Abstract
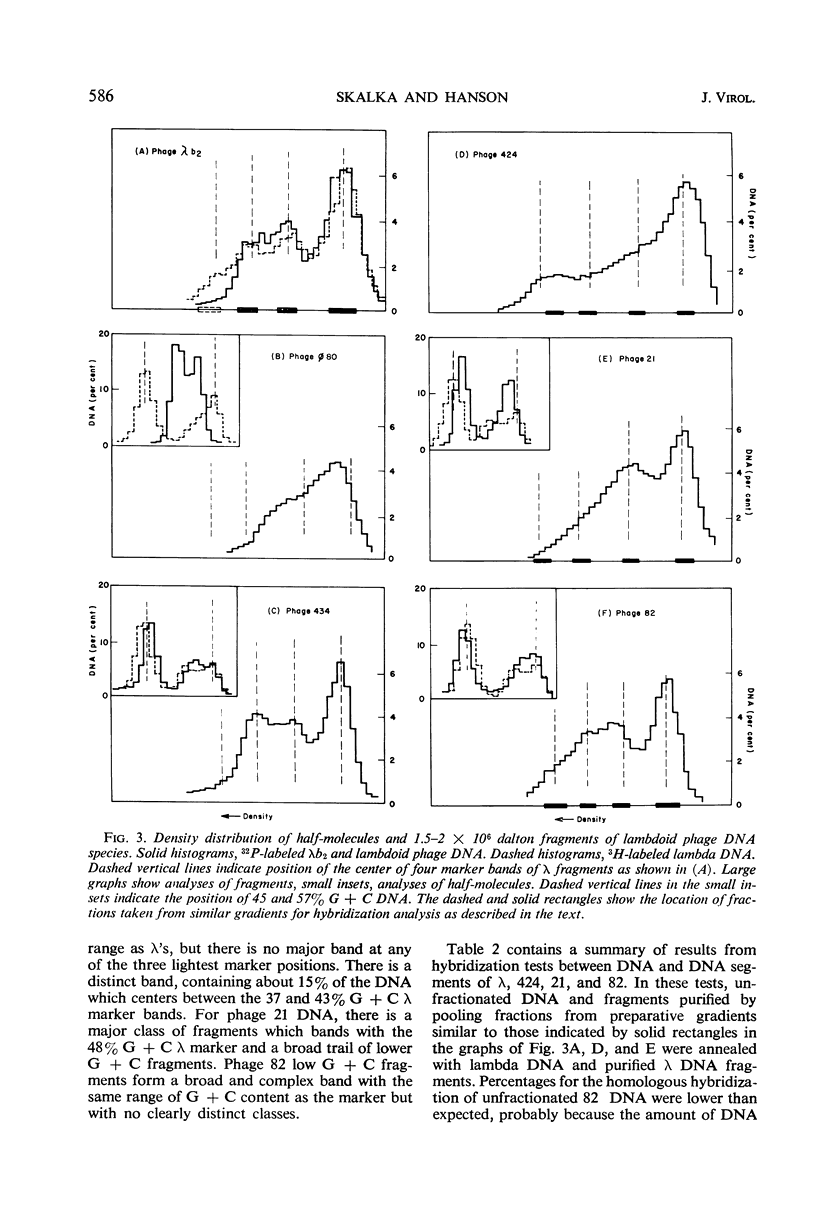
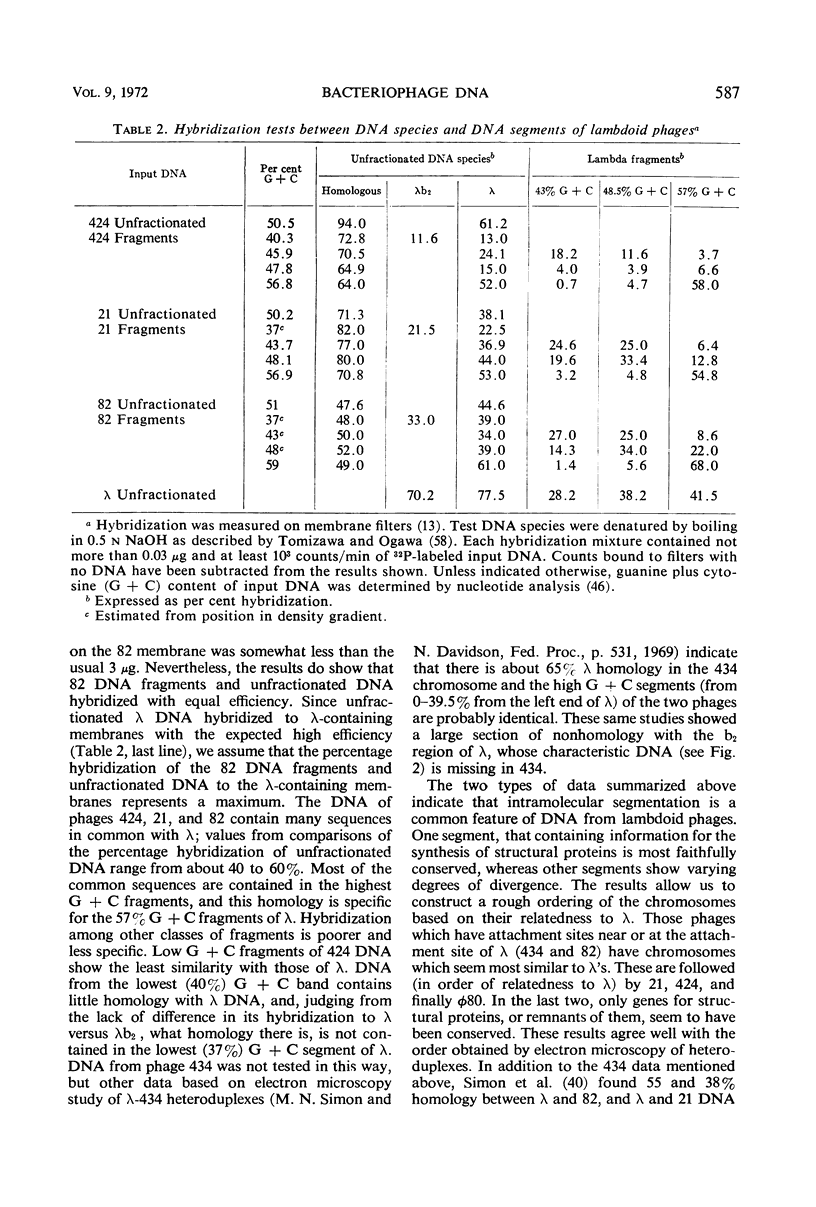
Results from comparisons of deoxyribonucleic acid (DNA) from several classes of bacteriophages suggest that most phage chromosomes contain either a homogeneous distribution of nucleotides or are made up of a few, rather large segments of different quanine plus cytosine (G + C) contents which are internally homogeneous. Among those temperate phages tested, most contained segmented DNA. Comparisons of sequence similarities among segments from lambdoid phage DNA species revealed the following order in relatedness to λ: 82 (and 434) > 21 > 424 > φ80. Most common sequences are found in the highest G + C segments, which in λ contain head and tail genes. Hybridization tests with λ and 186 or P2 DNA species verified that the lambdoids and 186 and P2 belong to two distinct groups. There are fewer homologous sequences between the DNA species of coliphages λ and P2 or 186 than there are between the DNA species of coliphage λ and salmonella phage P22.
Full text
PDF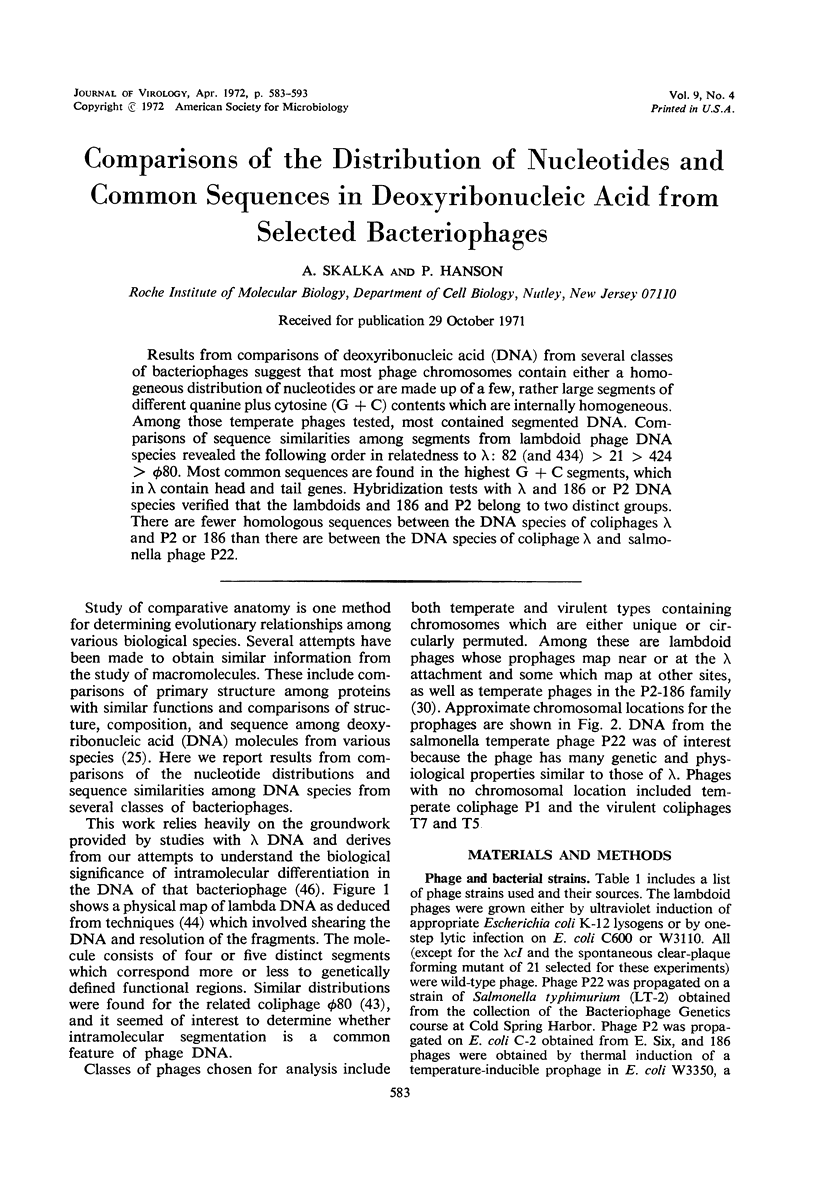




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BURGI E. Changes in molecular weight of DNA accompanying mutations in phage. Proc Natl Acad Sci U S A. 1963 Feb 15;49:151–155. doi: 10.1073/pnas.49.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin R. L., Barrand P., Fritsch A., Goldthwait D. A., Jacob F. Cohesive sites on the deoxyribonucleic acids from several temperate coliphages. J Mol Biol. 1966 Jun;17(2):343–357. doi: 10.1016/s0022-2836(66)80146-8. [DOI] [PubMed] [Google Scholar]
- Bear P. D., Skalka A. The molecular origin of lambda prophage mRNA. Proc Natl Acad Sci U S A. 1969 Feb;62(2):385–388. doi: 10.1073/pnas.62.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani L. E., Bertani G. Preparation and characterization of temperate, non-inducible bacteriophage P2 (host: Escherichia coli). J Gen Virol. 1970 Feb;6(2):201–212. doi: 10.1099/0022-1317-6-2-201. [DOI] [PubMed] [Google Scholar]
- Bram S. Secondary structure of DNA depends on base composition. Nat New Biol. 1971 Aug 11;232(2):174–176. doi: 10.1038/newbio232174a0. [DOI] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Cowie D. B., Szafranski P. Thermal chromatography of DNA-DNA reactions. Biophys J. 1967 Sep;7(5):567–584. doi: 10.1016/S0006-3495(67)86607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dove W. F. Action of the lambda chromosome. I. Control of functions late in bacteriophage development. J Mol Biol. 1966 Aug;19(1):187–201. doi: 10.1016/s0022-2836(66)80060-8. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Geisselsoder J., Mandel M. Physical properties of phage 299. Mol Gen Genet. 1970;180(2):158–166. [PubMed] [Google Scholar]
- Gough M., Levine M. The circularity of the phage P22 linkage map. Genetics. 1968 Feb;58(2):161–169. doi: 10.1093/genetics/58.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. I. Transducing fragments in generalized transduction by phage P1. I. Molecular origin of the fragments. J Mol Biol. 1965 Nov;14(1):85–109. doi: 10.1016/s0022-2836(65)80232-7. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- Inman R. B., Bertani G. Heat denaturation of P2 bacteriophage DNA: compositional heterogeneity. J Mol Biol. 1969 Sep 28;44(3):533–549. doi: 10.1016/0022-2836(69)90378-7. [DOI] [PubMed] [Google Scholar]
- Joyner A., Isaacs L. N., Echols H., Sly W. S. DNA replication and messenger RNA production after induction of wild-type lambda bacteriophage and lambda mutants. J Mol Biol. 1966 Aug;19(1):174–186. doi: 10.1016/s0022-2836(66)80059-1. [DOI] [PubMed] [Google Scholar]
- KELLY B. Localization of P2 prophage in two strains of Escherichia coli. Virology. 1963 Jan;19:32–39. doi: 10.1016/0042-6822(63)90021-7. [DOI] [PubMed] [Google Scholar]
- King J. L., Jukes T. H. Non-Darwinian evolution. Science. 1969 May 16;164(3881):788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- Kolstad R. A., Prell H. H. An amber map of Salmonella phage P22. Mol Gen Genet. 1969 Aug 15;104(4):339–350. doi: 10.1007/BF00334233. [DOI] [PubMed] [Google Scholar]
- Lodish H. F., Robertson H. D. Regulation of in vitro translation of bacteriophage f2 RNA. Cold Spring Harb Symp Quant Biol. 1969;34:655–673. doi: 10.1101/sqb.1969.034.01.076. [DOI] [PubMed] [Google Scholar]
- Mandel M., Berg A. Cohesive sites and helper phage function of P2, lambda, and 186 DNA's. Proc Natl Acad Sci U S A. 1968 May;60(1):265–268. doi: 10.1073/pnas.60.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Berg A. Melting temperatures of the cohesive ends of some non-inducible coliphage DNA's. J Mol Biol. 1968 Nov 28;38(1):137–139. doi: 10.1016/0022-2836(68)90135-6. [DOI] [PubMed] [Google Scholar]
- Mandel M. Infectivity of phage P2 DNA in presence of helper phage. Mol Gen Genet. 1967;99(1):88–96. doi: 10.1007/BF00306461. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Szpirer J., Thomas R. THE STRUCTURAL GENE FOR lambda EXONUCLEASE. Proc Natl Acad Sci U S A. 1967 Feb;57(2):277–283. doi: 10.1073/pnas.57.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M., MacHattie L. A., Thomas C. A., Jr The P22 bacteriophage DNA molecule. I. The mature form. J Mol Biol. 1968 Oct 14;37(1):21–40. doi: 10.1016/0022-2836(68)90071-5. [DOI] [PubMed] [Google Scholar]
- Schnös M., Inman R. B. Position of branch points in replicating lambda DNA. J Mol Biol. 1970 Jul 14;51(1):61–73. doi: 10.1016/0022-2836(70)90270-6. [DOI] [PubMed] [Google Scholar]
- Schnös M., Inman R. B. Starting point and direction of replication in P2 DNA. J Mol Biol. 1971 Jan 14;55(1):31–38. doi: 10.1016/0022-2836(71)90278-6. [DOI] [PubMed] [Google Scholar]
- Scott J. R. A defective P1 prophage with a chromosomal location. Virology. 1970 Jan;40(1):144–151. doi: 10.1016/0042-6822(70)90386-7. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Clear plaque mutants of phage P1. Virology. 1970 May;41(1):66–71. doi: 10.1016/0042-6822(70)90054-1. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Genetic studies on bacteriophage P1. Virology. 1968 Dec;36(4):564–574. doi: 10.1016/0042-6822(68)90188-8. [DOI] [PubMed] [Google Scholar]
- Skalka A., Burgi E., Hershey A. D. Segmental distribution of nucleotides in the DNA of bacteriophage lambda. J Mol Biol. 1968 May 28;34(1):1–16. doi: 10.1016/0022-2836(68)90230-1. [DOI] [PubMed] [Google Scholar]
- Skalka A., Butler B., Echols H. Genetic control of transcription during development of phage gamma. Proc Natl Acad Sci U S A. 1967 Aug;58(2):576–583. doi: 10.1073/pnas.58.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A. Multiple units of transcription in phage lambda. Cold Spring Harb Symp Quant Biol. 1966;31:377–379. doi: 10.1101/sqb.1966.031.01.048. [DOI] [PubMed] [Google Scholar]
- Skalka A. Nucleotide distribution and functional orientation in the deoxyribonucleic acid of phage phi 80. J Virol. 1969 Feb;3(2):150–156. doi: 10.1128/jvi.3.2.150-156.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalka A. Regional and temporal control of genetic transcription in phage lambda. Proc Natl Acad Sci U S A. 1966 May;55(5):1190–1195. doi: 10.1073/pnas.55.5.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O. Defective phage formation by lysogens of integration deficient phage P22 mutants. Virology. 1968 Feb;34(2):203–223. doi: 10.1016/0042-6822(68)90231-6. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Levine M. A phage P22 gene controlling integration of prophage. Virology. 1967 Feb;31(2):207–216. doi: 10.1016/0042-6822(67)90164-x. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Levine M. Gene order in prophage P22. Virology. 1965 Oct;27(2):229–231. doi: 10.1016/0042-6822(65)90166-2. [DOI] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Conformational changes of single-stranded DNA. J Mol Biol. 1969 Apr;41(2):189–197. doi: 10.1016/0022-2836(69)90384-2. [DOI] [PubMed] [Google Scholar]
- Studier F. W. Effects of the conformation of single-stranded DNA on renaturation and aggregation. J Mol Biol. 1969 Apr;41(2):199–209. doi: 10.1016/0022-2836(69)90385-4. [DOI] [PubMed] [Google Scholar]
- Studier F. W. The genetics and physiology of bacteriophage T7. Virology. 1969 Nov;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- Summers W. C. The process of infection with coliphage T7. IV. Stability of RNA in bacteriophage-infected cells. J Mol Biol. 1970 Aug;51(3):671–678. doi: 10.1016/0022-2836(70)90015-x. [DOI] [PubMed] [Google Scholar]
- Thomas C. A., Jr The rule of the ring. J Cell Physiol. 1967 Oct;70(2 Suppl):13–33. doi: 10.1002/jcp.1040700404. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Ogawa T. Replication of phage lambda DNA. Cold Spring Harb Symp Quant Biol. 1968;33:533–551. doi: 10.1101/sqb.1968.033.01.061. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Cyclization of coliphage 186 DNA. J Mol Biol. 1967 Sep 28;28(3):403–411. doi: 10.1016/s0022-2836(67)80089-5. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Schwartz H. Noncomplementarity in base sequences between the cohesive ends of coliphages 186 and lambda and the formation of interlocked rings between the two DNA's. Biopolymers. 1967;5(10):953–966. doi: 10.1002/bip.1967.360051008. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Nakamura K., Ozeki H. Cohesion occurring between DNA molecules of temperate phages phi 80 and lambda or phi 81. Biochem Biophys Res Commun. 1965 Sep 22;20(6):727–732. doi: 10.1016/0006-291x(65)90077-x. [DOI] [PubMed] [Google Scholar]
- Yamagishi H., Skalka A. Distribution of nucleotides in Escherichia coli DNA from lambda-transducing phages. J Mol Biol. 1971 Jun 14;58(2):417–430. doi: 10.1016/0022-2836(71)90360-3. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



