Abstract
During the course of the anti-infective drug discovery programme, actinomycete strain D25 was recovered from the Thar Desert soil, Rajasthan, India. Actinomycin type of compound isolated from the strain D25 showed promising activity against multi drug resistant and extensively drug resistant M. tuberculosis isolates. The present study reports the characteristics and phylogenetic status of the actinomycete strain D25. Phenotypic and cell wall characteristics revealed that the strain belongs to the genus Streptomyces. Further 16s rRNA analysis confined the genus Streptomyces with 97% similarity to the closely related species Streptomyces althioticus KCTC 9752. The 16s rRNA sequence was submitted to GenBank with the accession number JN604533.1. According to Bossard et al. (2003) strain D25 was found to be a novel species of the genus Streptomyces from Thar Desert soil, Rajasthan.
Keywords: Streptomyces, 16s rRNA, phylogenetic analysis, Thar Desert, antituberculous compound
Background
Actinomycetes are the well recognized as the richest source of bioactive compounds including clinically useful antibiotics, anti cancer agents and cell function modulators and hence of high pharmacological and commercial interest [1]. They are widely distributed in various normal and extreme ecosystems, due to their unparalleled ability to degrade wide range of complex substrates and withstand extreme physico-chemical conditions [2]. Based on the hypothesis “poorly researched habitats can offer better prospects for discovering new natural products”, actinomycetes from such habitats are currently in focus of considerable scientific interest [3]. Poorly explored ecosystems such as desert ecosystems have the potential to become a new resource for novel actinomycetes and chemical diversity [4, 5]. Identification of actinomycetes was sufficient earlier using morphological feature along with chemotaxonomy of many genera with few demerits [6, 7]. Now-a-days ribosomal DNA sequencing has been widely used in the identification of actinomycetes [8–10]. During our actinomycete bioprospecting programme, a pigmented anti-tuberculous compound producing actinomycete, designated strain D25, was isolated from Thar Desert soil, Rajasthan, India. A study, based on a combination of phenotypic and genotypic methods, reported here to determining the taxonomic position of strain D25.
Methodology
Phenotypic characterization:
The viability of the actinomycete strain D25 maintained in yeast extract malt extract (YEME) agar slants as well as in 30% glycerol broth. Micromorphological characteristics of strain D25 was studied by adopting slide culture method. The slides were observed under bright field microscope (Olympus) and Scanning electron microscope (JEOL model JSM5600LV). The recorded microscopic characteristics include presence of aerial mycelium, substrate mycelium, mycelial fragmentation, spore chain arrangement and spore surface morphology. Cultural characteristics of strain D25 were studied by inoculating the growth of actinomycete culture D25 into different ISP (International Streptomyces Project) medium. Cultural characteristics recorded include growth, consistency, aerial mass colour, presence of reverse side pigment and soluble pigment production. Effect of carbon and nitrogen sources were studied using basal medium supplemented with different sugars and amino acids respectively [11]. Effect of pH, temperature and sodium chloride on the growth of strain D25 was studied using ISP2 medium. Lipase, protease and amylase activities of the strain D25 were determined by adopting agar plate method using tween 80 agar, skim milk agar and starch agar, respectively. Antibiotic susceptibility pattern of strain D25 was determined against standard antibiotic discs by adopting disc diffusion method (Hi media).
Chemotaxonomy:
Isomers of diaminopimelic acid (DAP) and sugars in whole-cell hydrolysates of strain D25 were analyzed as described by Hasegawa et al. [12].
Molecular characterization:
The chromosomal DNA of actinomycete strain D25 was extracted using GENEI bacterial DNA purification kit. The DNA was pelleted and washed with 70% ethanol. The purified DNA obtained was suspended in TE buffer and stored at -200C. The PCR amplification of 16S ribosomal RNA gene was carried out at Kamini Research Foundation, Thuckalay, Nagercoil by following the methodology as described by Gothwal et al. [13] using Thermal cycler (Gene AMP 2720 – Applied Biosystem). Primers: Forward:'AACGGCTCACCAAGGCGACG 3'; Reverse: 5' GTACCGTCAAGGTGCCGCCC 3'. Reaction mixture used include sterile water - 38µl; 10x assay buffer -5µl; dNTPs mix (10mM each) - 3µl; Template DNA (20-30ng) - 1µl; Forward primer (100mM) - 1µl; Reverse primer (100mM) - 1µl; Taq poly (1U) -1µl. The amplified 16s rRNA gene was purified using 2% agarose gel prepared in TE buffer.
Sequencing of 16s rRNA:
Sanger Dideoxy chain Terminator sequencing was done in an automated DNA sequencer. The sequence detection was made by subjecting labeled ddNTP to a single capillary tube. DNA fragments of different colours were seperated by their respective sizes in a single electrophoretic gel. The sequences were read by determining the sequence of the colours emitted from specific peaks as they pass the detector. Colour of each dye represents the different nucleotides. Computer converted the data of emitted light in the nucleotide sequences.
16S rRNA sequence analysis and multiple sequence alignment:
The closely related homologs were identified through phylogenetic analysis by comparing the almost complete 16s rRNA gene sequence of strain D25 (1400 nt) with the nonredundant database of nucleotide sequences deposited at NCBI web server (www.ncbi.nlm.nih.gov), through Basic Local Alignment Tool (BLAST) program (http://www.ncbi.nim.nih.gov/blast/). Dataset of potential orthologs was prepared by considering those database sequences which had >98% sequence identity with the query sequence D25. Phylogenetic analysis and nucleotide conservation of the data set sequences were studied through multiple sequence alignment program viz., Mega 5.0 and clustering was calculated with the neighbour-joining method. The dendrogram/phylogram was prepared using method of phylogenetic tree construction. Distances between the studied sequences help in understanding the evolutionary distance among the species [14].
Criteria for species identification:
Identification of species was performed through sequence similarity basis following the criteria used by Bosshard et al. [15] which states the following selection parameters. 1) When the percentage similarity of the query sequence and the reference sequence is 99% or above, the unknown isolate is assigned to reference species; 2) when the percentage similarity is between 99-95%, the unknown isolate is assigned to the corresponding genus, and 3) when percentage similarity is less than 95%, the unknown isolate is assigned to a family.
Nucleotide accession number:
The 16s rRNA sequence of the potential actinomycete strain was submitted to Genbank to get the accession number.
Results
Phenotypic characteristics:
Under bright field microscopic observation, the vegetative substrate mycelium was lengthy and the reproductive aerial mycelium was dark and appeared in recti flexibile (RF) arrangement (Figure 1A). The aerial and substrate mycelium did not exhibit fragmentation. Smooth, oval shaped spores were borne in long straight to rectiflexibile chains with 40-50 spores in a filament (Figure 1B). Of a range of media tested, strain D25 grown well on ISP2, ISP3, ISP4, ISP6 and ISP7 medium and moderate growth was observed on ISP1, ISP5 medium. Strain D25 produced powdery colonies on all the ISP media. Growth pattern of actinomycete isolate D25 is as shown in Table 1 (see supplementary material). The strain D25 grown well utilising wide range of sugars and amino acids except on basal medium supplemented with sucrose, raffinose, cellulose and tyrosine. Growth of strain D25 was observed at a wide range of temperature (20°C – 40°C) at pH 7-11 and in the presence of 0 – 7.5% NaCl. Poor growth was observed at anaerobic condition. Strain D25 exhibited lipase, protease and amylase activity. Strain D25 was sensitive to streptomycin, tetracycline, chloramphenicol, gentamicin, vancomycin and erythromycin.
Figure 1.
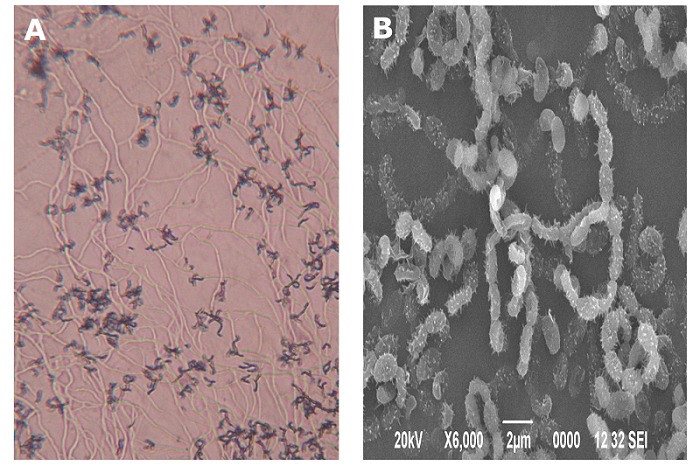
Micromorphology of actinomycete strain D25 under bright field (A) and scanning electron; (B) microscope
Chemotaxonomy:
The whole organism hydrolysates of strain D25 are rich in LLdiaminopimelic acid (DAP) and glycine. No diagnostic sugars are found in the cell wall constituents. Based on the studied phenotypic characteristics and cell wall analysis, actinomycete strain D25 was identified as the genus Streptomyces.
Molecular characterization:
The PCR amplification of 16s rRNA gene of actinomycete strain D25 yielded around 1400 base pair sequence. The BLAST analysis showed 97% similarity to Streptomyces althioticus KCTC 9752. Phylogenetic relationship of the strain D25 and related taxa are given in (Figure 2). Based on the criteria given by Bosshard et al. [15], the strain D25 was assigned to be a novel species of the genus Streptomyces though it showed sequence similarity between 95–99% with Streptomyces althioticus strain KCTC 9752. However, it is to be confirmed further by DNADNA hybridization tests. The 16s rRNA gene sequence of strain D25 is submitted to GenBank under the accession number JN604533.1.
Figure 2.
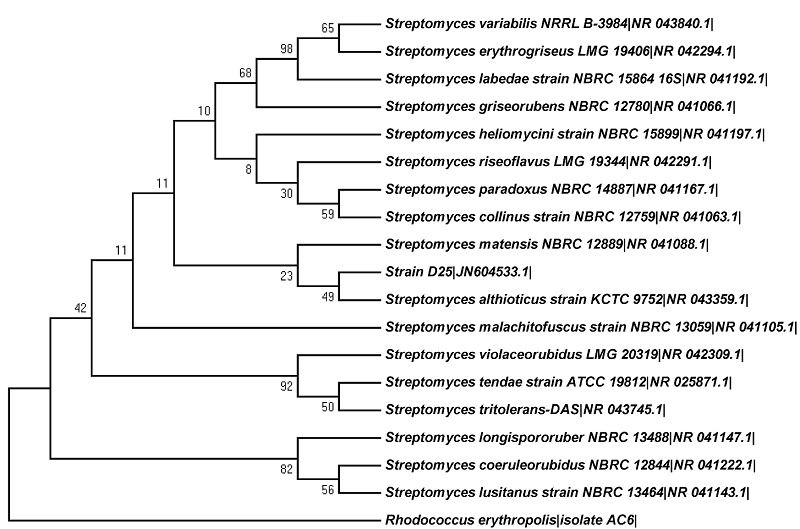
Phylogenetic relationship of the strain D25 and related taxa, based on 16s rDNA analysis. The evolutionary history was inferred using the Neighbor-Joining method [18]. The optimal tree with the sum of branch length = 0.19907131 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (100 replicates) is shown next to the branches [19]. The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The analysis involved 20 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1574 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [14].
Discussion
Actinomycete classification was originally based largely on morphological observations and physiological characteristics [11, 16]. The advent of chemotaxonomic criteria has provided reproducible and reliable tools to identify the genera at genus level. In the present study, the results of phenotypic characterisation and cell wall analysis revealed that the actinomycete strain D25 belongs to a species of the genus Streptomyces. But it is not adequate in itself to differentiate between genera. The analysis of 16s rRNA gene sequence has also revealed that the strain D25 belongs to a species of the genus Streptomyces. It showed only 97.0% similarity with the 16s rRNA gene sequence of its closely related species Streptomyces althioticus strain KCTC 9752 deposited in GenBank. However, the 16s rRNA gene offers limited resolution at the species level identification, since it uncovered more extensive genotypic differences than anticipated among many organisms with almost identical 16s rRNA gene sequences. At this level, whole genome sequencing and multiple gene derived parameters provide much more precise measurement of relatedness among organisms. One of the former parameters, the average nucleotide identity (ANI) [8] of all conserved genes between any two genomes shows potential to reform taxonomy because of its high precision and simplicity. Among several parameters tested, ANI was found to be the genome-derived parameter, that best co-relate with DNA: DNA hybridization (DDH) values; with 70% DDH, the most frequently used standard for species delineation [17], corresponding tightly to 95% ANI. Further, no organisms have been described to date that show less than 98.7% identity in their 16s rRNA gene and show less than 70% DDH or 95% ANI. These results enabled the substitution of cumbersome DDH and related procedures with portable and simple sequence based standard.
Based on the above literature evidence, the potential actinomycete strain D25 isolated from Thar desert is a novel species of the genus Streptomyces since its 16s rRNA gene sequence showed only 97.0% similarity with its phylogenetically close neighbour Streptomyces althioticus strain KCTC 9752. From the available literature, it was noticed that, till now there are no commercially available antibiotics, in particular the anti-TB compounds, produced from the strain Streptomyces althioticus strain KCTC 9752, the phylogenetically closely related species of strain D25. Hence, the actinomycete strain D25 isolated from Thar Desert soil may be a potential and novel source for the isolation of antituberculous compound.
Supplementary material
Acknowledgments
The authors thank the Department of Science & Technology, New Delhi for their support in the form of research grant (SR/SO/HS-42/2005). One of the authors (Dr. RB) thanks the Vice-chancellor of Periyar University for the laboratory facilities provided.
Footnotes
Citation:Radhakrishnan et al, Bioinformation 9(1): 018-022 (2013)
References
- 1.Butler MS. Nat Prod Rep. 2008;25:475. doi: 10.1039/b514294f. [DOI] [PubMed] [Google Scholar]
- 2.Berdy J. J Antibiot (Tokyo) 2005;58:1. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 3.Arumugam M, et al. Int J Syst Evol Microbiol. 2011;61:2664. doi: 10.1099/ijs.0.028258-0. [DOI] [PubMed] [Google Scholar]
- 4.Rateb ME, et al. J Nat Prod. 2011;74:1965. doi: 10.1021/np200470u. [DOI] [PubMed] [Google Scholar]
- 5.Nachtigall J, et al. J Antibiot (Tokyo) 2011;64:775. doi: 10.1038/ja.2011.96. [DOI] [PubMed] [Google Scholar]
- 6.Wang YM, et al. Int J Syst Evol Microbiol. 2001;51:467. doi: 10.1099/00207713-51-2-467. [DOI] [PubMed] [Google Scholar]
- 7.Yang R, et al. Int J Syst Evol Microbiol. 2008;58:1542. doi: 10.1099/ijs.0.65090-0. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinidis KT, JM Tiedje. Proc Natl Acad Sci. 2005;102:2567. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy TV, et al. Int J Syst Evol Microbiol. 2010;60:1755. doi: 10.1099/ijs.0.017749-0. [DOI] [PubMed] [Google Scholar]
- 10.Nimaichand S, et al. Int J Syst Evol Microbiol. 2012 [Google Scholar]
- 11.Shirling EB, Gottileb D. Int J Syst Bacteriol. 1966;16:313. doi:10.1099/00207713-16-3-313. [Google Scholar]
- 12.Hasegawa, et al. J Gen Appl Microbiol. 1983;29:319. doi.:10.2323/jgam.29.319. [Google Scholar]
- 13.Gothwal RK, et al. Electronic J Biotechnol. 2007;10:400. Doi:10.2225/vol10-issue3-fulltext-6. [Google Scholar]
- 14.Tamura, et al. Mol Biol Evol. 2011;28:2731. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosshard PP, et al. J Clin Microbiol. 2003;41:4134. doi: 10.1128/JCM.41.9.4134-4140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nonomura H. J Ferment Technol. 1974;52:78. [Google Scholar]
- 17.Stackbrandt E, et al. Int J Syst Evol Microbiol. 2002;52:1043. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 18.Saitou N, M Nei. Mol Biol Evol. 1987;4:406. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J, et al. Evolution. 1985;39:783. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


