goblet cell abnormalities could account for the increased incidence of IBD in individuals harbouring XBP1 variants.
Figure 4. Dysregulation of antimicrobial proteins in disease.
a Nucleotide oligomerization domain 2 (NOD2), transcription factor 4 (TCF4), X-box-binding protein 1 (XBP1) and autophagy related 16-like 1 (ATG16L1) promote antimicrobial protein (AMP) expression and secretion by Paneth cells. Polymorphisms in the corresponding genes are associated with an increased incidence of inflammatory bowel disease105–109. This could be due to compromised production of AMPs that normally control the microbiota and limit bacterial contact with the intestinal epithelial surface. b In the skin, excess production of T helper 2 (TH2) cell cytokines is associated with atopic dermatitis, which might be owing to inhibitory effects on the induction of β-defensins. Interleukin-4 (IL-4) and IL-13 induce the expression of suppressor of cytokine signalling 1 (SOCS1) and SOCS3 through signal transducer and activator of transcription 6 (STAT6). This inhibits the action of stimulatory signals for human β-defensin 2 (BD2) and BD3 expression that are transmitted by interferon-γ (IFNγ) through STAT1, or by tumour necrosis factor (TNF) through nuclear factor-κB (NF-κB). ER, endoplasmic reticulum; MDP, muramyl dipeptide.
These examples illustrate how genetic defects leading to decreased AMP production can be associated with intestinal inflammation and disease. However, it is important to note that in mice, defects in AMP production alone generally do not result in intestinal inflammation23,110. This suggests that in humans, IBD may require multiple genetic lesions that target other immune mechanisms in addition to AMPs.
Skin disease
Common human skin disorders such as atopic dermatitis, rosacea and psoriasis have been linked in part to the abnormal production of AMPs. In healthy individuals, cathelicidins and β-defensins are typically expressed at low levels in non-inflamed skin, but their expression is rapidly increased upon injury or inflammation (FIG. 4b). For example, infection with S. aureus has been shown to increase AMP production in the skin111,112. However, in patients with atopic dermatitis, the increased expression of AMPs during skin inflammation is partially suppressed113–116. Inhibition of AMP expression in atopic dermatitis seems to be partially due to excess production of the TH2 cell cytokines IL-4 and IL-13, which in turn induce the expression of suppressor of cytokine signalling 1 (SOCS1) and SOCS3 through signal transducer and activator of transcription 6 (STAT6)117. This relative defect in AMP induction correlates with the increased susceptibility of patients with atopic dermatitis to infection. By contrast, patients with psoriasis and rosacea are not more prone to infection but rather show inappropriate inflammatory reactions of the skin. In these patients, the skin surface contains excessive amounts of cathelicidin AMPs (whether produced by keratinocytes or infiltrating granulocytes) that may contribute to the excessive inflammation of the skin by mechanisms involving the inflammatory properties of cathelicidin48,50. In each of these disease associations, the defect in AMP expression is not inherent to the AMP genes themselves, but rather seems to be due to abnormalities in the specific systems that regulate both transcription and post-translational processing of AMPs117,118. For example, in rosacea, excess cathelicidin production may be influenced by abnormal TLR2 expression or polymorphisms in the vitamin D receptor that lead to increased transcription118,119. These recent observations implicating AMPs in skin disease pathogenesis have provided new opportunities for disease therapy.
Conclusions and future prospects
Body surface tissues continuously face complex microbial challenges that include maintaining homeostasis with indigenous microorganisms and limiting exposure to pathogens. The intestine and the skin each confront these challenges by producing a complex arsenal of AMPs that directly kill microorganisms and modulate immunity. It is becoming clear that endogenous AMPs are essential not only for protecting these sites from pathogenic microbial invasion but also for shaping the composition and location of indigenous microbial communities. However, many questions remain. For example, the complex regulatory networks that control the expression of several host-derived AMPs still need to be fully defined. In addition, future work should focus on gaining a precise understanding of how individual AMPs affect microbiota composition in the skin and the intestine. Finally, it will be crucial to gain a better understanding of how dysregulation of AMP expression and function contributes to diseases such as atopic dermatitis, psoriasis and IBD.
A major threat to human health is the increasing number of bacteria that have evolved resistance to available small-molecule antimicrobial therapeutics. This is in contrast to the AMPs produced by mammalian surface tissues, which for the most part have retained their antimicrobial efficacy over evolutionary timescales. Thus, it will be important to sustain efforts to understand the mechanistic basis of host-derived AMP function. Such efforts are likely to yield promising strategies for developing new antimicrobial therapeutic agents to combat disease. However, the therapeutic delivery of AMPs may be problematic owing to costs of production, potency and stability. It will thus also be important to continue to unravel the regulatory networks that dictate AMP expression. A better understanding of these regulatory networks should support strategies to augment endogenous production of AMPs during acute infections or during chronic inflammatory disease.
Acknowledgements
R.L.G. and L.V.H. thank their students and colleagues for the many discussions that contributed to the ideas in this manuscript. Work in R.L.G.'s laboratory is supported by US National Institutes of Health grants AR052728, AI052453, AI083358, contract HHSN272201000020C and a Merit Award from the Veterans Administration. Work in L.V.H.'s laboratory is supported by the Howard Hughes Medical Institute, the US National Institutes of Health (DK070855), the Burroughs Wellcome Foundation and the Crohn's and Colitis Foundation.
Glossary
- Commensal microorganisms
The microorganisms that are present in normal, healthy individuals. These microorganisms live in the gastrointestinal tract and at other body sites, and generally engage in mutually beneficial relationships with their hosts.
- C-type lectins
A large family of receptors that have carbohydrate recognition domains. The designation ‘C-type’ is based on the structure of the carbohydrate recognition domain. Several epithelial antimicrobial proteins, including regenerating islet-derived protein 3γ (REG3γ) and hepatointestinal pancreatic/pancreatitis-associated protein (HIP/PAP), are members of the C-type lectin family.
- Ribonucleases
(RNases). Enzymes that catalyse the breakdown of RNA. Several antimicrobial proteins (for example, RNase7 and angiogenin 4) have RNase activity, although the significance of this for the antibacterial activity of these proteins is not known.
- Cryptdins
Mouse α-defensins are frequently designated as ‘cryptdins’, which stands for ‘crypt α-defensins’.
- Peptidoglycan
A polymer of sugars, crosslinked by short peptides, that is a crucial component of the bacterial cell wall.
- Enterocytes
Absorptive columnar epithelial cells that are the major epithelial lineage of the intestine.
- Paneth cells
A specialized epithelial cell lineage that produces most of the antimicrobial proteins in the small intestine.
- Crypts of Lieberkühn
Invaginations of the small intestinal surface that contain both Paneth cells and intestinal stem cells.
- Bacterial resistance to antimicrobial proteins
The rapid development of bacterial resistance to commercially produced antibiotics has raised the question of how endogenous antimicrobial proteins (AMPs) have maintained their efficacy over mammalian evolutionary timescales. A partial answer to this question may lie in the fact that both skin and intestine deploy a diverse array of AMPs, making the development of combined resistance relatively infrequent. In addition, the targeting of essential cell wall or cell membrane structures might account for the continued effectiveness of endogenous AMPs, as bacteria cannot alter these structures without compromising fitness. However, several examples of the development of microbial resistance to mammalian AMPs have been described. Some successful human pathogens such as Staphylococcus aureus, Salmonella enterica and Legionella pneumophila modify their normally anionic cell walls with cationic substitutions to repulse cationic AMPs such as defensins120–122. Furthermore, many other systems have been described for the evasion of epithelial AMPs. These include the capacity of S. aureus and group A streptococcus to inactivate AMPs by proteolysis123,124, the efflux pumps that protect Neisseria gonorrhoeae from AMPs by active transport125 and the ability of Shigella spp. to evade cathelicidin and human β-defensin 1 (BD1) by inhibiting the enteric synthesis of these defence molecules126. Thus, although the AMPs are formidable inhibitors of microbial growth and survival, microorganisms have effectively used multiple mechanisms to evade their action. The interested reader is directed to two excellent reviews devoted to this subject127,128.
- Pattern recognition receptors
(PRRs). Host receptors (such as Toll-like receptors (TLRs) or NOD-like receptors (NLRs)) that can sense pathogen-associated molecular patterns and initiate signalling cascades that lead to an innate immune response. These can be membrane bound (for example, TLRs) or soluble cytoplasmic receptors (for example, RIG-I, MDA5 and NLRs).
- Germ-free animals
Animals that are reared in isolators, without exposure to microorganisms.
- WNT pathway
A signalling pathway that controls several physiological processes, including embryogenesis and cancer development. It also controls normal biological functions in adult animals and is essential for the expression of α-defensins in the small intestine.
- Conventionally raised mice
Mice that have been raised with normal exposure to microorganisms.
- Innate lymphoid cells
(ILCs). A diverse family of immune cells that produce cytokines and function to coordinate immunity and inflammation in body surface tissues such as the intestine and the lung. Although their developmental origins are still unclear, they phenotypically resemble natural killer cells.
- Climax community
A mature, stable community of organisms that develops through a process of ecological succession and remains in a steady state for an extended period of time.
- Anaerobes
Microorganisms that grow in the absence of oxygen.
- 16S ribosomal RNA gene sequencing
Determination of the sequences of the variable regions of bacterial ribosomal RNA genes, which are conserved within a species but differ between species. It is frequently used as a culture-independent technique for evaluating the composition of bacterial communities.
- Segmented filamentous bacteria
(SFB). A group of Gram-positive bacteria that are members of the intestinal microbiota of mice. They are characterized by their ability to adhere to the intestinal surface and are frequently immunostimulatory.
- The microbiota of the intestine and skin
The intestine and the skin are each home to large communities of indigenous bacteria. For decades, our understanding of these bacterial communities was based on identifying resident organisms by culture-based methods. However, many of the bacteria that reside at these sites are refractory to culture, making it difficult to assemble a complete catalogue of organisms residing at these tissue sites. In the past few years, improved culture-independent molecular profiling methods, such as 16S ribosomal RNA gene sequence analysis, have led to a revolution in the understanding of indigenous microbial communities. Microbial communities in the intestine and skin both have a high level of diversity at the species level but low phylum-level diversity. In all vertebrates, intestinal microbial communities are dominated by two phyla: the Firmicutes and the Bacteroidetes129,130. The Firmicutes are Gram-positive bacteria that include numerous species belonging to the Clostridia class, in addition to Enterococcaceae and Lactobacillaceae families and Lactococcus species. The Bacteroidetes are Gram-negative bacteria comprised of several Bacteroides species. Prominent intestinal Bacteroides species include Bacteroides thetaiotaomicron, Bacteroides fragilis and Bacteroides ovatus129. The remaining intestinal bacteria, comprising less than 10% of the total population, belong predominantly to the Proteobacteria and Actinobacteria phyla129. The skin microbiota is also composed predominantly of species from the Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria. However, the proportional representation of these phyla differs from the gastrointestinal tract, with Actinobacteria constituting the dominant bacterial phylum on the skin2. It is also important to note that there is variability in microbiota composition, even within an individual, depending on which skin site is sampled131.
- Quorum sensing
A system used by bacteria to coordinate gene expression as a function of population density.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION Richard L. Gallo's homepage: http://dermatology.ucsd.edu/research/gallo-lab.shtml
Lora V. Hooper's homepage: http://hooperlab.org/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Neish AS. The gut microflora and intestinal epithelial cells: a continuing dialogue. Microbes Infect. 2002;4:309–317. doi: 10.1016/s1286-4579(02)01543-5. [DOI] [PubMed] [Google Scholar]
- 2.Grice EA, Segre JA. The skin microbiome. Nature Rev. Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S, Vaishnava S, Hooper LV. Multi-layered regulation of intestinal antimicrobial defense. Cell. Mol. Life Sci. 2008;65:3019–3027. doi: 10.1007/s00018-008-8182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gläser R, et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nature Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 7.Schittek B, et al. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nature Immunol. 2001;2:1133–1137. doi: 10.1038/ni732. [DOI] [PubMed] [Google Scholar]
- 8.Ouellette AJ. Paneth cell α-defensins in enteric innate immunity. Cell. Mol. Life Sci. 2011;68:2215–2229. doi: 10.1007/s00018-011-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christa L, et al. HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and pancreatic cells. Am. J. Physiol. 1996;271:G993–G1002. doi: 10.1152/ajpgi.1996.271.6.G993. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa H, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm. Bowel Dis. 2003;9:162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehotzky RE, et al. Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc. Natl Acad. Sci. USA. 2010;107:7722–7727. doi: 10.1073/pnas.0909449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo RL, et al. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc. Natl Acad. Sci. USA. 1994;91:11035–11039. doi: 10.1073/pnas.91.23.11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harder J, Bartels J, Christophers E, Schröder JM. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 15.Larrick JW, et al. Structural, functional analysis and localization of the human CAP18 gene. FEBS Lett. 1996;398:74–80. doi: 10.1016/s0014-5793(96)01199-4. [DOI] [PubMed] [Google Scholar]
- 16.Gudmundsson GH, et al. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 1996;238:325–332. doi: 10.1111/j.1432-1033.1996.0325z.x. [DOI] [PubMed] [Google Scholar]
- 17.Gallo RL, et al. Identification of CRAMP, a cathelin-related antimicrobial peptide expressed in the embryonic and adult mouse. J. Biol. Chem. 1997;272:13088–13093. doi: 10.1074/jbc.272.20.13088. [DOI] [PubMed] [Google Scholar]
- 18.Di Nardo A, Vitiello A, Gallo RL. Cutting edge: mast cell antimicrobial activity is mediated by expression of cathelicidin antimicrobial peptide. J. Immunol. 2003;170:2274–2278. doi: 10.4049/jimmunol.170.5.2274. [DOI] [PubMed] [Google Scholar]
- 19.Murakami M, et al. Cathelicidin anti-microbial peptide expression in sweat, an innate defense system for the skin. J. Invest. Dermatol. 2002;119:1090–1095. doi: 10.1046/j.1523-1747.2002.19507.x. [DOI] [PubMed] [Google Scholar]
- 20.Frohm M, et al. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 1997;272:15258–15263. doi: 10.1074/jbc.272.24.15258. [DOI] [PubMed] [Google Scholar]
- 21.Dorschner RA, et al. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J. Invest. Dermatol. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 22.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl Acad. Sci. USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [This paper shows that the antibacterial protein REG3γ limits direct contact between the intestinal microbiota and host tissues, and thus helps to preserve the symbiotic nature of the host–microbiota relationship.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Neil DA, et al. Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- 25.Hase K, Eckmann L, Leopard JD, Varki N, Kagnoff MF. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect. Immun. 2002;70:953–963. doi: 10.1128/iai.70.2.953-963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 27.Johansson MEV, et al. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl Acad. Sci. USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer-Hoffert U, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 29.Emelianov VU, et al. Immunohistological pointers to a possible role for excessive cathelicidin (LL-37) expression by apocrine sweat glands in the pathogenesis of hidradenitis suppurativa/acne inversa. Br. J. Dermatol. 2012;166:1023–1034. doi: 10.1111/j.1365-2133.2011.10765.x. [DOI] [PubMed] [Google Scholar]
- 30.Frohm Nilsson M, et al. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect. Immun. 1999;67:2561–2566. doi: 10.1128/iai.67.5.2561-2566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sørensen OE, et al. Wound healing and expression of antimicrobial peptides/polypeptides in human keratinocytes, a consequence of common growth factors. J. Immunol. 2003;170:5583–5589. doi: 10.4049/jimmunol.170.11.5583. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, et al. Skin mast cells protect mice against vaccinia virus by triggering mast cell receptor S1PR2 and releasing antimicrobial peptides. J. Immunol. 2012;188:345–357. doi: 10.4049/jimmunol.1101703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harwig SS, et al. Bactericidal properties of murine intestinal phospholipase A2. J. Clin. Invest. 1995;95:603–610. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koprivnjak T, Peschel A, Gelb MH, Liang NS, Weiss JP. Role of charge properties of bacterial envelope in bactericidal action of human group IIA phospholipase A2 against Staphylococcus aureus. J. Biol. Chem. 2002;277:47636–47644. doi: 10.1074/jbc.M205104200. [DOI] [PubMed] [Google Scholar]
- 35.Kagan BL, Selsted ME, Ganz T, Lehrer RI. Antimicrobial defensin peptides form voltage-dependent ion-permeable channels in planar lipid bilayer membranes. Proc. Natl Acad. Sci. USA. 1990;87:210–214. doi: 10.1073/pnas.87.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bals R, Wilson JM. Cathelicidins — a family of multifunctional antimicrobial peptides. Cell. Mol. Life Sci. 2003;60:711–720. doi: 10.1007/s00018-003-2186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gennaro R, Zanetti M, Benincasa M, Podda E, Miani M. Pro-rich antimicrobial peptides from animals: structure, biological functions and mechanism of action. Curr. Pharm. Des. 2002;8:763–778. doi: 10.2174/1381612023395394. [DOI] [PubMed] [Google Scholar]
- 38.Harder J, Schroder J-M. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J. Biol. Chem. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 39.Corbin BD, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- 40.Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Yang, et al. LL-37, the neutrophil granule- and epithelial cell-derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J. Exp. Med. 2000;192:1069–1074. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurosaka K, Chen Q, Yarovinsky F, Oppenheim JJ, Yang D. Mouse cathelin-related antimicrobial peptide chemoattracts leukocytes using formyl peptide receptor-like 1/mouse formyl peptide receptor-like 2 as the receptor and acts as an immune adjuvant. J. Immunol. 2005;174:6257–6265. doi: 10.4049/jimmunol.174.10.6257. [DOI] [PubMed] [Google Scholar]
- 43.Yang D, Biragyn A, Kwak LW, Oppenheim JJ. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 44.Niyonsaba F, et al. A cathelicidin family of human antibacterial peptide LL-37 induces mast cell chemotaxis. Immunology. 2002;106:20–26. doi: 10.1046/j.1365-2567.2002.01398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang D, Chertov O, Oppenheim JJ. Participation of mammalian defensins and cathelicidins in antimicrobial immunity: receptors and activities of human defensins and cathelicidin (LL-37). J. Leukoc. Biol. 2001;69:691–697. [PubMed] [Google Scholar]
- 46.Yang D, et al. β-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 47.Tokumaru S, et al. Induction of keratinocyte migration via transactivation of the epidermal growth factor receptor by the antimicrobial peptide LL-37. J. Immunol. 2005;175:4662–4668. doi: 10.4049/jimmunol.175.7.4662. [DOI] [PubMed] [Google Scholar]
- 48.Yamasaki K, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nature Med. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 49.Di Nardo A, et al. Cathelicidin antimicrobial peptides block dendritic cell TLR4 activation and allergic contact sensitization. J. Immunol. 2007;178:1829–1834. doi: 10.4049/jimmunol.178.3.1829. [DOI] [PubMed] [Google Scholar]
- 50.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [This paper describes the discovery of a novel mechanism by which antimicrobial peptides can break tolerance to self-DNA.] [DOI] [PubMed] [Google Scholar]
- 51.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 1998;62:1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Es JH, et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nature Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- 53.Putsep K, et al. Germ-free and colonized mice generate the same products from enteric prodefensins. J. Biol. Chem. 2000;275:40478–40482. doi: 10.1074/jbc.M007816200. [DOI] [PubMed] [Google Scholar]
- 54.Hooper LV, et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 55.Abtin A, et al. Flagellin is the principal inducer of the antimicrobial peptide S100A7c (psoriasin) in human epidermal keratinocytes exposed to Escherichia coli. FASEB J. 2008;22:2168–2176. doi: 10.1096/fj.07-104117. [DOI] [PubMed] [Google Scholar]
- 56.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 58.Kinnebrew MA, et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schauber J, et al. Expression of the cathelicidin LL-37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52:735–741. doi: 10.1136/gut.52.5.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanos SL, Vonarbourg C, Mortha A, Diefenbach A. Control of epithelial cell function by interleukin-22-producing RORγt+ innate lymphoid cells. Immunology. 2011;132:453–465. doi: 10.1111/j.1365-2567.2011.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolk K, et al. IL-22 increases the innate immunity of tissues. Immunity. 2004;21:241–254. doi: 10.1016/j.immuni.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Sanos SL, et al. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nature Immunol. 2008;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogura Y, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn's ileitis. Gut. 2003;52:1591–1597. doi: 10.1136/gut.52.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Girardin SE. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 65.Petnicki-Ocwieja T, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl Acad. Sci. USA. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [This study shows that the PRR NOD2 controls microbiota load and composition in the small intestine.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kobayashi KS, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 67.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 68.Ménard S, et al. Developmental switch of intestinal antimicrobial peptide expression. J. Exp. Med. 2008;205:183–193. doi: 10.1084/jem.20071022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iimura M, et al. Cathelicidin mediates innate intestinal defense against colonization with epithelial adherent bacterial pathogens. J. Immunol. 2005;174:4901–4907. doi: 10.4049/jimmunol.174.8.4901. [DOI] [PubMed] [Google Scholar]
- 70.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl Acad. Sci. USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [This landmark paper describes the early identification of an antimicrobial peptide in frog skin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambert J, et al. Insect immunity: isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proc. Natl Acad. Sci. USA. 1989;86:262–266. doi: 10.1073/pnas.86.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aberg KM, et al. Co-regulation and interdependence of the mammalian epidermal permeability and antimicrobial barriers. J. Invest. Dermatol. 2008;128:917–925. doi: 10.1038/sj.jid.5701099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sørensen OE, et al. Injury-induced innate immune response in human skin mediated by transactivation of the epidermal growth factor receptor. J. Clin. Invest. 2006;116:1878–1885. doi: 10.1172/JCI28422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang T-T, et al. Cutting edge: 1,25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- 75.Gombart AF, Borregaard N, Koeffler HP. Human cathelicidin antimicrobial peptide (CAMP) gene is a direct target of the vitamin D receptor and is strongly up-regulated in myeloid cells by 1,25-dihydroxyvitamin D3. FASEB J. 2005;19:1067–1077. doi: 10.1096/fj.04-3284com. [DOI] [PubMed] [Google Scholar]
- 76.Liu PT, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [This paper linked control of antimicrobial peptide expression by vitamin D to susceptibility to tuberculosis.] [DOI] [PubMed] [Google Scholar]
- 77.Schauber J, et al. Injury enhances TLR2 function and antimicrobial peptide expression through a vitamin D-dependent mechanism. J. Clin. Invest. 2007;117:803–811. doi: 10.1172/JCI30142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hata TR, et al. Administration of oral vitamin D induces cathelicidin production in atopic individuals. J. Allergy Clin. Immunol. 2008;122:829–831. doi: 10.1016/j.jaci.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong SP, et al. Biopositive effects of low-dose UVB on epidermis: coordinate upregulation of antimicrobial peptides and permeability barrier reinforcement. J. Invest. Dermatol. 2008;128:2880–2887. doi: 10.1038/jid.2008.169. [DOI] [PubMed] [Google Scholar]
- 80.Lichtenstein A, Ganz T, Selsted ME, Lehrer RI. In vitro tumor cell cytolysis mediated by peptide defensins of human and rabbit granulocytes. Blood. 1986;68:1407–1410. [PubMed] [Google Scholar]
- 81.Wilson CL, et al. Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 82.Ghosh D, et al. Paneth cell trypsin is the processing enzyme for human defensin-5. Nature Immunol. 2002;3:583–590. doi: 10.1038/ni797. [DOI] [PubMed] [Google Scholar]
- 83.Mukherjee S, et al. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J. Biol. Chem. 2009;284:4881–4888. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sørensen O, Arnljots K, Cowland JB, Bainton DF, Borregaard N. The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood. 1997;90:2796–2803. [PubMed] [Google Scholar]
- 85.Yamasaki K, et al. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006;20:2068–2080. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 86.Schutte BC, McCray PB. β-defensins in lung host defense. Annu. Rev. Physiol. 2002;64:709–748. doi: 10.1146/annurev.physiol.64.081501.134340. [DOI] [PubMed] [Google Scholar]
- 87.Schroeder BO, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [This study provides new insights into the regulation of antimicrobial protein function by showing that the reducing environment of the intestine is required for the full expression of antimicrobial activity in a human β-defensin.] [DOI] [PubMed] [Google Scholar]
- 88.Ayabe T, et al. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nature Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 89.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–526. doi: 10.1038/nature01520. [This paper provides insights into the in vivo function of α-defensins by showing that DEFA5 limits pathogen colonization of the intestine.] [DOI] [PubMed] [Google Scholar]
- 90.Howell MD, et al. Selective killing of vaccinia virus by LL-37: implications for eczema vaccinatum. J. Immunol. 2004;172:1763–1767. doi: 10.4049/jimmunol.172.3.1763. [DOI] [PubMed] [Google Scholar]
- 91.Kulkarni MM, et al. Mammalian antimicrobial peptide influences control of cutaneous Leishmania infection. Cell. Microbiol. 2011;13:913–923. doi: 10.1111/j.1462-5822.2011.01589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nizet V, et al. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [This paper was the first direct demonstration that an antimicrobial peptide is essential for mammalian immune defence.] [DOI] [PubMed] [Google Scholar]
- 93.Chromek M, et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nature Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 94.Huang LC, Reins RY, Gallo RL, McDermott AM. Cathelicidin-deficient (Cnlp–/–) mice show increased susceptibility to Pseudomonas aeruginosa keratitis. Invest. Ophthalmol. Vis. Sci. 2007;48:4498–4508. doi: 10.1167/iovs.07-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salzman NH, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [This study showed for the first time that antimicrobial proteins can regulate the composition of intestinal microbial communities.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cogen AL, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iwase T, et al. Staphylococcus epidermidis Esp. inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 100.Otto M, Süssmuth R, Vuong C, Jung G, Götz F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999;450:257–262. doi: 10.1016/s0014-5793(99)00514-1. [DOI] [PubMed] [Google Scholar]
- 101.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 102.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koslowski MJ, et al. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn's disease. PLoS ONE. 2009;4:e4496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koslowski MJ, et al. Association of a functional variant in the Wnt co-receptor LRP6 with early onset ileal Crohn's disease. PLoS Genet. 2012;8:e1002523. doi: 10.1371/journal.pgen.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 106.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 107.Wehkamp J, et al. Reduced Paneth cell α-defensins in ileal Crohn's disease. Proc. Natl Acad. Sci. USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [This paper demonstrates that autophagy pathways regulate the exocytosis of Paneth cell secretory granules and provides insights into how perturbations in the autophagy pathway might lead to IBD.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J. Biol. Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 111.Zanger P, et al. Constitutive expression of the antimicrobial peptide RNase 7 is associated with Staphylococcus aureus infection of the skin. J. Infect. Dis. 2009;200:1907–1915. doi: 10.1086/648408. [DOI] [PubMed] [Google Scholar]
- 112.Zanger P, et al. Severity of Staphylococcus aureus infection of the skin is associated with inducibility of human β-defensin 3 but not human β-defensin 2. Infect. Immun. 2010;78:3112–3117. doi: 10.1128/IAI.00078-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ong PY, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 114.de Jongh GJ, et al. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J. Invest. Dermatol. 2005;125:1163–1173. doi: 10.1111/j.0022-202X.2005.23935.x. [DOI] [PubMed] [Google Scholar]
- 115.Kisich KO, Carspecken CW, Fiéve S, Boguniewicz M, Leung DYM. Defective killing of Staphylococcus aureus in atopic dermatitis is associated with reduced mobilization of human β-defensin-3. J. Allergy Clin. Immunol. 2008;122:62–68. doi: 10.1016/j.jaci.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 116.Hata TR, et al. History of eczema herpeticum is associated with the inability to induce human β-defensin (HBD)-2, HBD-3 and cathelicidin in the skin of patients with atopic dermatitis. Br. J. Dermatol. 2010;163:659–661. doi: 10.1111/j.1365-2133.2010.09892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Howell MD, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 118.Yamasaki K, et al. TLR2 expression is increased in rosacea and stimulates enhanced serine protease production by keratinocytes. J. Invest. Dermatol. 2011;131:688–697. doi: 10.1038/jid.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jansen T, Krug S, Kind P, Plewig G, Messer G. BsmI polymorphism of the vitamin D receptor gene in patients with the fulminant course of rosacea conglobata (rosacea fulminans). J. Dermatol. 2004;31:244–246. doi: 10.1111/j.1346-8138.2004.tb00665.x. [DOI] [PubMed] [Google Scholar]
- 120.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 121.Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar typhimurium. Infect. Immun. 2000;68:6139–6146. doi: 10.1128/iai.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Robey M, O'Connell W, Cianciotto NP. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sieprawska-Lupa M, et al. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 2004;48:4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nyberg P, Rasmussen M, Björck L. α2-macroglobulin-proteinase complexes protect Streptococcus pyogenes from killing by the antimicrobial peptide LL-37. J. Biol. Chem. 2004;279:52820–52823. doi: 10.1074/jbc.C400485200. [DOI] [PubMed] [Google Scholar]
- 125.Shafer WM, Qu X, Waring AJ, Lehrer RI. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl Acad. Sci. USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Islam D, et al. Downregulation of bactericidal peptides in enteric infections: a novel immune escape mechanism with bacterial DNA as a potential regulator. Nature Med. 2001;7:180–185. doi: 10.1038/84627. [DOI] [PubMed] [Google Scholar]
- 127.Koprivnjak T, Peschel A. Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci. 2011;68:2243–2254. doi: 10.1007/s00018-011-0716-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nizet V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J. Allergy Clin. Immunol. 2007;120:13–22. doi: 10.1016/j.jaci.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 129.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sumikawa Y, et al. Induction of β-defensin 3 in keratinocytes stimulated by bacterial lipopeptides through toll-like receptor 2. Microbes Infect. 2006;8:1513–1521. doi: 10.1016/j.micinf.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 133.Sass V, et al. Human β-defensin 3 inhibits cell wall biosynthesis in Staphylococci. Infect. Immun. 2010;78:2793–2800. doi: 10.1128/IAI.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stowell SR, et al. Innate immune lectins kill bacteria expressing blood group antigen. Nature Med. 2010;16:295–302. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 136.Raffatellu M, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Müller CA, Autenrieth IB, Peschel A. Innate defenses of the intestinal epithelial barrier. Cell. Mol. Life Sci. 2005;62:1297–1307. doi: 10.1007/s00018-005-5034-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Royet J, Gupta D, Dziarski R. Peptidoglycan recognition proteins: modulators of the microbiome and inflammation. Nature Rev. Immunol. 2011;11:837–851. doi: 10.1038/nri3089. [DOI] [PubMed] [Google Scholar]
- 139.Qu XD, Lehrer RI. Secretory phospholipase A2 is the principal bactericide for staphylococci and other gram-positive bacteria in human tears. Infect. Immun. 1998;66:2791–2797. doi: 10.1128/iai.66.6.2791-2797.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]