Abstract
The present study proposed a two-step drug repositioning method based on a protein-protein interaction (PPI) network of two diseases and the similarity of the drugs prescribed for one of the two. In the proposed method, first, lists of disease related genes were obtained from a meta-database called Genotator. Then genes shared by a pair of diseases were sought. At the first step of the method, if a drug having its target(s) in the PPI network, the drug was deemed a repositioning candidate. Because targets of many drugs are still unknown, the similarities between the prescribed drugs for a specific disease were used to infer repositioning candidates at the second step. As a first attempt, we applied the proposed method to four different types of diseases: hypertension, diabetes mellitus, Crohn disease, and autism. Some repositioning candidates were found both at the first and second steps.
Keywords: Drug repositioning, Disease related genes, Drug target, Drug interaction, Substructure, Side effect, Protein-Protein interaction network
Background
Drug discovery and design is a time consuming process, which often requires a lengthy period. A drug prescribed for a specific disease can be also effective for another disease if the two diseases share a common pathophysiologic mechanism. To identify a new use of existing drugs is called drug repositioning, and this approach is gathering momentum because it can markedly shorten the time to obtain drug approval [1]. Recent advancements of biomedical informatics enable systematic search for candidates for drug repositioning.
There exist many works on systematic drug repositioning [1–4]. Methods for systematic drug repositioning can be divided into groups based on the way they find repositioning candidates. One approach seeks candidates based on the similarity of diseases. Chiang and Butte devised a systematic method, in which if two diseases share some similar therapies, then other drugs used for only one of the two are deemed to also be therapeutic for the other [2]. Another approach is based on the drug similarity. Yang and Agarwal proposed a repositioning method based on clinical side effects [3]. In their method, if the side effects associated with a drug are also induced by many of drugs for another disease, then the drug is deemed a candidate for the disease [3]. Supervised inference methods such as network based inference, which constructs a drug-target bipartite network, were applied to predict drug-target interactions and infer repositioning candidates [4]. However, a combination of both approaches is rarely examined.
In this context, we proposed a two-step method which combines the above approaches. First, the method evaluated a similarity between two diseases by seeking the genes shared by them. A protein-protein interaction (PPI) network was generated based on the shared genes. Then drugs prescribed for the diseases were obtained from a drug database. At the first step, if the target gene(s) of each of the obtained drugs was involved in the PPI network, the drug was deemed a candidate for repositioning. However, because targets of many drugs were still unknown, the second step inferred repositioning candidates based on the similarities between the prescribed drugs.
Methodology
In the proposed two-step method, first, lists of disease related genes were obtained from a meta-database called Genotator [5]. As a first attempt, we obtained the lists of disease related genes for four different types of diseases: hypertension (a disease of the circulatory system), diabetes mellitus (an endocrine, nutritional and metabolic disease), autism (mental and behavioral disorder) and Crohn disease (non-infective enteritis and colitis). Genotator calculates a likelihood score for each disease related gene [5]. For the sake of simplicity, we used only the top 100 related genes for each disease. Then we sought the genes shared by a pair of diseases for all possible combinations of the four. Associations between the shared genes for each pair were analyzed using STRING [6]. The resulting PPI networks were used to investigate whether they included target proteins of the drugs for the diseases obtained from DrugBank [7]. If the PPI network of the shared genes from a pair included the target protein of a specific drug for one disease in the pair, the drug could interact with other proteins in the network and thus, was deemed a repositioning candidate for the other disease of the pair. This was the first step of the proposed method.
At the second step, repositioning candidates were inferred based on the similarities among the drugs prescribed for one of the diseases in the pair because targets of many drugs were still unknown. From DrugBank, we extracted information on drug targets, drug interactions, substructures and side effects. Then a drug-similarity network was generated using the extracted information for the drugs prescribed for a disease. A node in the drug-similarity network represented the drug itself, a target, another drug interacted with the drug, a substructure, or a side effect. Targets, drug interactions, substructures and side effects were included in the network only when two or more drugs shared the features. The resulting network was visualized with Cytoscape [8]. If multiple nodes connected to a specific drug were included in the drug-similarity network of the other disease in the pair, the drug is deemed a repositioning candidate. It should be noted that even though nodes representing substructures were included in the drug-similarity network, the drug was not deemed a candidate unless a node representing another type of evidence was also included. This was because the variety of substructures was limited and accordingly, many drugs shared the same substructures.
All information from Genotator, STRING and DrugBank was obtained between late December 2012 and early January 2013.
Results & Discussion
The number of the shared genes between autism and Crohn disease was two, and no drugs prescribed for them had any of the genes as target. Similarly, the number of the shared genes between autism and diabetes mellitus was four, and none of them were the targets of the drugs for the two diseases. Thus, for these two combinations, no candidate was found at the first step. In contrast, 7, 22, 11 and 43 out of the 100 genes were shared between autism and hypertension, Crohn disease and diabetes mellitus, Crohn disease and hypertension, and diabetes mellitus and hypertension, respectively. Figure 1 illustrates the PPI networks for a) autism–hypertension, b) Crohn disease–diabetes mellitus, c) Crohn disease– hypertension, and d) diabetes mellitus–hypertension. Interestingly, almost all shared genes were included in the PPI networks, suggesting that the shared genes form closely interacting networks and each pair of diseases share common molecular mechanism(s).
Figure 1.
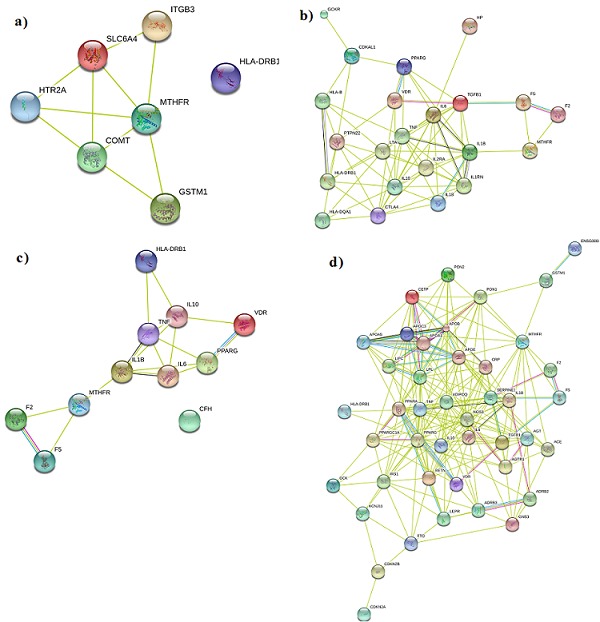
Protein-protein interaction networks of the disease related genes shared by pairs of diseases. a) autism-hypertension, b) Crohn disease-diabetes mellitus, c) Crohn diseaase-hypertension, and d) diabetes mellitus-hypertension
At DrugBank, we found three drugs used for both hypertension and diabetes mellitus, which shared the 43 genes. These three had the same drug target, ACE, which was included in the PPI network of the shared genes (Figure 1d). The drug-similarity network of the three is depicted in Figure 2a. The drugs were also connected to many common nodes representing other types of evidence. In this way, the three drugs shared many common features. These results suggested that the first and second steps could find repositioning candidates.
Figure 2.
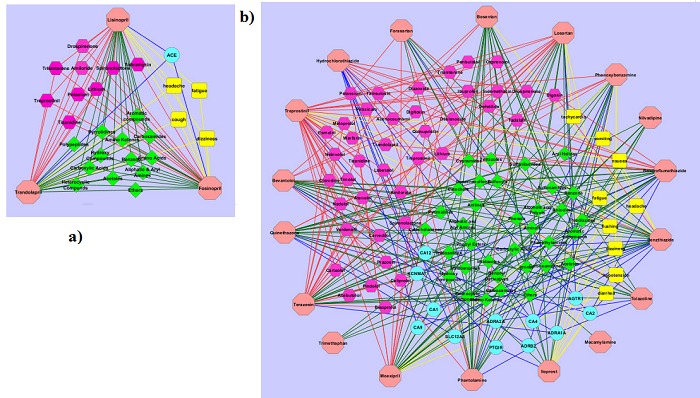
Drug-similarity networks. An octagon represents a drug prescribed for a specific disease. A circle, hexagon, diamond, and rectangle denote a drug target, a drug which interact the drug of interest, substructure, and side effect, respectively. a) Three drugs for both hypertension and diabetes mellitus, and b) drugs for hypertension.
Drugs extracted from DrugBank for diabetes mellitus included Voglibose, Pioglitazone, Gliquidone, Troglitazone, Tolbutamide, Glimepiride, Miglitol, Acarbose, Sitagliptin, Vildagliptin, and Rosiglitazone. Of these, Pioglitazone, Troglitazone, and Rosiglitazone had at least one drug target in the PPI network of the 43 shared genes. For example, one of the targets of Pioglitazone is peroxisome proliferator-activated receptor gamma (PPARG), which is involved in Figure 1d. Accordingly, this drug could be effective for hypertension. The drugs for hypertension included Bosentan, Treprostinil, Iloprost, Moexipril, Nilvadipine, Trimethaphan, Bevantolol, Benzthiazide, and Quinethazone. Of these, Moexipril and Bevantolol had at least one drug target in the network. In total, five drugs for diabetes mellitus had targets in the network while another five for hypertension had other targets in the network. These drugs were candidates for repositioning. Three, six, and one candidates were found for the combinations of autism–hypertension, Crohn disease–diabetes mellitus, and Crohn disease–hypertension, respectively at the first step.
Figure 2b depicts the drug-similarity network for hypertension. Drugs prescribed for autism, Crohn disease, and diabetes mellitus were examined whether they had multiple common features in the network as described above. At the second step, three, two, and nine candidates were found for the combinations of autism–hypertension, Crohn disease– hypertension, and diabetes mellitus–hypertension, respectively. None was found for the combination of Crohn disease and diabetes mellitus at the second step. Table 1 (see supplementary material) summarizes the repositioning candidates found at the first and second steps. If more stringent/weaker criteria were used to select candidates, the number of repositioning candidates would decrease/increase.
As shown in Figure 1d, the genes shared by diabetes mellitus and hypertension form a large, closely related network. This implied that drugs for one of them could be effective for the other. In fact, the number of the candidates found was the largest for this combination. Information on the functions of the shared genes could be useful to further narrow down the candidates. We used DAVID bioinformatics tools [9] to investigate the functions of the shared genes. The investigation revealed that the significantly enriched categories were: chemical homeostasis (GO BP term), positive regulation of lipid metabolic process (GO BP term), extracellular space (GO CC term), glycoprotein (UniProt keyword), regulation of foam cell differentiation (GO BP term), negative regulation of inflammatory response (GO BP term), positive regulation of catalytic activity (GO BP term), protein-lipid complex remodeling (GO BP term), regulation of steroid metabolic process (GO BP term), regulation of cholesterol transport (GO BP term), positive regulation of MAPKKK cascade (GO BP term), positive regulation of oxidoreductase activity (GO BP term), response to nutrient levels (GO BP term), regulation of cellular ketone metabolic process (GO BP term), and regulation of systemic arterial blood pressure mediated by a chemical signal (GO BP term). Drugs which interact these functions should be thoroughly investigated in the next step. However, it is beyond the scope of this study. Hypertension is occasionally found in patients with diabetes mellitus. Non-insulin dependent type diabetes mellitus is characterized by insulin resistance while hypertension is associated with abnormal glucose tolerance and an impairment of insulin action [10]. The enriched categories were related to lipid metabolism and glycoprotein, and these could be shared pathophysiological mechanisms. These facts also implied that drugs for one of them might be effective for the other.
Conclusion
In this study, we proposed a two-step method for drug repositioning based on the PPI network of genes shared by two diseases and the similarity of drugs prescribed for one of the two. At the first step, drugs having their targets in the PPI network of the shared genes were deemed repositioning candidates. Because targets of many drugs were still unknown, the similarities between the prescribed drugs for a specific disease were used to infer repositioning candidates at the second step.
As a first attempt, we applied the proposed two-step method to four different types of diseases: hypertension, diabetes mellitus, Crohn disease, and autism. Some repositioning candidates were found both at the first and second steps. Needless to say, further experimental investigations are required to verify whether the candidates can actually be repositioned. Because most of the systematic repositioning procedures were performed semi-automatically, we were able to investigate only four diseases. Also, the number of genes and drugs were limited because of the time consuming procedures. However, we believe that the basic idea of the proposed method was demonstrated through the investigation on the four diseases. We are planning to fully automatize the repositioning processes.
Supplementary material
Footnotes
Citation:Fukuoka et al, Bioinformation 9(2): 089-093 (2013)
References
- 1.Boguski MS, et al. Science. 2009;324:1394. doi: 10.1126/science.1169920. [DOI] [PubMed] [Google Scholar]
- 2.Chiang AP, Butte AJ. Clin Pharmacol Ther. 2009;86:507. doi: 10.1038/clpt.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Agarwal P. PLoS One. 2012;6:e28025. doi: 10.1371/journal.pone.0028025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng F, et al. PLoS Comput Biol. 2012;8:e1002503. doi: 10.1371/journal.pcbi.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wall DP, et al. BMC Med Genomics. 2010;3:article#50. doi: 10.1186/1755-8794-3-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szklarczyk D, et al. Nucleic Acids Res. 2011;39:D561. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knox C, et al. Nucleic Acids Res. 2011;39:D1035. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shannon P, et al. Genome Res. 2003;13:2498. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang da, et al. Nat Protoc. 2009;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 10.Simonson DC, et al. Postgrad Med J. 1988;64:39. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


