Abstract
Cubilin, (CUBN; also known as intrinsic factor-cobalamin receptor [Homo sapiens Entrez Pubmed ref NM_001081.3; NG_008967.1; GI: 119606627]), located in the epithelium of intestine and kidney acts as a receptor for intrinsic factor – vitamin B12 complexes. Mutations in CUBN may play a role in autosomal recessive megaloblastic anemia. The current study investigated the possible role of CUBN in evolution using phylogenetic testing. A total of 588 BLAST hits were found for the cubilin query sequence and these hits showed putative conserved domain, CUB superfamily (as on 27th Nov 2012). A first-pass phylogenetic tree was constructed to identify the taxa which most often contained the CUBN sequences. Following this, we narrowed down the search by manually deleting sequences which were not CUBN. A repeat phylogenetic analysis of 25 taxa was performed using PhyML, RAxML and TreeDyn softwares to confirm that CUBN is a conserved protein emphasizing its importance as an extracellular domain and being present in proteins mostly known to be involved in development in many chordate taxa but not found in prokaryotes, plants and yeast.. No horizontal gene transfers have been found between different taxa.
Keywords: Cubilin, CUBN, Amino acid sequences, Phylogeny, Sequence alignment
Background
Cubilin (CUBN, also known as intestinal intrinsic factor receptor or intrinsic factor-cobalamin receptor or intrinsic factor-vitamin B12 receptor), acts as a co-transporter and helps in the uptake of lipoprotein, vitamin and iron. It functions as a transporter in many absorptive epithelia (intestine, renal proximal tubules and embryonic yolk sac) [1]. A potential role of mutations in CUBN gene has been hypothesized to play a role in the etiology of autosomal recessive megaloblastic anemia [2]. This hereditary condition is also known as MGA1 Norwegian type or Imerslund-Grasbeck syndrome. The disease is characterized by defective absorption of vitamin B12 and impaired function of the enzyme thymidine synthase; therefore, DNA synthesis, particularly during erythropoiesis is affected [1, 3]. Cubilin interacts with megalin in a calcium dependent manner by forming a dual-receptor complex called cubam which facilitates the uptake of specific ligands like hemoglobin, uteroglobin etc. [4]. In addition, CUBN also controls and facilitates endocytosis of various ligands [5, 6]. In the proximal tubule cells, cubilin helps in reabsorption of vitamin D binding protein from glomerular filtrates and assists in the synthesis of 1α,25-dihydroxyvitamin D(3) [7, 8]. In addition, a missense mutation in CUBN gene was found to alter the levels of [9].
The cubilin protein has 27 CUB domains and 7 EGF-like domains and is coded by the human CUBN gene is located on chromosome 10 (10p12.31; 11933 bp mRNA; Entrez PubMed ref NM_001081.3; NG_008967.1; GI:119606627) and expressed as a 3623 amino acid, 460 kDa receptor [10]. The acquisition of CUBN in eukaryotes indicates one of the important events in evolution of cells. Because of the inherent role of CUBN as a transporter in many epithelia and its ability to facilitate absorption of hemoglobin and other important ligands, we aimed to understand if variance in this gene exists among in various organisms using a phylogenetic analysis of published protein sequences.
Methodology
Data Set, Sequence Alignment and Construction of Phylogenetic Tree:
We queried the GenBank database [11] for all available protein sequences of the CUBN. The retrieved sequences were saved in FASTA format. An initial first-pass phylogenetic tree was constructed using Neighbour Joining method [12] (maximum sequence difference of 0.85) using Domain Enhanced Lookup Time Accelerated Basic Local Alignment Search Tool [DELTA BLAST] pairwise alignments between the query and the database sequences searched [13]. Grishin computation was used by the software to calculate the evolutionary distance between any two sequences. This was modeled as expected fraction of amino acid substitutions per site [14].
The list of sequences obtained after DELTA BLAST were narrowed down to delete sequences which are not CUBN and which were hypothetical. The concerned retrieved sequences were then aligned using Kalign software 2.03 version [15] with the results being produced in the Clustal W [16] format and employing a gap extension penalty of 0.85. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v3.0 aLRT) [17, 18]. The Jones-Taylor-Thornton (JTT) substitution model was selected along with gamma-distributed rate categories to account for rate heterogeneity across sites. The reliability for internal branch was assessed using the aLRT test (SH-Like). The graphical representation and edition of the phylogenetic tree were performed with TreeDyn (v198.3) [19]. The RAxML (Randomized Axelerated Maximum Likelihood) program version 7.2.8 was then used for sequential and parallel Maximum Likelihood of the sequences [20]. An additional analysis of the presence of horizontal gene transfer was performed using the T-REX online software [21].
Results
From the NCBI GenBank database, 588 sequences of CUBN covering the CUB domain (Figure 1) were used to construct a first-pass phylogenetic tree. The sequences were mostly from arthropods, placozoans, nematodes, tunicates, rabbits, hares, primates, rodents, placentals, odd toed ungulates, even toed ungulates, bivalves, lancelets, hemichordates, sea urchins, bony fishes, amphibians, birds, lizards, marsupials and monotremes. This tree however had many repetitive and unrelated sequences which were deleted. A high degree of sequence similarity of CUBN enzyme in many of the selected sequences was observed during phylogeny reconstruction. Putative conserved domains were observed in many taxa at the CUB domain CUB domain (cd00041); extracellular domain; present in proteins mostly known to be involved in development; not found in prokaryotes, plants and yeast [22]. The actual alignment after DELTA BLAST was detected with cd00041, Cd Length: 113 Bit Score: 139.08, E-value: 4.18e-37. The final accession information for the tested sequences (n=25) are presented in Table 1 (see supplementary material); multiple sequence alignment is presented in Appendix 1. Using the PhyML program a tree was constructed for these sequences, the results of which are presented in (Figure 2). RAxML revealed that there were 3782 distinct alignment patterns and the proportion of gaps and completely undetermined characters in this alignment was 24.21%. RAxML rapid bootstrapping and subsequent ML search showed an ML estimate of 25 per site rate categories (Figure 3). No horizontal gene transfers have been observed in the selected taxa.
Figure 1.
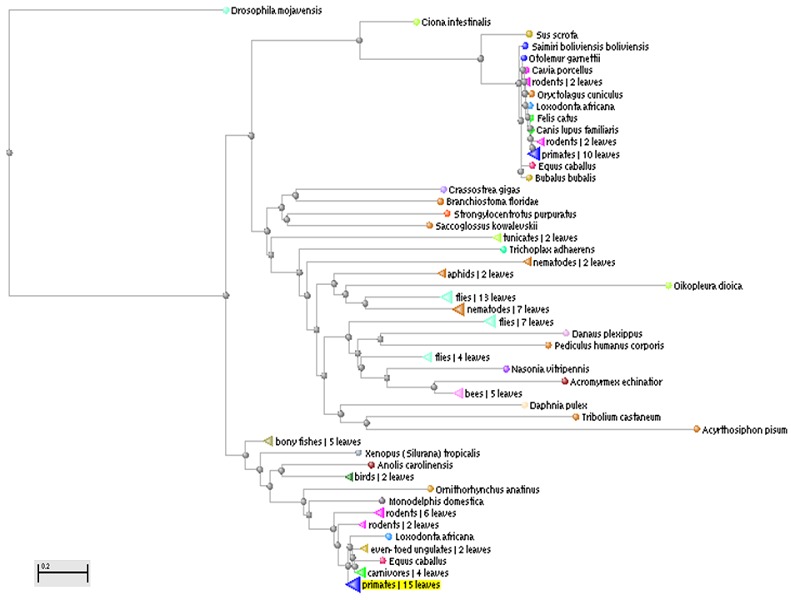
First pass phylogenetic tree constructed by multiple alignment using BLAST pair wise alignments: Results presented using Taxonomic name. First pass phylogenetic tree constructed showed that the CUBN sequences were mostly from arthropods, placozoans, nematodes, tunicates, rabbits, hares, primates, rodents, placentals, odd toed ungulates, even toed ungulates, bivalves, lancelets, hemichordates, sea urchins, bony fishes, amphibians, birds, lizards, marsupials and monotremes.
Figure 2.
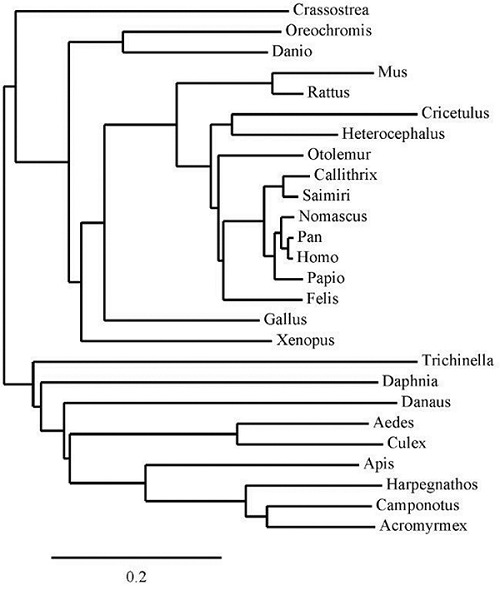
Phy ML: Phylogenetic tree of CUBN sequences. The final phylogenetic tree constructed using specific sequences revealed that the CUBN sequences were highly similar in most organisms. The closest similarity was observed in the primates.
Figure 3.
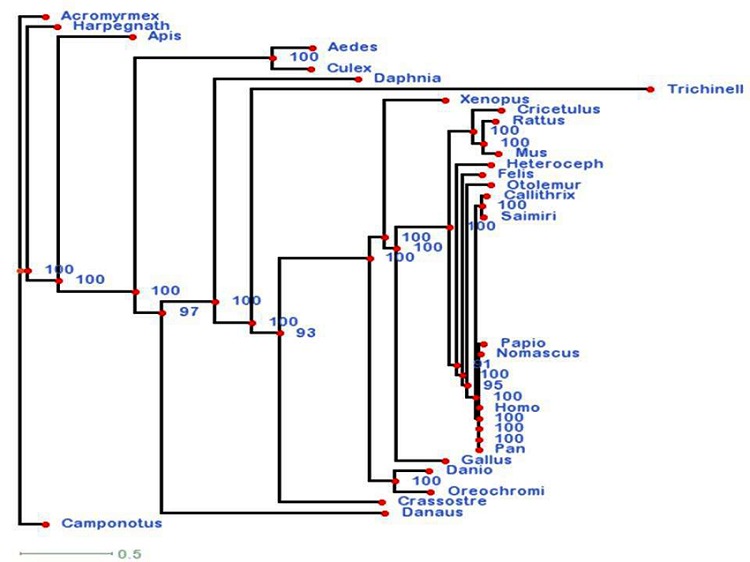
RAxML: Sequential and parallel maximum likelihood of sequences – best scoring ML tree. The values indicated here show the most scoring maximum likelihood tree constructed from the shortlisted organisms.
Discussion
The acquisition of cubilin and its importance in evolution was assessed by using a mix of programs to construct a phylogenetic tree [23, 24]. It is interesting to note that CUBN as a protein with receptor function has arisen only in the eukaryotes and thus not found in prokaryotes and plants. Cubilin is very helpful in the absorption of Vit B12 from the intestinal epithelia on which animals and protists are dependent for various biochemical pathways; however, they do not have the ability to synthesize it. It should be noted that plants and fungi neither produce Vit B12 nor use it but animals and protists use it but cannot synthesize it owing to the very complex de novo biosynthesis pathways [25]. The acquisition of cubilin in these phyla from an evolutionary stand point explains why mammalians have an efficient mechanism for uptake of extrinsic Vit B12.
We have used the neighbor-joining method to create trees based on multiple sequences in ClustalW; a JTT matrix method was used to cluster sequences at 85% identity level [26] followed by construction of a phylogenetic tree [18]. The PhyML software was used because it is an accurate but slightly faster than other phylogeny programs. DELTA-BLAST which uses a heuristic method to identify homologous sequences produced high scoring sequence alignment to generate a first pass phylogenetic tree from which relevant sequences were narrowed down. Kalign was used for multiple sequence alignment of shortlisted sequences because it is very fast, suitable for large alignments and concentrates on local regions providing insight to evolutionary relationships [18]. The RAxML software was used in inferring and validating the most scoring maximum likelihood tree [20]. In addition, using the T-Rex software, we have evaluated horizontal gene transfers in the selected organisms. It is well understood that this involves a transfer of genetic material from one lineage to another commonly found in prokaryotes as an adaptation mechanism to environment [21]. However, the selected organisms did not show the presence of this phenomenon. The final phylogenetic tree indicated that CUBN is a relatively new protein having evolved very late in evolution (Figure 2 & Figure 3). Also, no homologous gene transfer has been observed among the short-listed organisms which are indicative of the largely conserved domains within the amino acid sequences.
Conclusion
Acquisition of CUBN in relatively evolved organisms indicates its crucial role in the physiology of cells. A higher conservation at CUB domains indicates preserved function. The phylogenetic tree for CUBN revealed the presence of this gene in eukaryotes and indicated its importance in the biochemical pathways related to absorption of many ligands from the epithelial linings in cells re-emphasizing the importance of certain proteins in evolution.
Supplementary material
Acknowledgments
Authors would like to extend their appreciation to the Research Centre, College of Applied Medical Sciences and Deanship of Scientific Research at King Saud University for funding this research.
Footnotes
Citation:Shaik et al, Bioinformation 9(1): 029-036 (2013)
References
- 1.Kozyraki R, et al. Blood. 1998;91:3593. [PubMed] [Google Scholar]
- 2.Aminoff M, et al. Nat Genet. 1999;21:309. doi: 10.1038/6831. [DOI] [PubMed] [Google Scholar]
- 3.Moestrup SK, et al. J Biol Chem. 1998;273:5235. doi: 10.1074/jbc.273.9.5235. [DOI] [PubMed] [Google Scholar]
- 4.Fyfe JC, et al. Blood. 2004;103:1573. doi: 10.1182/blood-2003-08-2852. [DOI] [PubMed] [Google Scholar]
- 5.Kozyraki R, et al. Nat Med. 1999;5:656. doi: 10.1038/9504. [DOI] [PubMed] [Google Scholar]
- 6.Kozyraki R, et al. Proc Natl Acad Sci U.S.A. 2001;98:12491. doi: 10.1073/pnas.211291398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaseda R, et al. Ther Apher Dial. 2011;1:14. doi: 10.1111/j.1744-9987.2011.00920.x. [DOI] [PubMed] [Google Scholar]
- 8.Nykjaer A, et al. Proc Natl Acad Sci. U.S.A. 2001;98:13895. doi: 10.1073/pnas.241516998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Böger CA, et al. J Am Soc Nephrol. 2011;22:555. [Google Scholar]
- 10. http://ghr.nlm.nih.gov/gene/CUBN.
- 11.Benson DA, et al. GenBank Nucleic Acids Res. 2011;39:D32. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, et al. Mol Biol Evol. 1987;4:406. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Boratyn GM, et al. Biol Direct. 2012;7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grishin NV. J Mol Evol. 1995;41:675. doi: 10.1007/BF00175826. [DOI] [PubMed] [Google Scholar]
- 15.Lassmann T, et al. Nucleic acids Res. 2009;37:858. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomson JD, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dereeper A, et al. Nucleic Acids Res. 2008;1:W465. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dereeper A, et al. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevenet F, et al. BMC Bioinformatics. 2006;10:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stamatakis A, et al. Syst Biol. 2008;75:758. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 21.Boc A, et al. Nucleic Acids Res. 2012;40:W573. doi: 10.1093/nar/gks485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchler-Bauer A, et al. Nucleic Acids Res. 2011;39:225. [Google Scholar]
- 23.Wu CH, et al. Nucleic Acids Res. 2006;34:D191. [Google Scholar]
- 24.Strait DS, et al. J Hum Evol. 2004;47:399. doi: 10.1016/j.jhevol.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Roth JR, et al. Annu Rev Microbiol. 1996;50:137. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 26.Jones DT, et al. Comput Appl Biosci. 1992;8:275. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


