Abstract
Small- and intermediate-conductance Ca2+-activated potassium channels, activated by Ca2+-bound calmodulin, play an important role in regulating membrane excitability. These channels are also linked to clinical abnormalities. A tremendous amount of effort has been devoted to developing small molecule compounds targeting these channels. However, these compounds often suffer from low potency and lack of selectivity, hindering their potentials for clinical use. A key contributing factor is the lack of knowledge of the binding site(s) for these compounds. Here we demonstrate by X-ray crystallography that the binding pocket for the compounds of the 1-EBIO class is located at the calmodulin-channel interface. We show that, based on structure data and molecular docking, mutations of the channel can effectively change the potency of these compounds. Our results provide insight into the molecular nature of the binding pocket and its contribution to the potency and selectivity of the compounds of the 1-EBIO class.
Ca2+-activated potassium channels, such as small- and intermediate-conductance K+ channels (SK and IK), are widely expressed in excitable tissues, including the central nervous system and the cardiovascular system1–5. They play pivotal roles in regulating membrane excitability by Ca2+. Unlike voltage-gated K+ channels, activation of SK/IK channels is voltage-independent and is achieved exclusively by Ca2+6. Calmodulin (CaM), constitutively tethered to SK/IK channels, serves as the high-affinity Ca2+ sensor. Four canonical EF-hands, two located at the CaM N terminus (N-lobe) and the other two at the C terminus (C-lobe), are the high affinity Ca2+ binding domains7. Binding of Ca2+ to CaM results in changes in the CaM conformation and subsequent opening of the channel8–10. Both SK and IK channels belong to the same gene family, with four genes identified, KCNN1 for SK1, KCNN2 for SK2, KCNN3 for SK3 and KCNN4 for IK channels3,5. We have identified an SK2 channel splice variant, SK2-b, which is less sensitive to Ca2+ for its activation10. Activation of SK channels dampens firing of action potentials, and therefore contributes to regulation, by Ca2+, of neuronal excitability, dendritic integration, synaptic transmission and plasticity in the central nervous system1,3,5,11. Equally important are the roles of SK/IK channels in the cardiovascular system12–15. Vascular endothelial cells express both IK and SK3 channels, which contribute to the endothelium-derived hyperpolarizing factor (EDHF)-mediated vasodilation16–19. Increased SK/IK channel activities promote arteriolar vasodilation by enhancing agonist-evoked synthesis of nitric oxide in endothelial cells20.
The functional importance of SK/IK channels is further demonstrated by their potential involvement in clinical abnormalities14,21–23. CGA repeats of SK3 channels have been linked to schizophrenia and the bipolar disorder24–25. Single-nucleotide polymorphisms (SNPs) in the SK/IK genes have been implicated in cardiovascular abnormalities21. In the heart, genome-wide association studies reveal a correlation of SNPs of SK3 channels and atrial fibrillation26. Compromised SK channel activities may be a contributing factor of hypertension14. IK channels are implicated in atherogenesis in mice and human27. Recent studies have also suggested the potential link of SK channels and development of cancer, as abnormal expression of SK2 and SK3 channels might contribute to enhanced cell proliferation and cell migration23,28. Given the roles of SK/IK channels in physiological and pathophysiological conditions, a tremendous amount of effort has been devoted to developing small molecules targeting SK/IK channels11,28–32.
1-ethyl-2-benzimidazolinone (1-EBIO) is such a small molecule prototype, which potentiates the SK/IK channel activities and effectively decreases the neuronal excitability, thus potentially neuroprotective33. Indeed, studies on animal models have shown that 1-EBIO can reduce seizure incidence in seizure models34 and significantly improve survival of hippocampal neurons after cerebral ischemia22. SKA-31, a different channel modulator, enhances the EDHF-mediated responses and lowers the blood pressure35. In general, for compounds of the 1-EBIO class, problems include the low potency and lack of selectivity, which hamper their potential for evaluation in animal models and in clinical trials2,11. For instance, the EC50 of 1-EBIO is in the sub-millimolar range for its potentiation of SK2 channels11. NS309, although the most potent one of the 1-EBIO class, potentiates both IK and SK channels. CyPPA is relatively more selective, acting on SK2 and SK3 channels, but not SK1 or IK channels36. A key contributing factor is the lack of knowledge of the binding site(s) for these compounds in SK/IK channels, although the channel C-terminus is implicated for the action of 1-EBIO37. So far, it is not known where or how compounds of the 1-EBIO class interact with SK/IK channels.
Here we describe our discovery of the functional binding pocket at the CaM-channel interface of the CaM N-lobe for compounds of the 1-EBIO class, through the combined approaches of X-ray crystallography, molecular docking, mutagenesis and electrophysiology. We also show that phenylurea (PHU) is the most prelimitive structure of the SK channel modulators of the 1-EBIO class. Our results provide insight into the molecular nature of the binding pocket and its contribution to the potency and selectivity of the compounds of the 1-EBIO class. The knowledge will facilitate development of future generations of therapeutics targeting SK/IK channels.
RESULTS
Phenylurea is a weak SK channel modulator
We previously reported that CaM, when complexed with the CaM binding domain (CaMBD) of a SK2 channel splice variant SK2-b (CaMBD2-b), adopts a drastically different conformation at the CaM C-lobe10. Such conformational changes have restored the ability of the CaM C-lobe to bind Ca2+ ions with reduced affinity, which accounts for the reduced sensitivity of activation of SK2-b channels by Ca2+. In solving the structure of the CaM-CaMBD2-b complex (PDB accession code 3SJQ), it was discovered that phenylurea, an additive in the crystallization solution, is found at the CaM-CaMBD interface of the CaM N-lobe, but not the CaM C-lobe (Fig. 1a). The presence of PHU does not interfere with Ca2+-dependent formation of the 2×2 CaM-CaMBD2-b complex10. The interaction between PHU and the CaM complex is further confirmed by the protein crystal structure, at the resolution of 1.7 Å, obtained by soaking in the PHU solution of the CaM-CaMBD2-a complex crystals which had been grown in the absence of PHU.
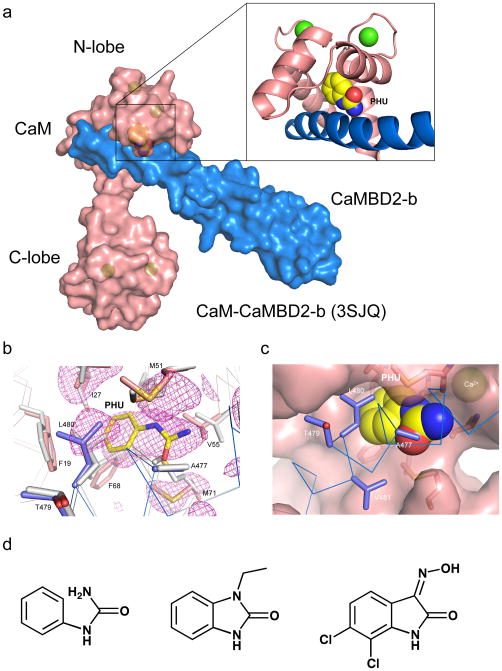
Figure 1. Phenylurea binds to the CaM-CaMBD interface at the CaM N-lobe.
(a) A space-filled model of the CaM-CaMBD2-b complex (3SJQ), depicting the location of PHU (yellow) between the interface of CaM (salmon) and CaMBD2-b (marine) at the CaM N-lobe. Only half of the 2×2 complex is shown for clarity. (b) Difference Fourier electron density map constructed using mFo–DFc coefficients calculated in PHENIX prior to modeling PHU and side chain rearrangements that occur in the binding site. The map is contoured at 2.5σ and displayed within a 6 Å sphere of PHU. The map is overlaid with the final coordinates for PHU and its surrounding amino acid residues in the CaM-CaMBD2-a complex. Marine are amino acid residues from CaMBD2-a and salmon are amino acid residues from CaM. Coordinates of the same amino acid residues (gray) from the original CaM-CaMBD2-a complex (without PHU, 1G4Y) are provided for comparison. (c) A space-filled model of PHU at the CaM-CaMBD interface. Marine represents CaMBD and salmon represents CaM. (d) Comparison of the chemical structure of PHU with that of 1-EBIO and NS309. Both 1-EBIO and NS309 are known SK/IK channel modulators.
Structural analysis shows extra electron density which corresponds to the structure of PHU in the CaM-CaMBD2-a complex (Fig. 1b and Supplementary Figs. S1a and 1b, Table 1). Like the CaM-CaMBD2-b complex (Fig. 1a), PHU is located at the interface of CaM-CaMBD2-a of the CaM N-lobe, forming close contacts with both CaM and CaMBD2-a. There are no significant global changes in the structure of the 2×2 configuration of the CaM-CaMBD2-a complex8,10. The PHU binding pocket includes A477, L480 and V481 of CaMBD, F19, I27, L32, M51, I52, V55, I63, F68 and M71 of CaM (within the 5 Å radius of PHU, Figs. 1b and 1c, and Supplementary Fig. S1). Compared to the original structure of CaM-CaMBD2-a (1G4Y)8, presence of PHU alters the spatial orientation of amino acids, including L480, F19, M51, V55 and M71 (Fig. 1b). Within the binding pocket, the urea-group of PHU can adopt two different conformations (flip-flop, Supplementary Fig. S1c). The most intriguing and unexpected discovery comes from the comparison of the chemical structure of PHU with the known SK/IK channel modulators, such as 1-EBIO and NS309. There is a striking resemblance of the PHU structure to those of 1-EBIO and NS309 (Fig. 1d).
Table 1.
Crystallographic statistics
| Data Collectiona | ||
|---|---|---|
| CaM-CaMBD2-a Complex with phenylurea | CaM-CaMBD2-a Complex with 1-EBIO | |
| Space Group Unit Cell Dimensions |
C2 a=76.5 Å, b=66.9 Å, c=65.1 Å α=90.0°, β=93.3°, γ=90.0° |
C2 a=77.0 Å, b=66.7 Å, c=64.9 Å α=90.0° β =94.0°, γ=90.0° |
| Wavelength (Å) | 1.12 | 1.12 |
| Resolution range (Å) | 50–1.65 (1.69–1.65) | 25–1.63 (1.67–1.63) |
| Completeness (%) | 99.5 (99.7) | 99.2 (99.5) |
| Total Observations | 197,980 (14,626) | 200,171 (15,060) |
| Unique Observations | 39,245 (2,902) | 40,567 (2,989) |
| Mean Redundancy | 5.0 (5.0) | 4.9 (5.0) |
| Mean I/σ(I) | 22.5 (2.3) | 12.3 (2.2) |
| Rmergeb | 0.035 (0.645) | 0.063 (0.595) |
| Rpimc | 0.017 (0.319) | 0.031 (0.288) |
| Model Refinementa | ||
| Resolution Range (Å) | 50–1.65 (1.69–1.65) | 50–1.63 (1.67–1.63) |
| Number of reflections | 39,243 (2,804) | 40,565 (2,886) |
| Rworkd | 0.191 (0.294) | 0.192 (0.265) |
| Rfreed | 0.228 (0.341) | 0.227 (0.304) |
| Number of atoms/Average B-factor (Å2) | 2,310/37.9 | 2,354/34.6 |
| protein | 2,069/37.2 | 2,069/33.5 |
| calcium ions | 2/38.8 | 2/36.1 |
| solvent waters | 208/42.0 | 250/41.1 |
| SK channel modulator | 10/36.4 | 12/45.0 |
| Phi/Psi angles most favored(%)/outliers(#) | 98.7/0 | 98.3/0 |
| r.m.s.d. bond angles (°) | 1.264 | 1.258 |
| r.m.s.d. bond lengths (Å) | 0.011 | 0.011 |
Values in parentheses refer to data in the highest resolution shell.
Rmerge = ΣhklΣj | Ij − 〈I〉 | /ΣhklΣj Ij. 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents.
Rpim (precision-indicating merge) = Σhkl (1/nhkl − 1)1/2 Σj | Ij − 〈I〉 | /ΣhklΣj Ij. n is the number of observations of reflection hkl.
Rcryst = Σhkl | Fobs − kFcalc | /Σhkl | Fobs |. Rfree = Rcryst for 5% of reflections excluded from crystallographic refinement.
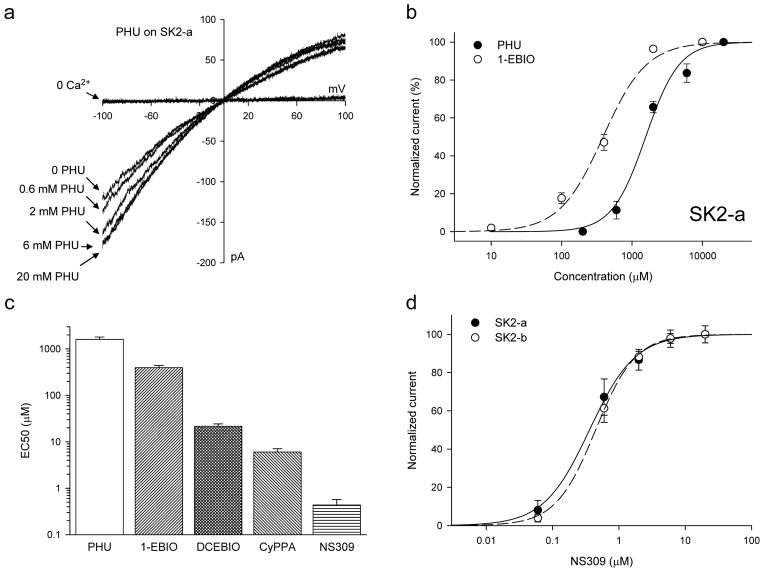
To explore whether PHU might act as an SK channel modulator, SK2-a channels were expressed in TsA cells and their responses to application of PHU were recorded in the presence of 200 nM Ca2+ in the bath solution10. SK2-a channels open upon application of a 200 nM Ca2+ solution (Fig. 2a). The channel activity is further potentiated with increasing PHU concentrations, from 0.2 mM up to 20 mM, with the EC50 of 1.61 ± 0.20 mM and a Hill coefficient of 1.81 ± 0.22 (n = 4). The same experiments were repeated using other known SK channel modulators, such as 1-EBIO, DCEBIO, CyPPA and NS30911. Compared to 1-EBIO (EC50 = 395.5 ± 45.2 μM, n = 5), PHU is much less potent in its potentiation of SK2 channels (p < 0.001, T-test, Fig. 2b). PHU is almost 75-fold less potent than DCEBIO (EC50 = 21.5 ± 2.7μM, n =8, Fig. 2c) and more than 3600-fold less potent compared to NS309 (EC50 = 0.44 ± 0.13 μM, n = 3). CyPPA, which selectively potentiates SK2 and SK3 channels36, is ~266-fold more potent than PHU (6.04 ± 1.04 μM, n = 6). SK2-b, a SK2 splice variant10, has the same responses as SK2-a to NS309, 0.50 ± 0.09 μM (n = 6, p = 0.713, T-test, Fig. 2d). This demonstrates that SK2-b, while less sensitive to Ca2+ for its activation10, has the same response to the SK channel modulator as SK2-a.
Figure 2. Phenylurea is a weak SK2 channel modulator.
(a) Raw current traces from an inside-out patch with SK2-a channels expressed. The SK2 channels are activated by 200 nM Ca2+, with subsequent potentiation by PHU at the concentrations indicated (all in the presence of 200 nM Ca2+). A voltage ramp, from −100 mV to +100 mV, was applied. (b) Dose-response curves for PHU (n = 4) and 1-EBIO (n = 5) for their potentiation of the SK2 channel activities (all in the presence of 200 nM Ca2+). The current amplitudes were measured at −90 mV and normalized to the maximal current obtained either with PHU or 1-EBIO. (c) EC50s of potentiation of the SK2 channel activities by PHU (n = 4), 1-EBIO (n = 5), DCEBIO (n = 8), CyPPA (n = 6) and NS309 (n = 3). All modulators were in a 200 nM Ca2+ solution. Note that the y-axis is in the log scale. (d) SK2-a (n = 3) and SK2-b (n = 6) have the same responses to application of NS309. Because SK2-b is less sensitive to Ca2+ for its activation, 500 nM Ca2+ was used instead of 200 nM Ca2+ for SK2-a channels. All data are mean ± s.e.m.
1-EBIO binds to the same pocket as PHU
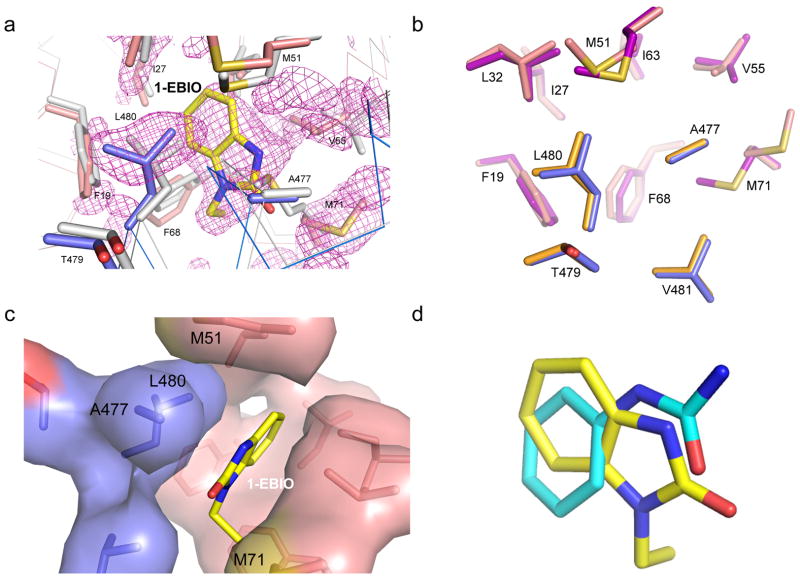
The above observation led us to test whether, like PHU, SK channel modulators of the 1-EBIO class might also bind to the same CaM-CaMBD interface at the CaM N-lobe. Protein crystals of CaM-CaMBD2-a, grown without SK channel modulators, were soaked in the crystallization solution supplemented with saturating amount of 1-EBIO. The structure of the protein complex with 1-EBIO was resolved at the resolution of 1.7 Å. Again, there exists extra electron density, at the same location of the CaM N-lobe (e.g. Fig. 1a), but not the CaM C-lobe (Fig. 3a and Supplementary Fig. S2a, Table 1). Likewise, no significant global changes are observed in the structure of the CaM-CaMBD2-a complex. The 1-EBIO binding pocket consists of A477, L480 and V481 of CaMBD2-a and F19, I27, L32, M51, I52, V55, I63, F68, M71 and M72 of CaM (within the 5 Å radius of 1-EBIO), virtually identical to the PHU binding pocket (Fig. 3b). Compared to the structure without 1-EBIO (PDB accession code 1G4Y), several of these residues display different spatial orientations, most noticeably A477 and L480 of CaMBD and F19, M51, V55 and M71 of CaM (Fig. 3a). For instance, in the absence of 1-EBIO or PHU, M71 of CaM forms close contacts with A477 and L480 of CaMBD (Supplementary Fig. S2b). Such interactions are completely disrupted by either 1-EBIO or PHU, leading to formation of the binding pocket for 1-EBIO or PHU (Fig. 3c and Supplementary Figs. S2c and S2d). Depending on whether the ligand is PHU or 1-EBIO, these amino acids may adopt different conformations, e.g. L480, F68 and M71, (Fig. 3b). Furthermore, the benzene ring of 1-EBIO adopts a different spatial orientation compared to that of PHU (Fig. 3d). Unlike PHU, 1-EBIO does not show flip-flop within its binding pocket.
Figure 3. The CaM-CaMBD interface harbors the binding pocket for 1-EBIO.
(a) Difference Fourier electron density map constructed using mFo–DFc coefficients calculated in PHENIX prior to modeling 1-EBIO and side chain rearrangements that occur in the binding site. The map is contoured at 2.2s and displayed within a 6 Å sphere of 1-EBIO. The map is overlaid with the final coordinates for 1-EBIO and its surrounding amino acid residues in the CaM-CaMBD2-a complex. Marine are amino acid residues from CaMBD2-a and salmon are amino acid residues from CaM. Coordinates of the same amino acid residues (gray) from the original CaM-CaMBD2-a complex (without 1-EBIO, 1G4Y) are provided for comparison. (b) Comparison of key amino acid residues which form the binding pocket for PHU (orange for CaMBD residues and purple for CaM residues) and that for 1-EBIO (marine for CaMBD residues and salmon for CaM residues). (c) Formation of the 1-EBIO binding pocket at the CaM-CaMBD interface. 1-EBIO disrupts the interaction between M71 of CaM (salmon) and A477/L480 of CaMBD (marine). (d) Overlay of the coordinates for 1-EBIO (yellow) and PHU (Cyan) obtained from their respective protein crystal structures.
Predicted binding pockets for other 1-EBIO compounds
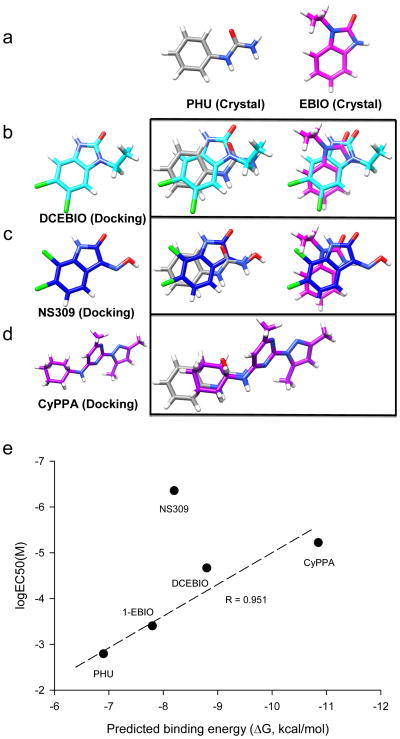
Poor solubility of NS309 and CyPPA in water made it difficult to get protein crystals with these compounds. We therefore turned to molecular docking and molecular dynamics (MD) simulations to test whether DCEBIO, NS309 and CyPPA might also fit into the same binding pocket for PHU and 1-EBIO. Both molecular docking and MD simulations were performed using protein coordinates from the PHU-bound CaM-CaMBD structure after removal of PHU. The Generalized-Born with molecular volume (GBMV) implicit solvent model was used to calculate the linear interaction energy (LIE(GBMV)) to score individual poses obtained from docking and MD modeling38–39. We started by testing whether molecular docking and MD simulations would correctly dock both PHU and 1-EBIO in their respective native conformations determined from the protein crystals (Figs. 1 and 3). Both rigid-receptor and “flexible-receptor” approaches were used to characterize putative binding modes for PHU and 1-EBIO. Rigid-receptor docking is able to successfully re-dock PHU into the PHU pocket with the difference between the experimental and predicted poses of 1.30 Å rmsd (root-mean square deviation, calculated over 10 heavy atoms). Likewise, 1-EBIO is able to dock into the PHU pocket with low energy unstrained ligand geometries and favorable LIE(GBMV) scores. The difference of the experimental and predicted poses of 1-EBIO is 2.44 Å rmsd (calculated over all 12 heavy atoms), which arises primarily from the symmetry related conformation of the ethyl group of 1-EBIO (5.50 Å rmsd), while the 10 atoms of the fused ring system only differ by 1.05 Å rmsd.
Unlike 1-EBIO, the rigid-receptor docking does not work well in docking both di-chloral compounds (DCEBIO and NS309) into the PHU pocket. The top-ranked binding geometries of these ligands exhibited strained ligand conformations, particularly, the clashes of both ligands with F68, M71 and M72 of CaM, consistent with the observation of the crystal structures that F68 and M71 change their conformation when the ligand is 1-EBIO instead of PHU (Fig. 3b). Receptor ensemble docking (RED) was performed using conformations generated from both MD simulations and from low energy rotamers built from the PHU-bound crystal structure40. Top-ranked ligand pose clusters were identified for each receptor conformation using the LIE(GBMV) scores. Comparison of top-ranked clusters across the ensemble of receptor conformations demonstrated a reasonable consensus ligand binding geometry for DCEBIO and NS309, such that at least three of the top five ranked clusters were within 1.0 Å heavy atom rmsd from each other. Compared to NS309 and DCEBIO, CyPPA is a much bigger molecule. Nevertheless, rigid-receptor docking works reasonably well and is able to dock CyPPA into the PHU binding pocket. Fig. 4 illustrates the predicted conformations for DCEBIO (Fig. 4b), NS309 (Fig. 4c) and CyPPA (Fig. 4d) and comparison of these conformations to those of PHU and 1-EBIO obtained from crystals (Fig. 4a). We also examined whether the NS309 tautomers might have different conformations when docked in the PHU pocket. All three tautomers are predicted to have the same conformation in the PHU pocket (Supplementary Fig. S3). The top-ranked poses of these compounds are further evaluated by a regression-based pair potential (S2 scoring) to predict their binding energy. Qualitatively, there is a positive correlation of the predicted binding energy and the EC50 for each compound in their potentiation of the SK2 channel activities (Fig. 4e). Among the five compounds, NS309 is clearly an exception in that it is over 100-fold more potent than the prediction (see discussion).
Figure 4. Other compounds dock into the same pocket as PHU and 1-EBIO.
(a) Conformations of PHU and 1-EBIO determined from the crystal structures. They serve as reference conformations. (b) The predicted conformation of DCEBIO in the binding pocket (left), which is superimposed with the conformations of PHU or 1-EBIO (right). (c) The predicted conformation of NS309 in the binding pocket (left), which is superimposed with the conformations of PHU or 1-EBIO (right). (d) The predicted conformation of CyPPA in the binding pocket (left), which is superimposed with the conformations of PHU or 1-EBIO (right). (e) Correlation of the predicted binding free energies of the five ligands using the S2 scoring function and the EC50s of these compounds in potentiation of SK2 channels. The EC50 for NS309 deviates from the prediction.
The CaM-CaMBD interface is the functional binding pocket
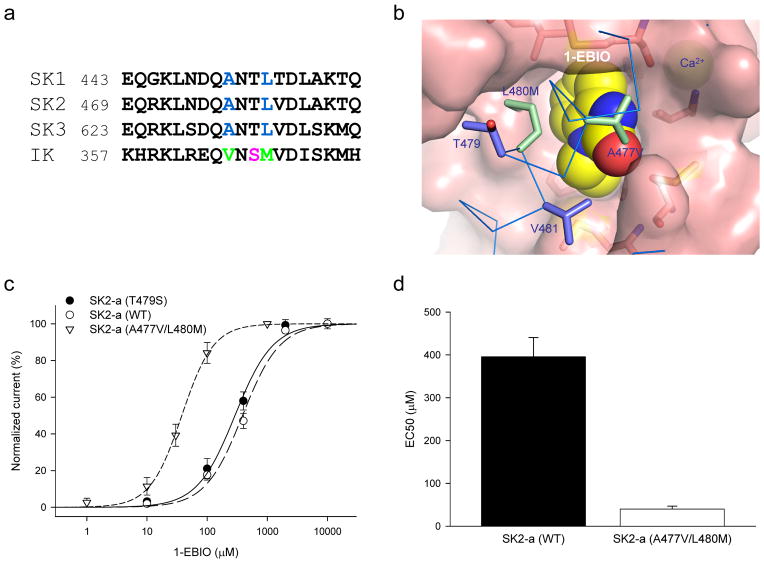
Site-directed mutagenesis was carried out to determine whether the CaM-CaMBD interface at the CaM N-lobe, identified from our structural data (Figs. 1 and 3) or predicted by molecular docking (Fig. 4), represents the functional binding pocket for compounds of the 1-EBIO class. Previous studies show that 1-EBIO is more potent in potentiating the IK channel activity than that of SK channels11,33. In the CaM-CaMBD2-a complex, the channel fragment, E469 – Q487, interacts with the CaM N-lobe in the presence of Ca2+8,10. Sequence alignment of IK and SK shows that of these 19 amino acids 11 are different between IK and SK2, including A477, L480 and V481, implicated in formation of the binding pocket for PHU or 1-EBIO (Figs. 1, 3, 5a and 5b). Mutagenesis studies were performed to test whether mutant channels might respond differently to potentiation by 1-EBIO or other channel modulators. A double mutant, A477V/L480M, was created and its responses to 1-EBIO were tested. A second mutant, T479S, was generated to test whether amino acids, which are different between IK and SK2 but are not involved in formation of the binding pocket for 1-EBIO (Fig. 3b), might have any impact on the effects of 1-EBIO.
Figure 5. A477V/L480M mutation increases the potency of 1-EBIO.
(a) Sequence alignment of the fragments (equivalent of E469 to Q487 of SK2-a) of CaMBD at the CaM N-lobe from SK1, SK2, SK3 and IK channels. A477, L480 and V481 are implicated in formation of the binding pocket for the channel modulators from the structure data. Used in the alignment are the human SK1, SK3 and IK channel sequences and the rat SK2 channel sequence. (b) A space-filled model of the A477V/L480M mutation (green) with 1-EBIO in the binding pocket. Changes are made based on the structure data using Pymol. Note that T479 points away from 1-EBIO. (c) Dose-response curves for potentiation by 1-EBIO of the SK2 channel activities from WT (n = 5), T479S (n = 4) and A477V/L480M (n = 4). 200 nM Ca2+ was present in all 1-EBIO solutions. (d) A477V/L480M mutation significantly increases the potency of 1-EBIO (10-fold). All data are mean ± s.e.m.
Both A477V/L480M and T479S mutants are activated in response to application of 200 nM Ca2+. Their responses to application of 1-EBIO, however, differ from each other significantly. The EC50 for potentiation of A477V/L480M by 1-EBIO is 40.0 ± 7.0 μM (n = 4), a 10-fold reduction compared to that of WT SK2-a (p < 0.001, T-test, Figs. 5c and 5d). In contrast, the EC50 for potentiation of T479S remains the same as that of WT (EC50 = 316.8 ± 40 μM, n = 4, p = 0.245, T-test, Fig. 5c), consistent with our structure data that T479 is not part of the binding pocket (Figs. 1, 3 and 5). While A477V/L480M becomes more sensitive to potentiation by 1-EBIO, there is no change in its Ca2+-dependent activation (EC50 = 0.32 ± 0.01 μM, n = 3, Supplementary Fig. S4). The unique role of A477 was further tested by additional mutants with the Ala residue replaced by much bulkier amino acid residues, such as Ile or Leu. We reasoned that bulkier residues at position 477 might clash with the channel modulators and therefore interfere with the interaction between the channel modulators and their binding pocket (e.g. Figs. 6a and 6b).
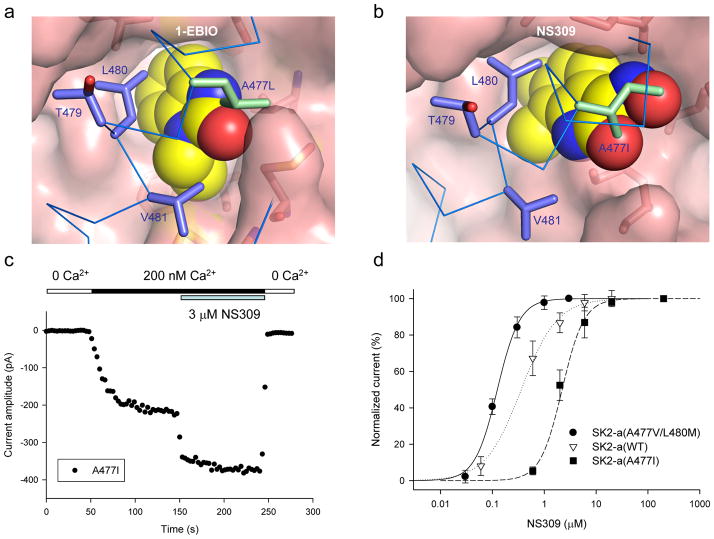
Figure 6. Effects of replacing A477 with bulky amino acid residues.
(a) A space-filled model of the A477L mutation (green) with 1-EBIO in the binding pocket. The Ala residue at position 477 is changed to Leu based on the structure data using Pymol. Introduction of such a bulky amino acid residue may interfere with the interaction of 1-EBIO and its binding pocket. (b) A space-filled model of the A477I mutation (green) with NS309 in the binding pocket. NS309 is docked to the binding pocket using MD modeling from the structure of the 1-EBIO-bound CaM-CaMBD2-a complex. The A477I change is then made using Pymol. (c) Responses of the A477I mutant, from an inside-out patch, to sequential applications of solutions containing (1) 0 Ca2+, (2) 200 nM Ca2+, (3) 3 μM NS309 with 200 nM Ca2+, and (4) 0 Ca2+ (as indicated). The current amplitudes were measured at −90 mV. (d) Dose-response curves for potentiation by NS309 of the SK2 channel activities from WT (n = 3), A477V/L480M (n = 4) and A477I (n = 3). 200 nM Ca2+ was present in all NS309 solutions. On average, the A477V/L480M mutation shifts the NS309 dose-response curve to the left by 3.4-fold, while the A477I mutation results in a 5-fold shift of the NS309 dose-response curve to the right. All data are mean ± s.e.m.
Both A477L and A477I were activated upon application of 200 nM Ca2+ (e.g. Fig. 6c). A477L was tested for its response to 1-EBIO in the presence of 200 nM Ca2+. Even at the saturating concentration of 1-EBIO, we were not able to obtain the maximal response by A477L, indicating that 1-EBIO becomes less potent in potentiation of the A477L mutant (data not shown). We next tested the effects of NS309, a more potent modulator, on these mutants. A477I becomes less responsive to potentiation by NS309 as compared to WT (Figs. 6c and 6d). On average, there is a 5-fold increase in the EC50 for A477I (2.21 ± 0.39 μM, n = 3), compared to that of WT (0.44 ± 0.13 μM, n = 3, p = 0.013, T-test). In contrast, when NS309 was tested on A477V/L480M, the EC50 is significantly reduced (Fig. 6d), at 0.13 ± 0.009 μM (n = 4, p = 0.039, T-test, compared to WT). Like A477V/L480M, Ca2+ dependent activation of A477I remains the same as WT (EC50 = 0.33 ± 0.03 μM, n = 3). The results suggest that NS309 binds to the same pocket as 1-EBIO, supporting the prediction by molecular docking (Fig. 4). Collectively, mutations, generated based on the structure data, can effectively increase or decrease the potency of both 1-EBIO and NS309, supporting the notion that the binding site, identified by our structure data, is indeed a functional binding pocket for the compounds of the 1-EBIO class.
Targeting the CaM-channel interface alters specificity
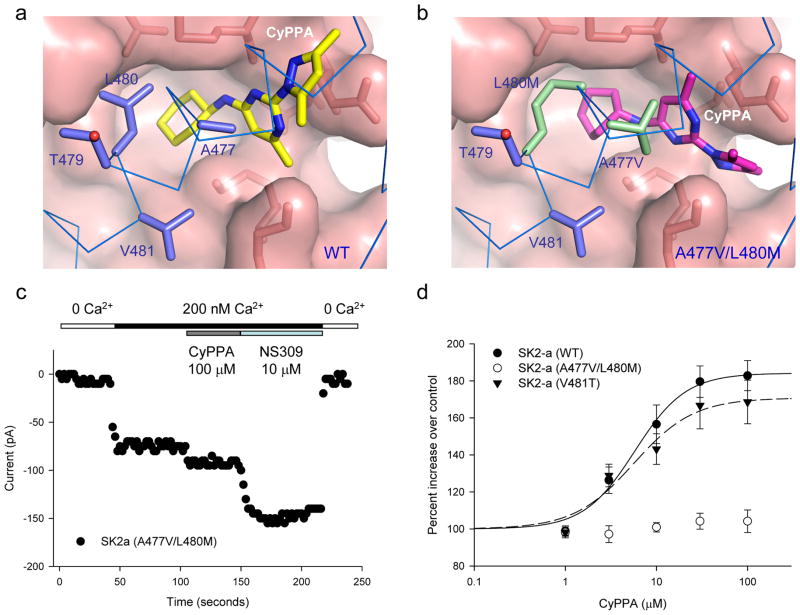
Compared to other compounds of the 1-EBIO class, CyPPA, a much bigger molecule (c.f. Fig. 1d and Supplementary Fig. S5a), is more selective, which potentiates SK2 and SK3 channels with little effect on SK1 or IK channels36. Molecular docking and MD simulations predicted that CyPPA could bind to the same binding pocket as 1-EBIO (Figs. 4 and 7a). It was further predicted that the A477V/L480M mutation might change the local environment of the binding pocket and force CyPPA to adopt a different conformation (Figs. 7b and Supplementary Fig. S5). Such predictions were tested by electrophysiology combined with mutagenesis. CyPPA is reasonably potent (EC50 = 6.04 ± 1.04 μM, n = 6, Fig. 2c). However, the response of A477V/L480M to CyPPA is distinctively different from that of 1-EBIO (Fig. 5c). A477V/L480M barely increases its current amplitude upon application of 100 μM CyPPA (Figs. 7c and 7d). Subsequent application of NS309 produces robust potentiation (Fig. 7c), indicating that A477V/L480M has virtually lost its response to CyPPA. In contrast, V481T yields an EC50 of 5.29 ± 1.01 μM (n = 3) for CyPPA, same as the WT (p = 0.666, T-test). The results again support the prediction, by molecular docking, that the binding site at the CaM-CaMBD interface, identified by our structure data, is the functional binding pocket for compounds of the 1-EBIO class. Furthermore, mutations A477V/L480M mimicks the selective effect of CyPPA on IK channels.
Figure 7. Responses of the A477V/L480M mutation to application of CyPPA.
(a) A space-filled model of CyPPA docked into the binding pocket. Molecular docking and MD simulation were performed to dock CyPPA into the PHU binding pocket based on the structure of PHU in the CaM-CaMBD2-a complex. (b) A space-filled model of CyPPA docked into the binding pocket with the A477V/L480M mutation (green). Molecular docking and MD simulations were performed to introduce the A477V/L480M mutation and then docking of CyPPA. Note that the A477V/L480M mutation is predicted to force CyPPA to adopt a different conformation in the pocket. (c) The A477V/L480M mutation on the effect of CyPPA. Responses of the A477V/L480M mutant, from an inside-out patch, to sequential application of solutions containing (1) 0 Ca2+, (2) 200 nM Ca2+, (3) 100 μM CyPPA with 200 nM Ca2+, (4) 10 μM NS309 with 200 nM Ca2+ and (5) 0 Ca2+. The current amplitudes were measured at −90 mV. Note that CyPPA, at 100 μM, barely potentiates the mutant channel activity. (d) Responses of SK2 channels, WT (n = 6), V418T (n = 3) and A477V/L480M (n = 5), to increasing concentrations of CyPPA. 200 nM Ca2+ was present in all CyPPA solutions. Note that there is little response from the A477V/L480M mutant at the CyPPA concentration range tested. All data are mean ± s.e.m.
DISSCUSSION
This study represents the first to demonstrate the location of the functional binding pocket for compounds of the 1-EBIO class. Several lines of evidence support this conclusion. First, PHU, a weak SK2 channel modulator, is found in the identical location at the CaM-CaMBD interface obtained from two CaMBD splice variants, CaMBD2-b and CaMBD2-a, under two different conditions to grow the protein crystals: co-crystallization or soaking of the naïve crystals in the PHU solution. Second, 1-EBIO is found to be at the same location as PHU in the CaM-CaMBD complex. Third, both PHU and 1-EBIO pockets are found at the CaM N-lobe, but not at the CaM C-lobe, despite the same hydrophobic nature of the CaM-CaMBD interface at the CaM C-lobe, indicating the specificity of the interaction between PHU/1-EBIO and their binding pockets. Forth, mutations of amino acids, which form the binding pocket, can change the potency of 1-EBIO, NS309 and CyPPA. Finally, mutations of amino acids not involved in formation of the binding pocket have the same responses to 1-EBIO or CyPPA as WT. The results are also consistent with a previous report that the channel C-terminus is responsible for the action of 1-EBIO37.
With few exceptions, compounds of the 1-EBIO class have low potency, with EC50s up to hundreds of micromolars in their potentiation of SK channels (see ref. 11 for a list of EC50s by different compounds). The low potency also contributes to the poor therapeutic index for these compounds2,34. Thus far, NS309 is the most potent SK/IK channel modulator, with the EC50 in the sub-micromolar range (Fig. 2c). Clearly, chemical modification of the compound has led to such a dramatic improvement in the potency41. Identification of the functional binding pocket allows us to evaluate whether the environment of the binding pocket may also play a role in determining the potency of compounds of the 1-EBIO class. Indeed, mutations, such as A477V/L480M and A477I, can significantly alter the potency of 1-EBIO or NS309 (e.g. Figs. 5 and 6). Molecular docking allows us to put DCEBIO, NS309 and CyPPA in the same binding pocket (Fig. 4), and the results of molecular docking are corroborated by effects of mutations on the effectiveness of NS309 and CyPPA (Figs. 6 and 7). Molecular docking and MD modeling further show, qualitatively, a correlation of improved interactions of the compounds with the binding pocket and their potency for PHU, 1-EBIO, DCEBIO and CyPPA (Fig. 4). NS309, however, is an exception, with its potency over 100-fold higher than the prediction (Fig. 4). NS309 is likely to bind to the same binding pocket as 1-EBIO, since both A477V/L480M and A477I mutants change the potency of NS309 (Fig. 6). Molecular docking puts NS309 in a somewhat different position from that of 1-EBIO (Fig. 4c). Three potential scenarios may explain why NS309 is an outlier in Fig. 4e. First, the NS309 binding pocket may include some additional amino acids from CaMBD and/or CaM, which do not contribute to formation of the PHU pocket or the 1-EBIO pocket used for molecular docking and MD simulations. Second, interactions of NS309 with its binding pocket may stabilize the CaM-CaMBD complex, as demonstrated by a recent study9. Such stabilizing effects are not reflected in our estimation of the binding energy (ΔG, Fig. 4e), which describes only the direct interaction between NS309 and the protein complex. Finally, it is also likely that different NS309 tautomers may have different effects on potentiation of the SK/IK channel activities, although this is a less likely scenario, since molecular docking shows that all three tautomers have the same conformation (Supplementary Fig. S3). Collectively, our results demonstrate that understanding of how compounds, such as 1-EBIO and NS309, interact with the binding pocket will shed light on how to improve the potency of the compounds of the 1-EBIO class.
Selectivity is another issue which hinders the development of more effective therapeutics targeting SK/IK channels11,28–32. With the exception of CyPPA, compounds of the 1-EBIO class as well as toxins, such as apamin, generally lack selectivity amongst SK1, SK2, SK3 and IK channels36. Lack of the knowledge of how compounds of the 1-EBIO class interact with their binding sites is likely to be a major roadblock to successful development of more selective therapeutics. The A477V/L480M mutation not only enhances the potency of 1-EBIO by 10-fold (Fig. 5), but also becomes almost insensitive to potentiation by CyPPA (Fig. 7), mimicking the profile of CyPPA on IK channels36. However, the results of the A477V/L480M mutation do not explain why SK1 channels fail to respond to CyPPA. Nevertheless, our results show the binding pocket, identified by our protein crystal structures, is at least partially responsible for the selectivity of CyPPA.
Protein-protein interactions and formation of macromolecular protein complexes are primary means of efficiency and specificity in cellular signaling42. Development of small molecular weight therapeutics targeting the interface of protein-protein interactions represents a new direction of drug discovery, with the biggest advantage of achieving selective actions on targets which are otherwise too common or widely distributed in cellular signaling43–46. Typically, the protein-protein interface is fairly large (~700–4000 Å2), and the size differences make it impossible for small molecules to completely block the protein-protein interactions like the traditional agonists/antagonists. Within the protein-protein interface, smaller regions, or “hotspots”, are responsible for a disproportionate contribution to the binding energy of the two interacting proteins and are targets for theraputics44–45. Our structure data show that the binding pocket for PHU and 1-EBIO includes L480 (Figs. 1 and 3), which is the anchor residue interacting with the CaM hydrophobic pocket8,10, an important hotspot for the CaM-CaMBD interaction47–48. Thus, compounds of the 1-EBIO class represent another example targeting the protein-protein interface with biological consequences. Understanding the nature of how 1-EBIO and its related compounds interact with the binding pocket will help develop compounds with better potency and selectivity targeting not only SK/IK channels but also other CaM-target protein complexes involved in Ca2+ dependent signaling47,49–51.
METHODS
Protein Expression and Purification
Briefly, rat CaM cDNA was cloned into pET-28b (Novagen), expressed in E. coli strain Rosetta2(DE3) (Novagen). CaM was purified using a low substitution phenyl sepharose fast flow column and an AKTA purifier (GE Healthcare). The codons of the CaMBD2-a were optimized for expression in E. coli and the synthetic gene was cloned into pET-28b. The sequence of CaMBD2-a used for expression is: MGRKLELTKAEKHVHNFMMDTQLTKRVKNAAANVLRETWLIYKNTKLVKKIDHAKVRKHQRKFLQAIHQLR SVKMEQRKLNDQANTLVDLAKTQLEHHHHHH. The His-tagged CaMBD2-a was expressed, solubilized with 0.2% w/v sarkosyl and purified on a nickel column (Qiagen). Both CaM and CaMBD2-a were subsequently purified a using Sephacryl S-100 high resolution gel filtration column (GE Healthcare). The protein concentrations were determined using predicted extinction coefficients. The CaM-CaMBD2-a protein complex was formed by slowly adding the CaMBD2-a to CaM (at a final ratio of 1:1). The complex was then purified using the gel filtration column (GE Healthcare) pre-equilibrated in a solution with 10 mM Tris-HCl, 150 mM NaCl, and 10 mM CaCl2 (pH 7.5). Fractions were collected and the purified proteins were concentrated to 1 mM and stored at −80°C.
Crystallization and structure determination
Protein crystals of the CaM-CaMBD2-a complex were grown in hanging drops by vapor diffusion at 20 °C 8,10. The complex (1mM) was mixed in a 1:1 ratio with the reservoir solution, which consists of 1.25 M Li2SO4, 0.5 M (NH4)2SO4, 0.1 M sodium citrate, pH 5.6. Monoclinic crystals grew within 3 weeks. These preformed protein crystals were then incubated with SK channel modulators at their saturating concentrations for two days. The channel modulator-soaked protein crystals were flash-cooled in liquid nitrogen for data collection, after a brief transfer to a suitable cryoprotectant (reservoir solution saturated with SK channel modulators, supplemented by 25% glycerol). X-ray diffraction data were collected from single protein crystals at Beamline of the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY) and at the SYBILS Beamline 12.3.1 at the Advanced Light Source (Berkeley, CA) and were processed using Xia252. The CaM-CAMBD2-a complex structure determined in the absence of SK channel agonists (1G4Y) 8 was used as a starting model to phase diffraction data. Solvent molecules were removed from the starting molecule prior to rigid body refinement against the channel modulator-soaked diffraction data. The crystallographic model was further constructed through iterative rounds of manual model building using Coot53 and crystallographic refinement using REFMAC554 and PHENIX55. The SK channel modulators were modeled in a hydrophobic pocket of the N-lobe of CaM based on strong electron density in difference Fourier maps and successful refinement of the coordinates. The current refined models contain CaM residues 2 to 147 with two Ca2+ ions, and SK channel residues 395 to 404 and 413 to 489. Crystallographic statistics for data collection and model refinement are summarized in Table 1 for both models. Structure graphics were created using PyMol (Schrödinger, LLC).
Molecular docking
CHARMM-based molecular docking is performed for an all-atom force field potential energy description of the protein-ligand complexes56–58. The Generalized-Born with molecular volume (GBMV) implicit solvent model was used in the calculation of linear interaction energy “LIE(GBMV)” scores to ensure physically rigorous evaluation of electrostatic potential energy components for the evaluation of top-ranked ligand poses as described previously38–39. A two-step scoring approach for CHARMM-based molecular docking was used, where the LIE(GBMV) scoring function was used for the identification of the top-ranked ligand pose geometry, and then a regression-based pair potential (S2) was used to predict binding affinities for ranking compounds59. Both rigid-receptor and “flexible-receptor” approaches were used to characterize putative binding modes for five ligands, PHU, 1-EBIO, DCEBIO, NS309 and CyPPA. Flexible receptor docking models were initially developed from the structure of PHU-bound CaM-CaMBD, and later compared to the 1-EBIO-bound structure. Short 100-ps molecular dynamics (MD) simulations were performed from the structure of PHU-bound CaM-CaMBD using the GBMV implicit solvent model38. Similar MD simulations were also performed from some top-ranked predicted binding modes of 1-EBIO, DCEBIO and NS309, to generate an ensemble of flexible receptor models. For the ensemble of flexible receptors derived from the PHU-bound structure and from MD simulations, 15 total conformations were used, each of which is generated from the final minimized structure from short 100-ps MD simulations of top-ranked binding posed of DCEBIO (5 conformations) and NS309 (10 conformations). An ensemble of rigid-receptor models were also constructed from the PHU-bound crystal structure using the most energetically favorable rotamers of M71 and M72, and F68 (backbone dependent DUNBRACK library)60. The rigid-receptor ensemble constructed from rotamers had 29 members, containing 9 rotamers of M71, 11 rotamers of M72, and 6 rotamers of F68, and finally, three low energy conformations formed from low energy optimized combinations of M71 and M72. Receptor ensemble docking (RED) was then performed assuming that each individual conformation was a rigid-receptor40. RED was performed using the ensemble of flexible receptor conformations generated from MD (15 conformations) and low-energy side chain rotamers built from the crystal structure (29 conformations). Molecular docking was also used to characterize the effect of binding of the ligands to mutations modeled from the PHU-bound structure using all energetically favorable rotamers (backbone dependent DUNBRACK library)60. Mutations, such as A477M/L480M, A477L and A477I, were also modeled using the PHU-bound crystal structure as previously described60.
Electrophysiology
Details can be found in our previous papers10. Briefly, both SK2-a and SK2-b channels (WT) along with CaM, were subcloned into the pCDNA3.1 expression vector (Invitrogen) respectively. Mutations were introduced into SK2-a using the QuickChange XL site-directed mutagenesis kit (Stratagene-Agilent) and subsequently confirmed by DNA sequencing. All SK channel modulators were from Tocris, except CyPPA which was from Sigma-Aldrich. WT and mutant channels, along with CaM and GFP, were expressed in TsA201 cells cultured in the Dulbecco’s modified Eagle’s medium (DMEM), with 10% fetal bovine serum and penicillin and streptomycin. A calcium phosphate method was used for transfection of SK2 cDNA (WT or mutants), together with CaM and GFP at a ratio of 5:2.5:1 (weight). Channel activities were recorded 1 – 2 days after transfection, with a Multiclamp 700B or a Axon200B amplifier (Molecular Devices) at room temperature. pClamp 10.2 (Molecular Devices) was used for data acquisition and analysis. The resistance of the patch electrodes ranged from 3–7 MΩ. The pipette solution contained: 140 mM KCl, 10 mM HEPES, 1 mM MgSO4, at pH 7.4. The bath solution contained: 140 mM KCl, and 10 mM HEPES, at pH 7.2. EGTA (1 mM), HEDTA (1 mM) were mixed with Ca2+ to obtain 0.2 μM free Ca2+ calculated using the software by Chris Patton of Stanford University (http://www.stanford.edu/~cpatton/maxc.html). The Ca2+ concentrations were verified using Fluo-4 and standard Ca2+ buffers (Molecular Probes). Currents were recorded using an inside-out patch configuration. For SK2-a and its mutants, the intracellular face was initially exposed to a zero-Ca2+ bath solution, and subsequently to bath solutions with 0.2 μM Ca2+. Currents were recorded by repetitive 1 sec-voltage ramps from −100 mV to +100 mV from a holding potential of 0mV. One minute after switch of bath solutions, ten sweeps, with a one-second interval, were recorded at concentrations for the channel modulators in the presence of 0.2 μM Ca2+. For SK2-b channel, a 0.5 μM Ca2+ solution was used instead of 0.2 μM, because of its reduced Ca2+ sensitivity for channel activation10. The integrity of the patch was examined by switching the bath solution back to the zero-Ca2+ buffer. Data from patches, which did not show significant changes in the seal resistance after solution changes, were used for further analysis. To construct the dose dependent potentiation of channel activities, the current amplitudes at −90mV in response to various concentrations of a channel modulator were normalized to that obtained at maximal concentration. The normalized currents were plotted as a function of the concentrations of each channel modulator. EC50s and Hill coefficients were determined by fitting the data points to a standard dose-response curve (Y = 100/(1 + (X/EC50)^−Hill)).
Statistical analysis
All data are presented in mean ± s.e.m. The Student’s T-test is used for data comparison.
Supplementary Material
Acknowledgments
We would like to thank Drs. Spike Horn and Pat Loll for their encouragement and helpful discussions; the structural facility of the Kimmel Cancer Center of Thomas Jefferson University for access of equipment in initial protein crystal screening and initial in-house X-ray diffraction; staff at the Beamline facility (X29A and X6A) of the Brookhaven National labs and the SYBILS Beamline 12.3.1 at the Advanced Light Source (Berkeley, CA) for assistance with collection of X-ray diffraction data. The work is supported by grants from NIH to JFZ (R01MH073060 & R01NS39355).
Footnotes
Author contributions
M.Z. and J.M.P. did experiments of X-ray crystallography and the structure determination. M.S. and R.S.A performed molecular docking and MD modeling. M.Z. and J.F.Z. carried out experiments of electrophysiology and mutagenesis. M.Z., J.M.P., R.S.A. and J.F.Z. prepared the manuscript and the figures.
Competing financial interests
The authors declare no competing financial interests.
Accession codes
The atomic coordinates have been deposited in the Protein Data Bank under accession codes 3SJQ, 4G27 and 4G28 for the CaM-CaMBD2-b complex, PHU-bound CaM-CaMBD2-a complex and the 1-EBIO-bound CaM-CaMBD2-a complex, respectively.
Additional information
Supplementary information is available online.
REFFERENCES
- 1.Kohler M, et al. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 2.Faber ESL, Sah P. Functions of SK channels in central neurons. Clin Exp Pharmacol Physiol. 2007;34:1077–1083. doi: 10.1111/j.1440-1681.2007.04725.x. [DOI] [PubMed] [Google Scholar]
- 3.Stocker M. Ca2+-activated K+ channels: molecular determinants and function of the SK family. Nat Rev Neurosci. 2004;5:758–770. doi: 10.1038/nrn1516. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, et al. Molecular Identification and functional roles of a Ca2+ activated K+ channel in human and mouse hearts. J Biol Chem. 2003;278:49085–49094. doi: 10.1074/jbc.M307508200. [DOI] [PubMed] [Google Scholar]
- 5.Adelman JP, Maylie J, Sah P. Small-conductance Ca2+-activated K+ channels: Form and function. Annu Rev Physiol. 2012;74:245–269. doi: 10.1146/annurev-physiol-020911-153336. [DOI] [PubMed] [Google Scholar]
- 6.Xia XM, et al. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature. 1998;395:503–507. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- 7.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher MA, Rivard AF, Bachinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 9.Li W, Halling DB, Hall AW, Aldrich RW. EF hands at the N-lobe of calmodulin are required for both SK channel gating and stable SK-calmodulin interaction. J Gen Physiol. 2009;134:281–293. doi: 10.1085/jgp.200910295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, et al. Structural basis for calmodulin as a dynamic calcium sensor. Structure. 2012;20:911–923. doi: 10.1016/j.str.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedarzani P, Stocker M. Molecular and cellular basis of small- and intermediate-conductance, calcium-activated potassium channel function in the brain. Cell Mol Life Sci. 2008;65:3196–3217. doi: 10.1007/s00018-008-8216-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor MS, et al. Altered expression of small-conductance Ca2+-activated K+ (SK3) channels modulates arterial tone and blood pressure. Circ Res. 2003;93:124–131. doi: 10.1161/01.RES.0000081980.63146.69. [DOI] [PubMed] [Google Scholar]
- 13.Feng J, et al. Calcium-activated potassium channels contribute to human coronary microvascular dysfunction after cardioplegic arrest. Circulation. 2008;118:S46–51. doi: 10.1161/CIRCULATIONAHA.107.755827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garland CJ. Compromised vascular endothelial cell SKCa activity: a fundamental aspect of hypertension? Br J Pharmacol. 2010;160:833–835. doi: 10.1111/j.1476-5381.2010.00692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy N, et al. Role of Ca2+-sensitive K+ currents in controlling ventricular repolarization: possible implications for future antiarrhytmic drug therapy. Curr Med Chem. 2011;18:3622–3639. doi: 10.2174/092986711796642463. [DOI] [PubMed] [Google Scholar]
- 16.Marrelli SP, Eckmann MS, Hunte MS. Role of endothelial intermediate conductance KCa channels in cerebral EDHF-mediated dilations. Am J Physiol Heart Circ Physiol. 2003;285:H1590–1599. doi: 10.1152/ajpheart.00376.2003. [DOI] [PubMed] [Google Scholar]
- 17.Si H, et al. Impaired endothelium-derived hyperpolarizing factor-mediated dilations and increased blood pressure in mice deficient of the intermediate-conductance Ca2+-activated K+ channel. Circ Res. 2006;99:537–544. doi: 10.1161/01.RES.0000238377.08219.0c. [DOI] [PubMed] [Google Scholar]
- 18.McNeish AJ, Dora KA, Garland CJ. Possible role for K+ in endothelium-derived hyperpolarizing factor-linked dilatation in rat middle cerebral artery. Stroke. 2005;36:1526–1532. doi: 10.1161/01.STR.0000169929.66497.73. [DOI] [PubMed] [Google Scholar]
- 19.Brahler S, et al. Genetic deficit of SK3 and IK1 channels disrupts the endothelium-derived hyperpolarizing factor vasodilator pathway and causes hypertension. Circulation. 2009;119:2323–2332. doi: 10.1161/CIRCULATIONAHA.108.846634. [DOI] [PubMed] [Google Scholar]
- 20.Sheng J-z, Ella S, Davis MJ, Hill MA, Braun AP. Openers of SKCa and IKCa channels enhance agonist-evoked endothelial nitric oxide synthesis and arteriolar vasodilation. FASEB J. 2009;23:1138–1145. doi: 10.1096/fj.08-120451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler R. Single-nucleotide polymorphisms in vascular Ca2+-activated K+-channel genes and cardiovascular disease. Pflügers Arch Eur J Physiol. 2010;460:343–351. doi: 10.1007/s00424-009-0768-6. [DOI] [PubMed] [Google Scholar]
- 22.Allen D, et al. SK2 channels are neuroprotective for ischemia-induced neuronal cell death. J Cereb Blood Flow Metab. 2011;31:2302–2312. doi: 10.1038/jcbfm.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chou CC, Lunn CA, Murgolo NJ. KCa3.1: target and marker for cancer, autoimmune disorder and vascular inflammation? Expert Rev Mol Diagn. 2008;8:179–187. doi: 10.1586/14737159.8.2.179. [DOI] [PubMed] [Google Scholar]
- 24.Chandy KG, et al. Isolation of a novel potassium channel gene hSKCa3 containing a polymorphic CAG repeat: a candidate for schizophrenia and bipolar disorder? Mol Psychiatry. 1998;3:32–37. doi: 10.1038/sj.mp.4000353. [DOI] [PubMed] [Google Scholar]
- 25.Ritsner M, et al. An association of CAG repeats at the KCNN3 locus with symptom dimensions of schizophrenia. Biol Psychiatry. 2002;51:788–794. doi: 10.1016/s0006-3223(01)01348-8. [DOI] [PubMed] [Google Scholar]
- 26.Ellinor PT, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyama K, et al. The intermediate-conductance calcium-activated potassium channel KCa3.1 contributes to atherogenesis in mice and humans. J Clin Invest. 2008;118:3025–3037. doi: 10.1172/JCI30836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girault A, et al. Targeting SKCa channels in cancer: potential new therapeutic approaches. Curr Med Chem. 2011;19:697–713. doi: 10.2174/092986712798992039. [DOI] [PubMed] [Google Scholar]
- 29.Blank T, Nijholt I, Kye MJ, Spiess J. Small conductance Ca2+-activated K+ channels as targets of CNS drug development. Curr Drug Targets - CNS & Neurol Disorders. 2004;3:161–167. doi: 10.2174/1568007043337472. [DOI] [PubMed] [Google Scholar]
- 30.Liegeois J, et al. Modulation of small conductance calcium-activated potassium (SK) channels: A new challenge in medicinal chemistry. Curr Med Chem. 2003;10:625–647. doi: 10.2174/0929867033457908. [DOI] [PubMed] [Google Scholar]
- 31.Wulff H, Zhorov BS. K+ channel modulators for the treatment of neurological disorders and autoimmune diseases. Chem Rev. 2008;108:1744–1773. doi: 10.1021/cr078234p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wulff H, et al. Design of a potent and selective inhibitor of the intermediate-conductance Ca2+-activated K+ channel, IKCa1: A potential immunosuppressant. Proc Natl Acad Sci. 2000;97:8151–8156. doi: 10.1073/pnas.97.14.8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devor DC, Singh AK, Frizzell RA, Bridges RJ. Modulation of Cl- secretion by benzimidazolones. I Direct activation of a Ca2+-dependent K+ channel. Am J Physiol. 1996;271:L775–784. doi: 10.1152/ajplung.1996.271.5.L775. [DOI] [PubMed] [Google Scholar]
- 34.Anderson NJ, Slough S, Watson WP. In vivo characterisation of the small-conductance KCa (SK) channel activator 1-ethyl-2-benzimidazolinone (1-EBIO) as a potential anticonvulsant. Eur J Pharmacol. 2006;546:48–53. doi: 10.1016/j.ejphar.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Sankaranarayanan A, et al. Naphtho[1,2-d]thiazol-2-ylamine (SKA-31), a new activator of KCa2 and KCa3.1 potassium channels, potentiates the endothelium-derived hyperpolarizing factor response and lowers blood pressure. Mol Pharmacol. 2009;75:281–295. doi: 10.1124/mol.108.051425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hougaard C, et al. Selective positive modulation of the SK3 and SK2 subtypes of small conductance Ca2+-activated K+ channels. Br J Pharmacol. 2007;151:655–665. doi: 10.1038/sj.bjp.0707281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedarzani P, et al. Control of electrical activity in central neurons by modulating the gating of small conductance Ca2+-activated K+ channels. J Biol Chem. 2001;276:9762–9769. doi: 10.1074/jbc.M010001200. [DOI] [PubMed] [Google Scholar]
- 38.Taufer M, Armen R, Chen J, Teller P, Brooks C. Computational multiscale modeling in protein--ligand docking. IEEE Eng Med Biol Mag. 2009;28:58–69. doi: 10.1109/MEMB.2009.931789. [DOI] [PubMed] [Google Scholar]
- 39.Armen RS, Chen J, Brooks CL., 3rd An evaluation of explicit receptor flexibility in molecular docking using molecular dynamics and torsion angle molecular dynamics. J Chem Theory Comput. 2009;5:2909–2923. doi: 10.1021/ct900262t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cavasotto CN, Kovacs JA, Abagyan RA. Representing receptor flexibility in ligand docking through relevant normal modes. J Am Chem Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 41.Pedarzani P, et al. Specific enhancement of SK channel activity selectively potentiates the afterhyperpolarizing current IAHP and modulates the firing properties of hippocampal pyramidal neurons. J Biol Chem. 2005;280:41404–41411. doi: 10.1074/jbc.M509610200. [DOI] [PubMed] [Google Scholar]
- 42.Scott JD, Pawson T. Cell Signaling in Space and Time: Where Proteins Come Together and When They’re Apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadek TR, Nicholas JB. Cell Signaling in space and time: Where proteins come together and when they’re apart. Science. 2003;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schön A, Lam SY, Freire E. Thermodynamics-based drug design: strategies for inhibiting protein-protein interactions. Future Med Chem. 2011;3:1129–1137. doi: 10.4155/fmc.11.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fry DC. Protein–protein interactions as targets for small molecule drug discovery. Peptide Sci. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 46.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein–protein interactions. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 47.Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA. The NMDA receptor NR1 C1 region bound to calmodulin: Structural insights into functional differences between homologous domains. Structure. 2007;15:1603–1617. doi: 10.1016/j.str.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osawa M, et al. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 49.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 50.Clapham DE. Calcium Signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 51.Hoeflich KP, Ikura M. Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 52.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 53.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 54.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks BR, et al. Charmm - a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 57.Roche O, Kiyama R, Brooks CL. Ligand-Protein DataBase: Linking protein-ligand complex structures to binding data. J Med Chem. 2001;44:3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 58.Vieth M, Hirst JD, Kolinski A, Brooks CL. Assessing energy functions for flexible docking. J Comput Chem. 1998;19:1612–1622. [Google Scholar]
- 59.Rahaman O, et al. Evaluation of several two-step scoring functions based on linear interaction energy, effective ligand size, and empirical pair potentials for prediction of protein-ligand binding geometry and free Energy. J Chem Inf Model. 2011 doi: 10.1021/ci1003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Armen RS, Schiller SM, Brooks CL., 3rd Steric and thermodynamic limits of design for the incorporation of large unnatural amino acids in aminoacyl-tRNA synthetase enzymes. Proteins. 2010;78:1926–1938. doi: 10.1002/prot.22706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.