Abstract
Relative insulin deficiency, in response to increased metabolic demand (obesity, genetic insulin resistance, pregnancy and aging) lead to Type2 diabetes. Susceptibility of the type 2 diabetes has a genetic basis, as a subset of people with risk factors (obesity, Insulin Resistance, pregnancy), develop Type2 Diabetes. We aimed to identify ‘cluster’ of overexpressed genes, underlying increased beta cell survival in diabetes resistant C57BL/6J ob/ob mice (compared to diabetes susceptible BTBR ob/ob mice). We used ‘consensus’ overexpression status to identify ‘cluster’ of 11 genes consisting of Aldh18a1, Rfc4, Dynlt3, Prom1, H13, Psen1, Ssr4, Dad1, Anpep, Fam111a and Plk1. Information (biological processes, molecular functions, cellular components, protein-protein interactions/associations, gene deletion/knockout/inhibition studies) of all the genes in ‘cluster’ were collected by text mining using different literature search tools, gene information databases and protein-protein interaction databases. Beta cell specific function of these genes were also inferred using meta analysis tool of Beta Cell Biology Consortium, by studying the expression pattern of these genes in microarray studies related to beta-cell stimulation/injury, pancreas development and growth and cell differentiation. In the ‘clusters’, 6 genes (Dad1, Psen1, Ssr4, Rfc4, H13, Plk1) have a role in cell survival. Only Psen1 was previously identified to have role in successful beta cell compensation. We advocate these genes to be potentially involved in successful beta cell compensation and prevent T2D in humans, by conferring protection against diabetogenic insults.
Keywords: Obesity, Diabetes, metabolic load, Dad1, Psen1
Background
Insulin Secretion by beta cells, is essential for maintenance of glucose homeostasis. Higher insulin demand in response to metabolic stress (obesity, aging, pregnancy and genetic insulin resistance) requires an increased functional beta cell mass and/or enhanced insulin secretion capacity [1–4]. Beta cell mass is directly proportional to beta cell size and number. Beta cells number is dynamic and depends on balance of gain (proliferation, neogenesis) and loss (apoptosis) of beta cells [5]. Inadequate compensation of functional beta cell mass results in insulin insufficiency and Type 2 diabetes (T2D) [6]. Susceptibility of the T2D also depend on genetic factors, a subset of people with risk factors (obesity, Insulin Resistance) develop T2D, while others do not [7].
T2D at present is incurable and global prevalence is increasing every year [8]. There is an urgent need to identify new therapeutic targets for treatment as well as improvement of glycemic control in T2D. Increased functional beta cells, as well as ex vivo expansion of islet and prevention of apoptosis for better islet transplantation regimen are some of the newer approaches for T2D treatment [9]. Animal models of Obesity and T2D, have advanced understanding of the beta cell proliferation as well as survival, in response to insulin resistance (IR) and T2D [10]. C57BL/6J ob/ob mice and BTBR ob/ob mice are mouse models of obesity with different T2D susceptibility. BTBR ob/ob mice develop severe T2D (fasting blood glucose of 400mg/dL at 10 wks of age) while C57BL/6J ob/ob mice has mildly elevated blood glucose levels (<250mg/dL at all ages) [11]. These mice offer promises of novel insights into human T2D and thus are valuable model system to study link between Obesity/IR and T2D [12].
Role of growth factors (GLP-1, PTHrP, Lactogens, HGF, IGFs/Insulin), insulin signaling pathways and its downstream effectors (IRS1/2, P85, PI3K, AKT, FOXO1) and cell cycle proteins (CDK4, Cyclin D2) in positive regulation of proliferation and survival of beta cells have been well documented, on contrary, genes involved in JNK1 pathway, ER stress response and cytokine induced nitric oxide (NO) production and BCL2 family (apoptosis) induces beta cell death and decreased survival of beta cells [13]. Islet gene expression patterns at different physiological stresses (IR, obesity, pregnancy) provide us the clues for the reconfiguration of islets to pathogenesis of T2D and IR [14]. Deriving a ‘consensus’ expression status of each gene in a genelist, across multiple studies provide a clear expression status, under specific physiological conditions and/or cell-types and have previously been used for different physiological conditions in uterus and testis as well as development of two new databases (Uterus and testis) [15–18]. In this study we aimed to identify a ‘cluster’ of significantly upregulated genes in C57BL/6J ob/ob islets (as compared to BTBR ob/ob islets) using ‘consensus’ status of these genes across other studies, involved in successful beta cell compensation (islets of C57BL/6J ob/ob obese vs lean at 4 months and 10 months of age, islets of pregnant mice vs non pregnant mice). Information (biological processes, Molecular function, cellular component, protein-protein Interaction/associations, gene deletion/knockout/inhibition studies) of all the genes in ‘cluster’ were collected by text mining using different literature search tools, gene information databases and protein-protein interaction databases. Beta cell specific function of these genes were also inferred using meta analysis tool of Beta Cell Biology Consortium by studying the expression pattern of these genes in microarray studies related to beta-cell stimulation/injury, pancreas development and growth and cell differentiation. We believe this approach would help us identify novel genes involved in preventing beta cell dysfunction in response to metabolic stress.
Methodology
Data retrieval and gene expression analysis:
Published datasets archived in publicly available Gene Expression Omnibus (GEO) repository was used for re-analysis using GEO2R (NCBI online gene expression tool) [19]. Datasets published by Lan et al (2003) in GEO reference series GSE2899 and data sets GDS1443, was used for gene expression analysis of islets in diabetic resistant C57BL/6J ob/ob mice and diabetic BTBR ob/ob mice, using Affymetrix MGU74A array platform [11]. Other datasets were from GSE10785 series (Islets of C57BL/6J ob/ob mice vs Lean at 4 wks and 10wks of age respectively, Rosetta/Merck Mouse 44k 1.0 microarray) and GSE22125 (12.5 dpc pregnant mice vs non pregnant using Affymetrix Mouse Genome 430 2.0 Array platform) [12, 20]. p< 0.05 was taken for a gene to be called significant, with no conflict between different reporters.
A ‘cluster’ of genes overexpressed in non diabetic C57BL/6J ob/ob mice (as compared to diabetic BTBR ob/ob mice) were selected, based on ‘consensus’ over expression status approach i.e., the shortlisted genes were significantly upregulated in C57BL/6J ob/ob mice as well as also significantly overexpressed in two other studies involving successful beta cell compensation (C57BL/6J ob/ob vs lean 4 wks and 10 wks old respectively, 12.5 dpc pregnant vs non pregnant mice). Information (biological processes, Molecular function, cellular component, protein-protein Interaction/associations, gene deletion/knockout/inhibition) of all the genes in ‘cluster’ were collected by text mining usinsg different literature search tools (PubMed , HighWire Press, Google Scholar and iHOP) [21, 22], gene information databases (entrez gene, rat genome database, mouse genome database) [23–25] and protein protein interaction databases (HPRD, MINT, STRING, Genemania) [26–29]. Beta cell specific information was prioritized, when not available, the information on these genes in other cell types were extracted. Meta analysis tool of beta cell biology consortium (www.betacell.org) funded by NIDDK U01DK072473, was also used for analyzing the expression pattern of these genes (cluster) in studies related to beta-cell stimulation/injury, pancreas development and growth and Cell differentiation, tissue expression and survey (pancreatic) [30]. For a gene to be called significant, fold cutoff of at least 1.5 was taken and default significance parameter of the database was used (at least one reporter with confidence greater than 80% and no conflicts between two reporters).
Discussion
By using ‘consensus’ overexpression approach we identified a ‘cluster’ of genes with probable role in diabetes resistance in response to diabetogenic insult (Figure 1). The function of other genes in beta cell compensation was inferred by their role (biological process and molecular function) in other cell types using literature mining, databases and meta analysis tools. Expression pattern analysis of selected genes in studies related to cell differentiation, beta-cell stimulation/injury, pancreas development revealed their probable role in regulating these biological processes. Literature mining revealed role of PSEN1 in inhibiting apoptosis in beta cells, while the function of other genes in beta cells remain unknown. We have inferred their probable role in beta cells by extracting information on these genes in other cell types. Majority of the genes in the ‘cluster’ were related to cell survival and prevention of apotosis.
Figure 1.

Gene expression patterns. The figure shows the expression pattern of ‘cluster’ of genes across different studies (successful beta cell compensation C57bl6 mice). Only those genes which were significantly overexpressed in C57BL/6 ob/ob mice as compared to BTBR ob/ob mice ,and also show ‘consensus’ overexpression in other studies were considered. Genes considered to be significant at p<0.05.
Information (biological processes, Molecular function, cellular component, protein-protein interaction/associations, gene deletion/knockout/inhibition) of all the genes in ‘cluster’ were collected by text mining using different literature search tools. DAD1 and PSEN1 are known to inhibit apotosis [31, 32]. DAD1 interacts with MCL1 (BCL2 family) and prevents apoptosis [32]. Deletion of PSEN1 as well as DAD1 leads to embryonic death in mice [33, 34]. SSR4 and H13 are known to promote protein quality control and thus involved in protective unfolded response. SSR4 interacts with sec61 translocon, and also forms TRAP complex with SSR1, SSR2 and SSR3 subunits. Deletion of SSR4 results in disruption of the TRAP complex and delay of endoplasmic reticulum-associated degradation of misfolded glycoproteins and simultaneous overexpression of all the subunits (SSR1, SSR2, SSR3, SSR4) led to accelerated ER associated degradation [35]. PROM1 (also known as CD133), initially identified as stem cell marker (hemopoeitic stem cells and progenitor cells). PROM1 is known to be immunolocalized in Ductal epithelial cells of human, mouse and rat pancreas. PROM1 positive cells were also observed as small cells within pancreatic islets in young diet fed obese as well as control mice. However, at higher age (150 days), these cells were exclusively observed in islet peripheries and ducts of obese mice [36]. PROM1 is known to be involved in plasma membrane organization and play a role in AKT and MAPK pathway [37]. DNYLT3 interacts with BUB3 and regulate mitotic cell cycle, deletion of DNYLT3 increases mitotic index in NRK cells [38] PLK1 is involved in cell cycle progression and DNA replication in S phase and DNA damage repair [39]. ALDH18A1 and ANPEP are known to be involved in metabolic processes and role of FAM111A is unknown [25, 40].
The role of nutrients (especially glucose) and cytokines on beta cell survival and function is well known. Rat Islets cultured in presence of intermediate glucose concentration (10mM/L) showed optimum beta cell function, while beta cell impairment was observed in islet cultured at low concentration ( 2 mM/L and 5mM/L) and high concentration (30mM/L), with little effect on beta cell survival [41]. Beta cell impairment and apoptosis is induced by pro inflammatory cytokines [42]. Meta analysis tool of Beta Cell Biology Consortium revealed expression pattern of these genes across studies related to beta cell stimulation/injury (glucose, cytokines) [30]. At high glucose concentration (30mM vs 5mM/2mM glucose) Dad1, Aldh18A, H13, Fam111a and Ssr4 were up regulated while Prom1 was down regulated. H13 and Ssr4 genes were upregulated when cultured on intermediate levels of glucose as compared to low level of glucose (10mM vs. 5mM/2mM glucose). These genes may have a functional significance in beta cell survival and insulin secretion response to prolonged plasma glucose levels (hyperglycemia/hypoglycemia). Treatment of cytokine (IL-1β + IFN-γ) to Ins1 cells resulted in upregulation of Psen1, Ssr4, Dad1, Fam111a and plk1 while treatment of inducible NO synthase (iNOS) blocker NGmonomethyl- l-arginine (NMA) and cytokines led to overexpression of Ssr4 , Fam111a and Plk1 (Figure 2).
Figure 2.
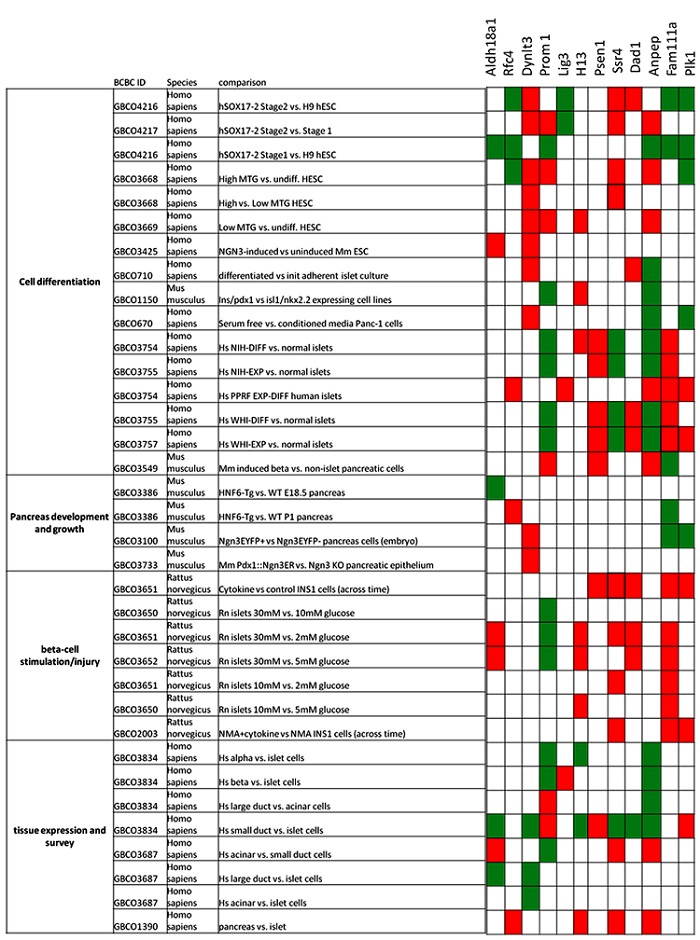
Gene expression patterns. The figure shows the expression pattern of selected genes in studies related to cell differentiation, beta-cell stimulation/injury, pancreas development and growth and tissue expression and survey (pancreatic). Meta analysis tool (http://genomics.betacell.org) of Beta Cell Biology Consortium (www.betacell.org) for analyzing the expression pattern of these genes. Fold cutoff of at least 1.5 was taken and default significance parameter of the database was used (at least one reporter with confidence greater than 80% and no conflicts between two reporters). Red block: significantly upregulated; Green block: significantly downregulated; White block: not significant/ no evidence.
Understanding the process of pancreatic organogenesis and beta cell differentiation/islet regeneration in vitro and in vivo, will benefit the patients with T2D, in restoration of β cell mass [43]. Sox17 is essential for endoderm development [44]. Meta analysis tool of Beta Cell Biology Consortium revealed expression pattern of these genes across microarray studies related to beta cell differentiation [30]. Aldh18a1, Prom1, Rfc4, Anpep, Fam111a and Plk1 were downregulated in hSOX17positive ‘stage 1’ cells (hESCs treated with activin A and Wnt3a to form definite endoderm) as compared to uninduced ‘H9’ hESCs. Dad1, Ssr4 and Dynlt3 were upregulated and Rfc4, lig3, Fam111a and Plk1 were downregulated in hSOX17positive ‘stage 2’ cells (hESCs induced to form cells resembling primitive gut tube-stage endoderm) as compared to uninduced ‘H9’ hESC cells. Comparative gene expression analysis between hSOX17positive ‘stage 1’ and hSOX17positive ‘stage 2’ revealed upregulation of Dynlt3, Prom1, Ssr4 and Anpep and downregulation of lig3 gene. Neurogenin3 (Ngn3) directs embryonic stem cells towards endocrine lineage and also regulate beta cell regeneration and differentiation [45–47]. Dynlt3 was upregulated, and Fam111a and Plk1 were downregulated in Purified Ngn3-positive progenitor cells in mice embryo (as compared to Ngn3- negative cells at 15 days of development). Aldh18a1 and Dynlt3 may be a potential Ngn3 target genes, as they were upregulated in Ngn3 induced potential endocrine pancreas progenitors (ESC differentiation using successive Activin A and Retinoic acid treatment and induction of Ngn3) as compared to uninduced ESCs. In a model of islet dysmorphogenesis (Pdx1PBHnf6 transgenic mice) at late gestation (e18.5) stage underexpressed Aldh18a1 when compared to wild type mice. These transgenic mice at postnatal day one (P1) overexpressed Rfc4 and underexpressed Fam111a when compared to wild type mice (Figure 2).
With some genes been quoted more frequently than the others, a high data divide exists in literature. The frequently quoted genes may not be the most important genes for the cell [48–50]. Further characterizing less quoted genes may help us in understanding the biological processes as well as pathophysiology of metabolic disorders like T2D. Previous study by Keller et al [12], the involvement of cell cycle regulating genes in predicting diabetes susceptibility,C57BL/6J ob/ob mice has obesity dependent increase in beta cell replication on the other hand BTBR ob/ob beta cells failed to increase proliferation in response to obesity [12]. In adult humans, beta cell proliferation is limited while increase in β-cell mass is primarily by beta cell expansion [51]. Prevention of β- cell apoptosis may lead to restoration of β-cell mass in T2D patients [51, 52]. Among ‘cluster’ of 11 genes, 6 genes (Dad1, Psen1, Ssr4, Rfc4, H13, Plk1) have a role in cell survival. We advocate these genes to be potentially involved in successful beta cells compensation and prevent T2D in humans, by conferring protection against diabetogenic insults.
Competing Interests
The authors have declared that no competing interest exists
Acknowledgments
We acknowledge Mrs. Mangala Kudali, (IBAB, Bangalore), Akhilesh B (IBAB, Bangalore) and Sravanthi D (IBAB, Bangalore) for their inputs. We also acknowledge Startbioinfo (http://www.startbioinfo.com/) for compiled resource lists.
Footnotes
Citation:Singh et al, Bioinformation 9(1): 037-041 (2013)
References
- 1.Matveyenko AV, et al. Am J Physiol Endocrinol Metab. 2008;295:E832. doi: 10.1152/ajpendo.90451.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorenson RL, TC Brelje. Horm Metab Res. 2008;29:301. doi: 10.1055/s-2007-979040. [DOI] [PubMed] [Google Scholar]
- 3.Kloppel G, et al. Surv Synth Pathol Res. 2008;4:110. doi: 10.1159/000156969. [DOI] [PubMed] [Google Scholar]
- 4.Bruning JC, et al. Cell. 1997;88:561. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 5.Rieck S, KH Kaestner. Trends Endocrinol Metab. 2010;21:151. doi: 10.1016/j.tem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karaca M, et al. Diabetes Metab. 2009;35:77. doi: 10.1016/j.diabet.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Neeland IJ, et al. JAMA. 2012;308:1150. doi: 10.1001/2012.jama.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimmet P, et al. Nature. 2001;414:782. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 9.Mishra PK, et al. Front Biosci. 2010;15:461. doi: 10.2741/3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cefalu WT. ILAR J. 2006;47:186. doi: 10.1093/ilar.47.3.186. [DOI] [PubMed] [Google Scholar]
- 11.Lan H, et al. Diabetes. 2003;52:688. doi: 10.2337/diabetes.52.3.688. [DOI] [PubMed] [Google Scholar]
- 12.Keller MP, et al. Genome Res. 2008;18:706. doi: 10.1101/gr.074914.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prentki M, CJ Nolan. J Clin Invest. 2006;116:1802. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller MP, AD Attie. Annu Rev Nutr. 2010;30:341. doi: 10.1146/annurev.nutr.012809.104747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bajpai AK, et al. PLoS One. 2012;7:e36776. doi: 10.1371/journal.pone.0036776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acharya KK, et al. BMC Genomics. 2010;11:467. doi: 10.1186/1471-2164-11-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. http://resource.ibab.ac.in/MGEx-Tdb/
- 18. http://resource.ibab.ac.in/MGEx-Udb/
- 19.Edgar R, et al. Nucleic Acids Res. 2002;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, et al. Nat Med. 2010;16:804. doi: 10.1038/nm.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manconi A, et al. Adv Bioinformatics. 2012;2012:573846. doi: 10.1155/2012/573846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann R, A Valencia. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 23.Dwinell MR, et al. Nucleic Acids Res. 2009;37:D744. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eppig JT, et al. Nucleic Acids Res. 2012;40:D881. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maglott D, et al. Nucleic Acids Res. 2011;39:D52. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goel R, et al. Mol Biosyst. 2012;8:453. doi: 10.1039/c1mb05340j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Licata L, et al. Nucleic Acids Res. 2012;40:D857. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szklarczyk D, et al. Nucleic Acids Res. 2011;39:D561. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warde-Farley D, et al. Nucleic Acids Res. 2010;38:W214. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. http://genomics.betacell.org.
- 31.Dror V, et al. Diabetologia. 2007;50:2504. doi: 10.1007/s00125-007-0835-5. [DOI] [PubMed] [Google Scholar]
- 32.Makishima T, et al. J Biochem. 2000;128:399. doi: 10.1093/oxfordjournals.jbchem.a022767. [DOI] [PubMed] [Google Scholar]
- 33.Brewster JL, et al. Genesis. 2000;26:271. [PubMed] [Google Scholar]
- 34.Shen J, et al. Cell. 1997;89:629. [Google Scholar]
- 35.Nagasawa K, et al. EMBO Rep. 2007;8:483. doi: 10.1038/sj.embor.7400933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thole AA, et al. Tissue Cell. 2012;44:238. doi: 10.1016/j.tice.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Wang YK, et al. Carcinogenesis. 2010;31:1376. doi: 10.1093/carcin/bgq120. [DOI] [PubMed] [Google Scholar]
- 38.Lo KW, et al. J Biol Chem. 2007;282:11205. doi: 10.1074/jbc.M611279200. [DOI] [PubMed] [Google Scholar]
- 39.Lu LY, et al. Mol Cell Biol. 2008;28:6870. doi: 10.1128/MCB.00392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu CA, et al. J Biol Chem. 1999;274:6754. doi: 10.1074/jbc.274.10.6754. [DOI] [PubMed] [Google Scholar]
- 41.Bensellam M, et al. Diabetologia. 2009;52:463. doi: 10.1007/s00125-008-1245-z. [DOI] [PubMed] [Google Scholar]
- 42.Kutlu B, et al. Diabetes. 2003;52:2701. doi: 10.2337/diabetes.52.11.2701. [DOI] [PubMed] [Google Scholar]
- 43.Guney MA, M Gannon. Birth Defects Res C Embryo Today. 2009;87:232. doi: 10.1002/bdrc.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi E, et al. Stem Cells. 2012;30:2297. doi: 10.1002/stem.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soyer J, et al. Development. 2010;137:203. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rukstalis JM, JF Habener. Islets. 2009;1:177. doi: 10.4161/isl.1.3.9877. [DOI] [PubMed] [Google Scholar]
- 47.Serafimidis I, et al. Stem Cells. 2008;26:3. doi: 10.1634/stemcells.2007-0194. [DOI] [PubMed] [Google Scholar]
- 48.Hoffmann R, A Valencia. Trends Genet. 2003;19:79. doi: 10.1016/s0168-9525(02)00014-8. [DOI] [PubMed] [Google Scholar]
- 49.Ramasamy A, et al. PLoS Med. 2008;5:e184. doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wren JD. Bioinformatics. 2009;25:1694. doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler AE, et al. Diabetes. 2008;52:102. [Google Scholar]
- 52.Meier JJ, et al. Diabetes. 2008;57:1584. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]


