Abstract
Dengue and West Nile viruses (WNV) are mosquito-borne members of flaviviruses that cause significant morbidity and mortality. There is no approved vaccine or antiviral drugs for human use to date. In this study, a series of functionalized meta and para aminobenzamide derivatives were synthesized and subsequently screened in vitro against Dengue virus and West Nile virus proteases. Four active compounds were identified which showed comparable activity toward the two proteases and shared in common a meta or para(phenoxy)phenyl group. The inhibition constants (Ki) for the most potent compound 7n against Dengue and West Nile virus proteases were 8.77 and 5.55 μM, respectively. The kinetics data support a competitive mode of inhibition of both proteases by compound 7n. This conclusion is further supported by molecular modeling. This study reveals a new chemical scaffold which is amenable to further optimization to yield potent inhibitors of the viral proteases via the combined utilization of iterative medicinal chemistry/structure-activity relationship studies and in vitro screening.
Introduction
The Dengue virus (DENV) and West Nile Virus (WNV) are members of the family Flaviviridae which comprise several human and animal pathogens, including Yellow fever virus (YFV), Hepatitis C virus (HCV), and Japanese encephalitis virus (JEV).1 Infection by DENV1-4 serotypes results in Dengue fever which can progress to life-threatening Dengue hemorrhagic fever (DHF) and Dengue shock syndrome (DSS).2 Dengue infection is a global health threat for which there are currently no small molecule antiviral therapeutics or effective vaccines. Inhibition of DENV replication constitutes a promising avenue of investigation for the development of therapeutics against DENV.3-5
Both Dengue and West Nile viruses are small, enveloped viruses containing a positive-stranded 11-kb RNA genome which encodes a polyprotein precursor which is co- and post-translationally processed by host cell proteases and the viral encoded trypsin-like protease, NS2B/NS3, into three structural proteins (C, prM, E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).6-7 NS3 is a multi-functional protein containing a serine protease with a prototypical catalytic triad (H51, D75, S135) within the N-terminal 185 amino acid residues, an NTPase/RNA helicase, and a 5’-RNA triphosphatase within the C-terminal (161-618 amino acid residues) region of NS3.4,5-6 Activation of the serine protease domain of NS3 requires NS2B cofactor, a ~130 amino acid hydrophobic integral membrane protein in the endoplasmic reticulum (ER). A ~45 amino acid conserved hydrophilic segment from the NS2B sequence is sufficient for interaction with and activation of the NS3 protease domain for protease activity (hereafter referred to as “NS2B/NS3pro”) in vitro.5,7-8 Processing of the polyprotein by NS2B/NS3pro is essential for viral replication, consequently NS2B/NS3pro has emerged as an attractive target for the discovery and development of small molecule therapeutics for DENV infection.3-6
DENV NS2B/NS3pro has a substrate specificity for an -X-K-R-R-G/S-sequence corresponding to the subsites -S4-S3-S2-S1-S1’-.9 Cleavage is at the P1-P1’ (R-G/S) scissile bond. The hydrophilic β-hairpin segment of the NS2B protein cofactor wraps around the NS3 protease core and is required for optimal catalytic efficiency.10-12 Crystallographic12-13 and high-field NMR studies14 have illuminated greatly our understanding of the role played by the NS2B cofactor and the nature of the interaction of NS2B/NS3pro with small-molecule inhibitors.
DENV NS2B/NS3pro plays a vital role in virus replication, consequently, orally-bioavailable drug-like agents that inhibit NS2B/NS3pro are of value as potential therapeutics for DENV infection. Inhibitors of NS2B/NS3pro containing a highly charged peptidyl recognition element with a dibasic motif at P1-P2 coupled to an array of electrophilic warheads (aldehydes, trifluoromethyl ketones, and boronic acids),15-18 non-peptidyl α-ketoamides,19 phthalazine-based derivatives,20 arylcyanoacrylamides,21 retro peptide-hybrids,22 benz[d]isothiazol-3(2H)-one derivatives,23 and others24-25 have been reported. We describe herein the results of synthetic and biochemical studies related to the inhibition of DENV and WNV NS2B/NS3pro by functionalized para- and meta-substituted aminobenzamide derivatives represented by structure (I) (Figure 1).
Figure 1.
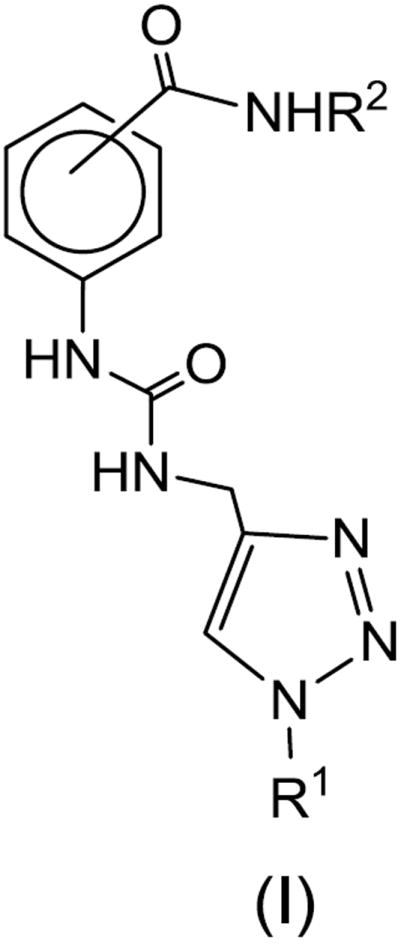
General structure of aminobenzamide derivatives (I).
Chemistry
Compounds 7a-t (Table 1) and 8a-f (Table 2) were synthesized starting with methyl p-aminobenzoate or methyl m-aminobenzoate, respectively, as illustrated in Scheme 1. Thus, treatment with trichloromethyl chloroformate yielded the corresponding isocyanate which was reacted with propargylamine to form urea derivatives 2a-b. Click chemistry26 with an array of structurally diverse azides yielded the corresponding functionalized urea derivatives which were further elaborated to form the final compounds.
Table 1.
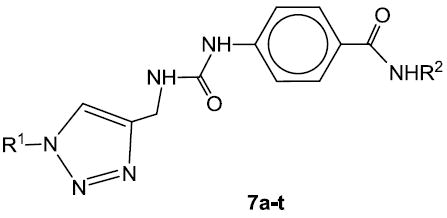
| |||||
|---|---|---|---|---|---|
| Compound | R1 | R2 | Compound | R1 | R2 |
| 7a | phenylthiomethyl | 4-phenoxyphenyl | 7k | benzyl | 2-furanylmethyl |
| 7b | phenylthiomethyl | benzyl | 7l | benzyl | 2-morpholinoethyl |
| 7c | phenylthiomethyl | phenylethyl | 7m | benzyl | 3-phenoxyphenyl |
| 7d | phenylthiomethyl | phenyl | 7n | 4-fluorobenzyl | 4-phenoxyphenyl |
| 7e | phenylthiomethyl | 2-furanylmethyl | 7o | 4-fluorobenzyl | benzyl |
| 7f | phenylthiomethyl | 2-morpholinoethyl | 7p | 4-fluorobenzyl | phenylethyl |
| 7g | benzyl | 4-phenoxyphenyl | 7q | 4-fluorobenzyl | phenyl |
| 7h | benzyl | benzyl | 7r | 4-fluorobenzyl | 2-furanylmethyl |
| 7i | benzyl | phenylethyl | 7s | 4-fluorobenzyl | 2-morpholinomethyl |
| 7j | benzyl | phenyl | 7t | 4-fluorobenzyl | 3-phenoxyphenyl |
Table 2.
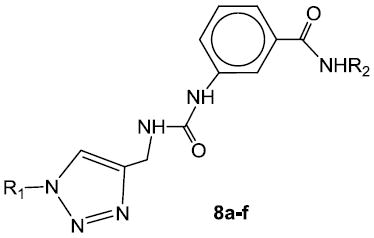
| ||
|---|---|---|
| Compound | R1 | R2 |
| 8a | phenylthiomethyl | benzyl |
| 8b | phenylthiomethyl | phenylethyl |
| 8c | phenylthiomethyl | 3-phenoxyphenyl |
| 8d | benzyl | benzyl |
| 8e | benzyl | phenylethyl |
| 8f | benzyl | 3-phenoxyphenyl |
Scheme 1.
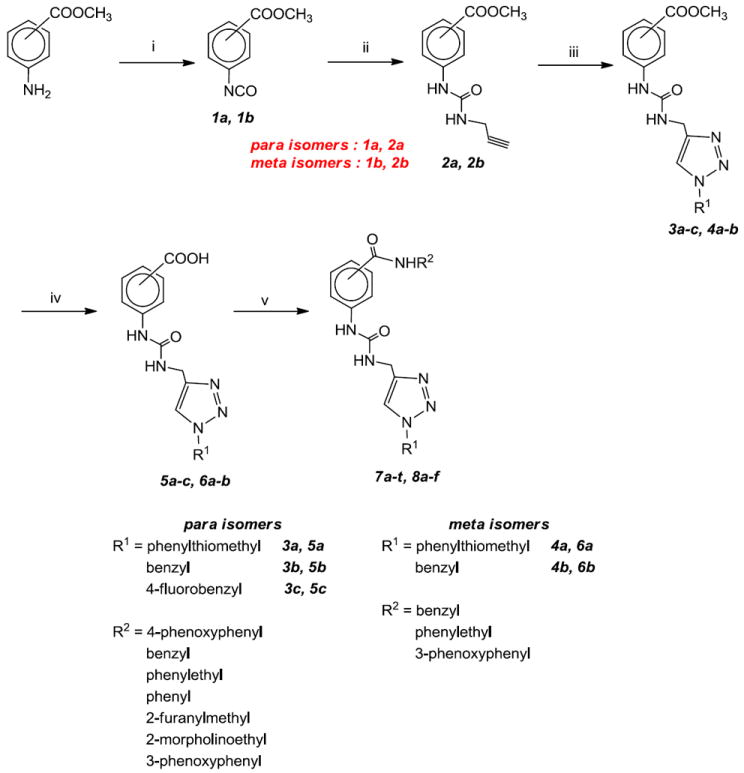
Reagents and conditions: i) Trichloromethyl chloroformate/THF, Δ; ii) Propargyl amine/THF; iii) Arylalkyl azide, t-BuOH and H2O (1:1), CuSO4 .5H2O, sodium ascorbate; iv) aqLiOH/1,4-dioxane; v) CDl/THF, followed by amine
Biochemistry
The expression and purification of DENV NS2B/NS3pro and WNV NS2B/NS3pro have been previously described.27-29 Enzyme assays and inhibition studies were carried out as previously described30-32 and the results are summarized in Figures 2-4 and Tables 3 and 4.
Figure 2. Inhibition of DENV2 and WNV NS2B/NS3pro by selected compounds at 25 μM.
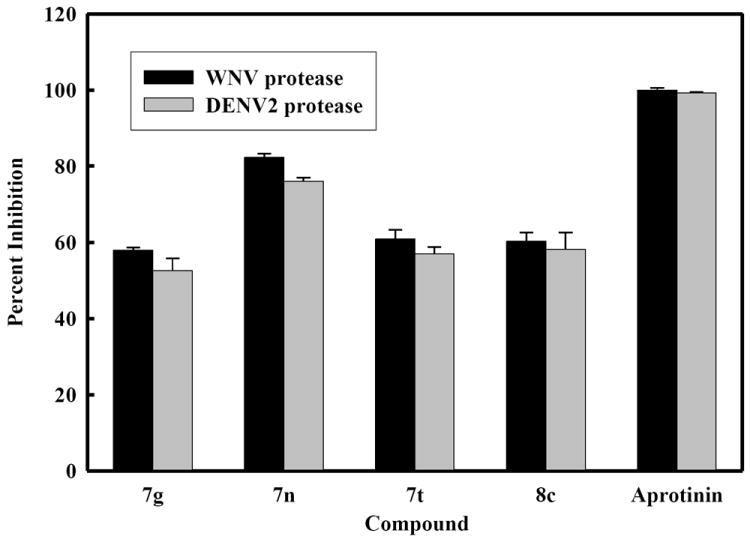
The concentrations of WNV and DENV2 NS2B/NS3pro protease were 28 nM and 25 nM, respectively. The buffer used contained 200 mM Tris HCl, 6.0 mM NaCl, 30 % glycerol, and 0.1% CHAPS, pH 9.5. The percent values were calculated from the relative fluorescence units obtained in the presence and absence of tested compound. BPTI (Aprotinin) and DMSO were used as a positive and negative control, respectively. All assays were performed in triplicate and the average values are shown.
Figure 4. Inhibition of WNV NS2B/NS3pro protease activity by compound 7n.
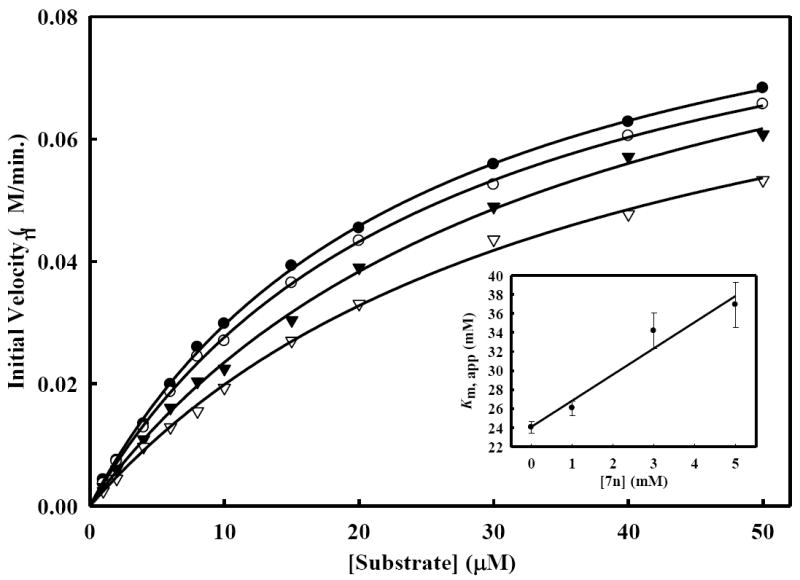
Initial reaction rates of the substrate (Bz-Nle-Lys-Arg-Arg-AMC) cleavage catalyzed by WNV NS2B/NS3pro protease (28 nM) in 200 mM Tris·HCl (pH 9.5), 6.0 mM NaCl, 30% glycerol and 0.1% CHAPS at 37 °C were determined by varying the substrate concentrations in the range of 0, 1, 2, 4, 6, 8, 10, 15, 20, 30, 40 and 50 μM at each concentration of inhibitor fixed at 0 (solid circle), 1.0 μM (open circle), 3.0 μM (solid triangle) and 5.0 μM (open triangle). The reactions were initiated by the addition of WNV NS2B/NS3pro protease and the fluorescence intensity at 460 nm was monitored with an excitation at 380 nm. Reactions were less than 5% completion in all cases to maintain valid steady-state measurements. The solid lines are fitted lines using the Michaelis-Menten equation. Inset: Secondary plot of Km, app against the concentration of compound 7n. Kinetics studies were carried out as described utilizing substrate concentrations of 0-50 μM Bz-Nle-Lys-Arg-Arg-AMC. Each experiment was performed in duplicate and repeated three times. Data were analyzed using SigmaPlot 2001 v7.0 software41 to determine values for apparent Km and kcat.
Table 3.
Inhibition of DENV2 and WNV NS2B/NS3pro protease by compounds 7g, 7n, 7t and 8c.
| Inhibitors | DENV2 | WNV | ||
|---|---|---|---|---|
| % inhibition at 10 μM | % inhibition at 25 μM | % inhibition at 10 μM | % inhibition at 25 μM | |
| 7g | 39.41 ± 2.69 | 52.54 ± 3.21 | 46.51 ± 1.75 | 57.95 ± 0.71 |
| 7n | 55.31 ± 1.39 | 76.01 ± 0.96 | 58.78 ± 1.62 | 82.26 ± 1.04 |
| 7t | 41.06 ± 1.83 | 56.98 ± 1.81 | 44.45 ± 2.08 | 60.85 ± 2.46 |
| 8c | 40.27 ± 1.11 | 58.21 ± 4.31 | 49.28 ± 1.87 | 60.27 ± 2.36 |
[WNV protease] = 28 nM; [DENV2 protease] = 25 nM
Table 4.
Kinetics parameters for the tetra-peptide substrate and compound 7n against WNV NS2B/NS3pro at 37 °C
| [7n] | Km | kcat | kcat/Km |
|---|---|---|---|
|
| |||
| μM | μM | s-1 | M-1·s-1 |
| 0 | 24.01 ± 0.62 | 0.060 ± 0.0013 | 2499 ± 99 |
| 1 | 26.04 ± 0.75 | 0.059 ± 0.0014 | 2274 ± 100 |
| 3 | 34.16 ± 1.88 | 0.062 ± 0.0018 | 1811 ± 163 |
| 5 | 36.90 ± 2.36 | 0.055 ± 0.0020 | 1503 ± 162 |
Results and Discussion
The world-wide health problem stemming from infection by Dengue virus and related flaviviruses, as well as the paucity of small-molecule therapeutics for combating flavivirus infection, have provided the impetus for the research described herein. The aminobenzamide scaffold was utilized in the synthesis of a series of structurally-diverse meta and para-substituted derivatives, represented by general structure (I), (Figure 1). The design of (I) rested on the following considerations: (a) the shallow active site of DENV NS2B/NS3pro presents a formidable challenge in terms of the design of potent inhibitors of the enzyme, necessitating the use of a suitably-embellished multifunctional molecule capable of engaging in multiple favorable binding interactions with the enzyme in order to attain high potency, without compromising oral bioavailability and PK characteristics.33-40 The problem is further compounded by the stringent substrate specificity requirements of the protease for positively charged substrates/inhibitors; (b) based on insights gained from examining the X-ray crystal structures of DENV2 NS2B/NS3pro with bound ligands12-13 and molecular modeling studies, it was hypothesized that the utilization of a planar platform capable of orienting appended non-peptidyl recognition elements in a precisely-defined vector relationship would lead to agents capable of interacting with multiple active site residues. Thus, the aminobenzamide platform was chosen for generating the desired compounds and for conducting exploratory studies. An added advantage is the flexibility afforded by the two points of diversity in the chosen scaffold, augmenting synthetic tractability; (c) a urea functionality was initially employed to lessen the conformational flexibility of the appended recognition elements. The latter included an electron-rich heterocycle having multiple hydrogen bond acceptor sites linked to an aryl alkyl group and, (d) it was furthermore envisaged that the initial attachment of an array of structurally-diverse aliphatic and aromatic amines to the carboxyl group of the aminobenzamide scaffold would provide additional binding sites.
The desired compounds were readily obtained as shown in Scheme 1. These were subsequently screened against DENV NS2B/NS3pro and WNV NS2B/NS3pro. Four of the compounds based on (I) were found to exhibit activity against both proteases and the results are summarized in Table 3 and Figure 2. It can be generally inferred from the SAR studies that the nature of both R1 and R2 influence activity. Furthermore, activity is manifested when R1 is a meta or p-(phenoxy)phenyl group. All other amides were inactive, suggesting that the meta or p-(phenoxy)phenyl group is accommodated in a hydrophobic cleft. It should be noted that some of the compounds were inactive despite the presence of a meta or p-(phenoxy)phenyl group (compounds 7a and 8f).
The percent inhibition shown by compound 7n against both DENV2 and WNV was found to be higher than any of the other selected compounds (Figure 2). The apparent IC50 values of compound 7n (Figure 3) were determined to be 6.82 ± 0.09 and 5.51 ± 0.08 μM against DENV2 and WNV protease, respectively. We also performed kinetics analyses to determine the Km, kcat and Vmax values in the presence and absence of compound 7n at four different concentrations (Figure 4 and Table 4). The apparent Michaelis-Menten constants (Km, app) increased and kcat/Km decreased proportionally with increasing concentration of compound 7n. The results support a competitive mode of inhibition. Next, we employed molecular modeling to identify a plausible binding mode for 7n. Based on the molecular docking simulations, a high affinity bound conformer that is similar in both the DENV and WNV proteases was identified. Docking poses analysis yielded only one high potency conformer (i.e., one of the top five scoring poses) that conserved most of the key interactions in both DENV and WNV (Figures 5A and 5B, respectively). Key features of this pose that are conserved across the two receptors include π-π stacking interactions between the ligand fluorobenzyl group and the side chain of Tyr161, and hydrophobic interactions between the ligand phenoxyphenyl group and Val72. Visual inspection suggests that these interactions are not only conserved but are also likely to comprise most of the favorable binding features in both cases. Furthermore confidence in this proposed pharmacophore is attained through comparison with predicted binding modes for inactive compounds 7a and 8f. In the case of DENV NS2B/NS3pro in Figure 5A, neither of the inactive compounds is predicted to exploit either of the two lipophiles listed above with the same effectiveness as 7n: compound 8f has poor overlap with both lipophiles, and while compound 7a is predicted to achieve some π-π stacking with Tyr161, its furan group is not situated anywhere near any lipophiles. In the case of WNV NS2B/NS3pro (Figure 5B), reasonable π-π stacking is achieved with Tyr161 in all cases, but the furan group of 7a is again poorly situated, while the phenoxyphenyl group of 8f is not predicted to derive nearly as much a hydrophobic interaction as is achieved by the corresponding phenoxyphenyl group of 7n, whose terminal phenyl group is predicted to wedge favorably between Val72 and the lipophilic portion of neighboring Lys73.
Figure 3. Determination of IC50 value of the inhibitor 7n against DENV-2 and WNV protease.
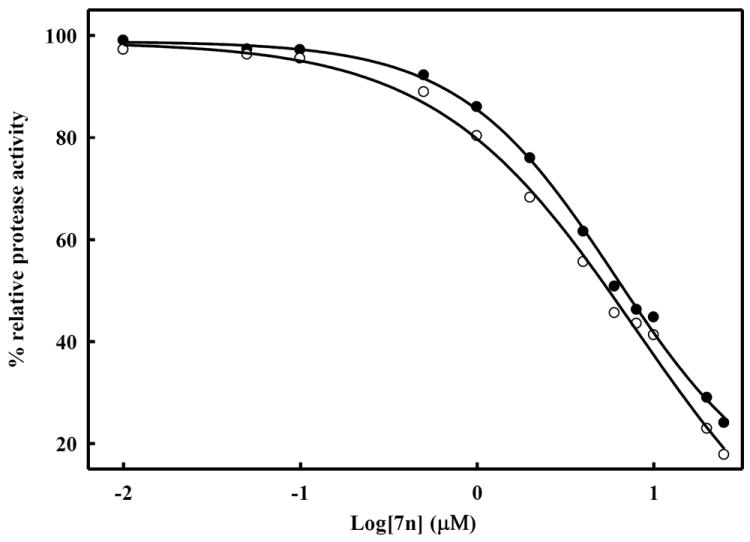
The inhibitor was incubated with DENV2 NS2B/NS3pro (25 nM) or WNV NS2B/NS3pro (28 nM) in buffer (200 mM Tris HCl, 6 mM NaCl and 30% glycerol, pH 9.5) for 15 min. Bz-Nle-Lys-Arg-Arg-AMC (5.0 μM) was added to the mixture in a final volume of 100 μL. The fluorescence intensity was measured at 460 nm with excitation at 380 nm and converted to the percentage of protease activity in the absence and presence of inhibitors. The solid line is the theoretical fitting curve based on the Sigmoidal Equation.41 The apparent IC50 values for compound 7n were 6.82 ± 0.09 and 5.51 ± 0.08 μM against DENV-2 (solid circle) and WNV (open circle), respectively.
Figure 5.
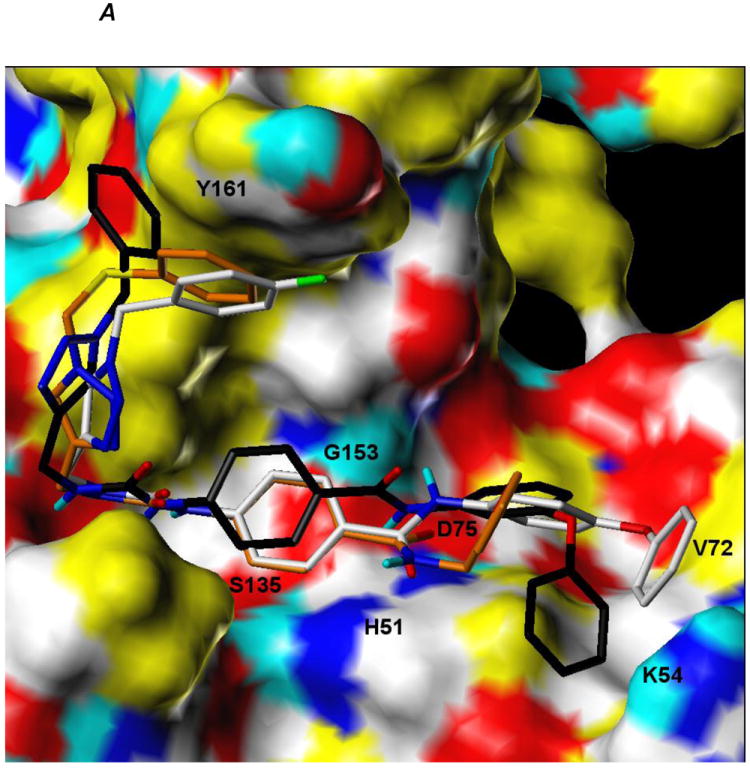
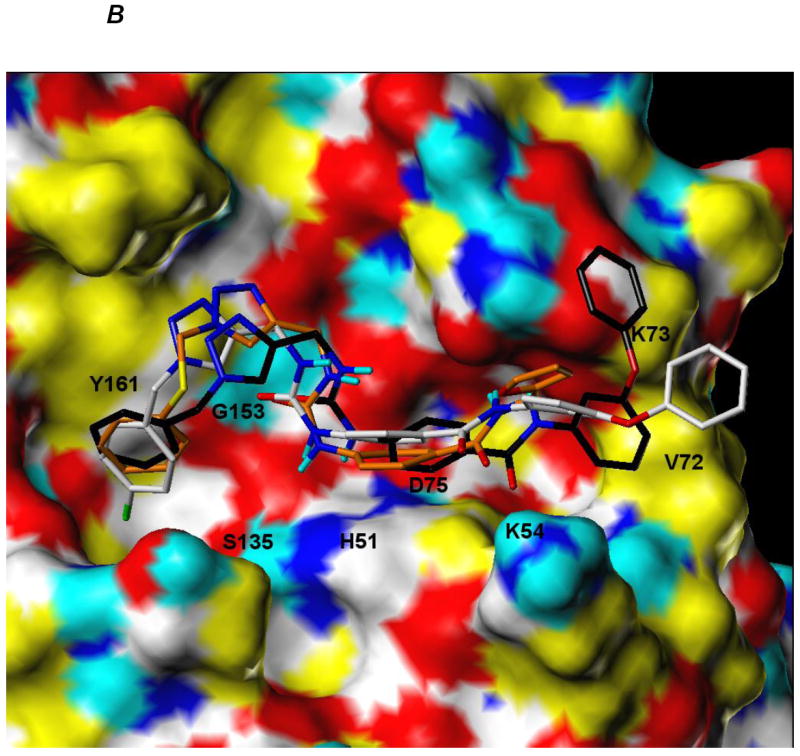
A. Computationally predicted conformation of compounds 7a, 7n and 8f bound to the catalytic site of DENV2 NS2B/NS3 protease
Ligands are rendered as CPK-colored sticks with the exception of carbon atoms (7a: orange, 7n: white, 8f: black), while the receptor surface is colored as follows: yellow = hydrophobic, white = polarized alkyl or aryl groups, cyan = polar hydrogens, blue = polar nitrogens and red = polar oxygens.
B. Computationally predicted conformation of compound 7n bound to the catalytic site of WNV NS2B/NS3 protease. Ligands are rendered as CPK-colored sticks with the exception of carbon atoms (7a: orange, 7n: white, 8f: black), while the receptor surface is colored as follows: yellow = hydrophobic, white = polarized alkyl or aryl groups, cyan = polar hydrogens, blue = polar nitrogens and red = polar oxygens.
In summary, the studies described herein have demonstrated that the aminobenzamide scaffold can be employed in the synthesis of inhibitors of DENV and WNV NS2B/NS3pro.
Experimental Section
General
The 1H spectra were recorded on a Varian XL-300 or XL-400 NMR spectrometer. Melting points were determined on a Mel-Temp apparatus and are uncorrected. High resolution mass spectra (HRMS) were performed at the University of Kansas Mass Spectrometry Lab. Reagents and solvents were purchased from various chemical suppliers (Aldrich, Acros Organics, TCI America, and Bachem). Silica gel (230-450 mesh) used for flash chromatography was purchased from Sorbent Technologies (Atlanta, GA). Thin layer chromatography was performed using Analtech silica gel plates. The TLC plates for the final compounds were eluted using two different solvent systems and were visualized using iodine and/or UV light. Each individual compound was identified as a single spot on TLC plate (purity was greater than 95% by 1H NMR). DENV2 NS2B/NS3 pro (or WNV NS2B/NS3 pro) substrate Bz-Nle-Lys-Arg-Arg-AMC was purchased from Bachem, Torrance, CA or custom synthesized by NeoBioScience, Cambridge, MA.
Representative syntheses
Compound 1a
To methyl 4-aminobenzoate (30.23 g; 200 mmol) in anhydrous 1,4-dioxane (500 mL) was added trichloromethyl chloroformate (59.35 g; 300 mmol) dropwise and the reaction mixture was refluxed for 8 h. The solvent was removed on the rotary evaporator to yield 1a as a yellow solid, which was purified by vacuum distillation to give a yellow solid (34 g, 96% yield). IR (neat) νNCO 2268 cm-1. 1H NMR (CDCl3): δ 3.88 (s, 3H), 7.14-7.18 (m, 2H), 7.96-8.02 (m, 2H).
Compound 1b
Brown solid (94% yield). IR (neat) νNCO 2257 cm-1. 1H NMR (CDCl3): δ 3.93 (s, 3H), 7.28-7.29 (m, 1H), 7.37-7.44 (m, 1H), 7.76-7.78 (1H), 7.85-7.89 (m, 1H).
Compound 2a
To a solution of 1a (15.06 g; 85 mmol) in anhydrous THF (150 mL) was added propargylamine (4.68 g; 85 mmol). The reaction mixture was stirred at room temperature overnight. The precipitate formed was collected by suction filtration to give 2a as a white solid (18.5 g; 94% yield), mp 175-177 °C. 1H NMR (DMSO-d6): δ 3.13 (t, J = 2.45 Hz, 1H), 3.80 (s, 3H), 3.88 (dd, J = 5.70, 2.48 Hz, 2H), 6.71 (t, J = 5.69 Hz, 1H), 7.46-7.59 (m, 2H), 7.78-7.91 (m, 2H), 9.11 (s, 1H).
Compound 2b
White solid (90% yield), mp 158-160 °C. 1H NMR (DMSO-d6): δ 3.11 (t, J = 2.46 Hz, 1H), 3.84 (s, 3H), 3.88 (dd, J = 5.72, 2.46 Hz, 2H), 6.61 (t, J = 5.73 Hz, 1H), 7.37 (t, J = 7.87 Hz, 1H), 7.49-7.52 (m, 1H), 7.58-7.62 (m, 1H), 8.12 (t, J = 1.80 Hz, 1H), 8.94 (s, 1H).
Compound 3a
Compound 2a (11.61 g; 50 mmol) and phenylthiomethyl azide (8.26 g; 50 mmol) were suspended in a 1:1 mixture of t-butanol and water (100 mL). Sodium ascorbate (1.00 g; 5 mmol) and CuSO4·5H2O (0.12 g; 0.5 mmol) were then added and the heterogeneous mixture was stirred vigorously at room temperature for 2 days. The progress of the reaction was monitored by TLC. The reaction mixture was diluted with water (200 mL) and cooled in an ice bath. The precipitate formed was collected by suction filtration to give 3a as an off-white powder (19.47 g, 98% yield), mp 155-157 °C. 1H NMR (CDCl3): δ 3.88 (s, 3H), 4.48 (d, J = 5.84 Hz, 2H), 5.58 (s, 2H), 6.86 (t, J = 4.92 Hz, 1H), 7.21-7.34 (m, 5H), 7.39-7.42 (m, 2H), 7.66 (s, 1H), 7.91 (d, J = 8.73 Hz, 3H).
Compound 3b
Light green solid (93% yield), mp 175-177 °C. 1H NMR (DMSO-d6): δ 3.79 (s, 3H), 4.34 (d, J = 5.51 Hz, 2H), 5.57 (s, 2H), 6.79 (br s, 1H), 7.28-7.40 (m, 5H), 7.50-7.53 (m, 2H), 7.82-7.84 (m, 2H), 8.02 (s, 1H), 9.03 (s, 1H).
Compound 3c
Light green solid (100% yield), mp 194-195 °C. 1H NMR (DMSO-d6): δ 3.79 (s, 3H), 4.33 (d, J = 5.55 Hz, 2H), 5.56 (s, 2H), 6.78 (t, J = 5.61, Hz, 1H), 7.17-7.23 (m, 2H), 7.36-7.41 (m, 2H), 7.50-7.54 (m, 2H), 7.82-7.84 (m, 2H), 8.03 (s, 1H), 9.03 (s, 1H).
Compound 4a
White solid (80% yield). 1H NMR (DMSO-d6): δ 3.83 (s, 3H), 4.29 (d, J = 5.67 Hz, 2H), 5.92 (s, 2H), 6.69 (t, J = 5.67 Hz, 1H), 7.25-7.42 (m, 6H), 7.48-7.51 (m, 1H), 7.56-7.59 (m,1H), 7.91 (s, 1H), 8.13 (t, J = 1.92 Hz, 1H), 8.89 (s, 1H).
Compound 4b
White solid (66% yield), mp 158-160 °C. 1H NMR (DMSO-d6): δ 3.83 (s, 3H), 4.33 (d, J = 5.47 Hz, 2H), 5.57 (s, 2H), 6.66 (t, J = 5.37Hz, 1H), 7.29-7.40 (m, 6H), 7.48-7.50 (m, 1H), 7.56-7.59 (m, 1H), 8.02 (s, 1H), 8.13 (t, J = 1.76 Hz, 1H), 8.84 (s, 1H)
Compound 5a
To a solution of ester 3a (17 g; 42.5 mmol) in 1,4-dioxane (500 mL) was added a solution of LiOH (12.21 g; 510 mmol) in water (150 mL) and the reaction mixture was stirred at room temperature overnight. The reaction mixture was cooled in an ice bath and a 5% HCl solution was added dropwise until the pH was ~3. The precipitate formed was collected by suction filtration to give acid 5a as a white solid (18.64 g, 97% yield), mp >230 °C. 1H NMR (DMSO-d6): δ 4.30 (d, J = 5.61 Hz, 2H), 5.93 (s, 2H), 6.77 (t, J = 5.67 Hz, 1H), 7.23-7.42 (m, 5H), 7.48-7.51 (m, 2H), 7.80-7.83 (m, 2H), 7.92 (s, 1H), 9.01 (s, 1H), 12.49-12.72 (br s, 1H).
Compound 5b
White solid (87% yield), mp >230 °C. 1H NMR (DMSO-d6): δ 4.33 (s, 2H), 5.57 (s, 2H), 6.76-6.90 (br s, 1H), 7.28-7.40 (m, 5H), 7.47-7.51 (m, 2H), 7.78-7.82 (m, 2H), 8.02 (s, 1H), 9.10 (s, 1H).
Compound 5c
White solid (99% yield), mp >230 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.61 Hz, 2H), 5.56 (s, 2H), 6.80 (t, J = 5.65 Hz, 1H), 7.16-7.24 (m, 2H), 7.35-7.42 (m, 2H), 7.46-7.50 (m, 2H), 7.78-7.82 (m, 2H), 8.03 (s, 1H), 9.03 (s, 1H), 12.25-12.84 (br s, 1H).
Compound 6a
Off white solid (82% yield), mp 185-189 °C. 1H NMR (DMSO-d6): δ 4.29 (d, J = 5.56 Hz, 2H), 5.92 (s, 2H), 6.74 (t, J = 5.72 Hz, 1H), 7.24-7.49 (m, 7H), 7.57-7.61 (m,1H), 7.91 (s, 1H), 8.06 (t, J = 1.78 Hz, 1H), 8.96 (s, 1H), 12.78-13.15 (br s, 1H).
Compound 6b
Light green powder (88% yield), mp 192-194 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.60 Hz, 2H), 5.57 (s, 2H), 6.65 (t, J = 5.63 Hz, 1H), 7.25-7.39 (m, 6H), 7.45-7.49 (m, 1H), 7.55-7.60 (m, 1H), 8.02 (s, 1H), 8.04-8.06 (m,1H), 8.80 (s, 1H), 12.64-13.06 (br s, 1H).
Compound 7a
To a solution of acid 5a (0.5 g; 1.30 mmol) in anhydrous THF (20 mL) was added CDI (0.21 g; 1.304 mmol) and the mixture was refluxed for 20 min. 4-Phenoxyaniline (0.24 g; 1.30 mmol) was added and the mixture was stirred at room temperature for 2 h. The precipitate formed was collected by suction filtration to give 7a as an off-white solid (0.23 g; 33% yield). 1H NMR (DMSO-d6): δ 4.32 (d, J = 5.60 Hz, 2H), 5.93 (s, 2H), 6.81 (t, J = 5.67Hz, 1H), 6.96-7.14 (m, 5H), 7.23-7.43 (m, 8H), 7.50-7.57 (m, 2H), 7.74-7.81 (m, 2H), 7.93 (s, 2H), 9.01 (s, 1H), 10.09 (s, 1H). HRMS (ESI): Calculated for C30H26N6O3SNa (M+Na) 573.1685; found 573.1689.
Compound 7b
White solid (89% yield), mp 182-183 °C. 1H NMR (DMSO-d6): δ 4.30 (d, J = 5.63 Hz, 2H), 4.45 (d, J = 5.99 Hz, 2H), 5.93 (s, 2H), 6.90 (t, J = 5.40 Hz 1H), 7.20-7.49 (m,12H), 7.79 (d, J = 8.83 Hz, 2H), 7.92 (s, 1H), 8.86 (d, J = 5.98 Hz, 1H), 9.04 (s, 1H). HRMS (ESI): Calculated for C25H24N6O2SNa (M+Na) 495.1579; found 495.1596.
Compound 7c
White solid (68% yield), mp 201-202 °C. 1H NMR (DMSO-d6): δ 2.82 (t, J = 7.45 Hz, 2H), 3.41-3.49 (m, 2H), 4.30 (d, J = 5.59 Hz, 2H), 5.92 (s, 2H), 6.83 (t, 5.64 Hz, 1H), 7.19-7.35 (m, 8H), 7.38-7.46 (m, 4H), 7.70-7.73 (m, 2H), 7.91 (s, 1H), 8.36 (t, J = 5.64 Hz, 1H), 8.96 (s, 1H). HRMS (ESI): Calculated for C26H26N6O2SNa (M+Na) 509.1736; found 509.1730.
Compound 7d
Off white solid (88% yield), mp 190-191 °C. 1H NMR (DMSO-d6):δ 4.29-4.35 (m, 2H), 5.94 (s, 2H), 6.76 (t, J = 5.71 Hz, 1H), 7.04-7.10 (m, 2H), 7.24-7.36 (m, 5H), 7.39-7.43 (m, 2H), 7.48-7.55 (m, 2H), 7.74-7.93 (m, 4H), 8.98 (s, 1H), 10.05 (s, 1H). HRMS (ESI): Calculated for C24H22N6O2SNa (M+Na) 481.1423; found 481.1439.
Compound 7e
Light brown solid (90% yield), mp 146-148 °C. 1H NMR (DMSO-d6): δ 4.31 (d, J = 5.55 Hz, 2H), 4.44 (d, J = 5.62 Hz, 2H), 5.93 (s, 1H), 6.86 (t, J = 5.50 Hz, 1H), 7.04 (br s, 2H), 7.22-7.92 (m, 11H), 8.79 (t, J = 5.71Hz, 1H), 9.02 (s, 1H). HRMS (ESI): Calculated for C23H22N6O3SNa (M+Na) 485.1372; found 485.1373
Compound 7f
Off white solid (46% yield), mp 184-185 °C. 1H NMR (DMSO-d6): δ 2.40-2.45 (m, 8H), 3.56 (t, J = 4.45 Hz, 4H), 4.30 (d, J = 5.57 Hz, 2H), 5.93 (s, 2H), 6.89 (t, J = 5.18 Hz, 1H), 7.24-7.47 (m, 7H), 7.71-7.73 (m, 2H), 7.92 (s, 1H), 8.21 (t, J = 5.80 Hz, 1H), 9.02 (s, 1H). HRMS (ESI): Calculated for C24H30N3O7S (M+H) 496.2131; found 496.2114.
Compound 7g
Light pink solid (47% yield), mp > 230 °C. 1H NMR (DMSO-d6): δ 4.35 (d, J = 5.56 Hz, 2H), 5.58 (s, 2H), 6.77 (t, J = 5.67 Hz, 1H), 6.96-7.05 (m, 4H), 7.08-7.14 (m, 1H), 7.31-7.41 (m, 7H), 7.53 (d, J = 8.78 Hz, 2H), 7.75-7.80 (m, 2H), 7.87 (d, J = 8.77 Hz, 2H), 8.03(s, 1H), 8.95 (s, 1H), 10.08 (s, 1H). 13C NMR (DMSO-d6): δ 164.71, 157.27, 154.63, 151.71, 145.55, 143.43, 136.04, 135.17, 129.85, 128.62, 128.00, 127.85, 126.88, 122.84, 122.70, 122.57, 121.88, 119.18, 117.75, 116.53, 52.61, 34.74. HRMS (ESI): Calculated for C30H26N6O3SNa (M+Na) 541.1964; found 541.1956.
Compound 7h
White solid (54% yield), mp 213-214 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.56 Hz, 2H), 4.45 (d, J = 5.96 Hz, 2H), 5.57 (s, 2H), 6.74 (t, J = 5.53 Hz, 1H), 7.30-7.40 (m, 10H), 7.44-7.48 (m, 2H), 7.76-7.82 (m, 2H), 8.02 (s, 1H), 8.78-8.94 (m, 2H). HRMS (ESI): Calculated for C25H24N6O2SNa (M+Na) 463.1858; found 463.1844.
Compound 7i
White solid (62% yield), mp 230 °C. 1H NMR (DMSO-d6): δ 2.82 (t, J = 7.0 Hz, 2H), 3.41-3.48 (m, 2H), 4.33 (d, J = 5.57 Hz, 2H), 5.57 (s, 2H), 6.74 (t, J = 5.59 Hz, 1H), 7.16-7.40 (m, 10H), 7.42-7.47 (m, 2H), 7.70-7.73 (m, 2H), 8.02 (s, 1H), 8.38 (t, J = 5.65 Hz,1H), 8.87 (s, 1H). HRMS (ESI): Calculated for C26H26N6O2SNa (M+Na) 477.2015; found 477.2036.
Compound 7j
White solid (35% yield), mp > 230 °C. 1H NMR (DMSO-d6): δ 4.35 (d, J = 5.56 Hz, 2H), 5.58 (s, 2H), 6.75 (t, J = 5.56 Hz, 1H), 7.07 (t, J = 7.37 Hz, 1H), 7.29-7.40 (m, 7H), 7.50-7.54 (m, 2H), 7.74-7.78 (m, 2H), 7.85-7.90 (m, 2H), 8.03 (s, 1H), 8.94 (s, 1H), 10.04 (s, 1H). HRMS (ESI): Calculated for C24H22N6O2SNa (M+Na) 449.1702; found 449.1722.
Compound 7k
White solid (35% yield), mp 207-208 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.56 Hz, 2H), 4.43 (d, J = 5.66 Hz, 2H), 5.57 (s, 2H), 6.24-6.25 (m, 1H), 6.38-6.39 (m, 1H), 6.76 (t, J = 5.59 Hz, 1H), 7.28-7.40 (m, 5H), 7.42-7.46 (m, 2H), 7.56-7.57 (m, 1H), 7.74-7.78 (m, 2H), 8.02 (s, 1H), 8.76(t, J = 5.80 Hz, 1H), 8.89 (s, 1H). HRMS (ESI): Calculated for C23H22N6O3SNa (M+Na) 453.1651; found 453.1651.
Compound 7l
White solid (45% yield), mp 204-206 °C. 1H NMR (DMSO-d6): δ 2.36-2.45 (m, 6H), 3.33-3.37 (m, 2H), 3.56 (t, J = 4.59 Hz, 4H), 4.33 (d, J = 5.57 Hz, 2H), 5.57 (s, 2H), 6.77 (t, J = 5.63 Hz, 1H), 7.29-7.39 (m, 5H), 7.42-7.45 (m, 2 H), 7.70-7.73 (m, 2H), 8.02 (s, 1H), 8.21 (t, J = 5.67 Hz, 1H), 8.91(s, 1H). HRMS (ESI): Calculated for C24H30N7O3 (M+H) 464.2410; found 464.2422.
Compound 7m
Light pink solid (25% yield), mp 199-200 °C. 1H NMR (DMSO-d6): δ 4.34 (d, J = 5.55 Hz, 2H), 5.57 (s, 2H), 6.72-6.77 (m, 2H), 7.02-7.07 (m, 2H), 7.12-7.18 (m, 1H), 7.30-7.44 (m, 8H), 7.49-7.58 (m, 4H), 7.82-7.85 (m, 2H), 8.02 (s, 1H), 8.94 (s, 1H), 10.10 (s, 1H). HRMS (ESI): Calculated for C30H26N6O3Na (M+Na) 541.1964; found 541.1932.
Compound 7n
White solid (49% yield), mp >230 °C. 1H NMR (DMSO-d6): δ 4.32 (d, J = 5.59 Hz, 2H), 5.55 (s, 2H), 6.71 (t, J = 5.62 Hz, 1H), 6.93-7.04 (m, 4H), 7.05-7.12 (m, 1H), 7.14-7.24 (m, 2H), 7.32-7.42 (m, 4H), 7.47-7.52 (m, 2H), 7.72-7.80 (m, 2H), 7.82-7.90 (m, 2H), 8.02 (s, 1H), 8.91 (s, 1H), 10.06 (s, 1H). 13C NMR (DMSO-d6): δ 164.72, 162.97, 160.54, 157.28, 154.64, 151.73, 145.62, 143.44, 135.18, 132.34, 130.24, 129.87, 128.54, 126.90, 122.86, 122.59, 121.85, 119.17, 117.77, 116.51, 115.58, 115.37, 51.80, 34.74. HRMS (ESI): Calculated for C30H25FN6O3Na (M+Na) 559.1870; found 559.1859.
Compound 7o
White solid (91% yield), mp 208-209 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.56 Hz, 2H), 4.45 (d, J = 5.94 Hz, 2H), 5.56 (s, 2H), 6.98 (t, J = 5.58 Hz, 1H), 7.15-7.26 (m, 3H), 7.28-7.33 (m, 4H), 7.34-7.43 (m, 2H), 7.46-7.49 (m, 2H), 7.77-7.80 (m, 2H), 8.03 (s, 1H), 8.85 (t, J = 5.95 Hz, 1H), 9.10 (s, 1H). HRMS (ESI): Calculated for C25H23FN6O2Na (M+Na) 481.1761; found 481.1754.
Compound 7p
White solid (90% yield), mp 207-209 °C. 1H NMR (DMSO-d6): δ 2.82 (t, J = 7.46 Hz, 2H), 3.43 (m, 2H), 4.33 (d, J = 5.55 Hz, 2H), 5.56 (s, 2H), 7.05 (t, J = 5.56 Hz, 1H), 7.16-7.41 (m, 9H), 7.46 (d, J = 8.79 Hz, 2H), 7.71 (d, J = 8.75 Hz, 2H), 8.03 (s, 1H), 8.38 (t, J = 5.55 Hz, 1H), 9.17 (s, 1H). HRMS (ESI): Calculated for C26H25FN6O2Na (M+Na) 495.1921; found 495.1921.
Compound 7q
White solid (46% yield), mp >230 °C. 1H NMR (DMSO-d6): δ 4.35 (d, J = 5.55 Hz, 2H), 5.57 (s, 2H), 6.86 (t, J = 5.62 Hz, 1H), 7.04-7.09 (m, 1H), 7.17-7.43 (m, 7H), 7.53 (d, J = 8.78 Hz, 1H), 7.75-7.79 (m, 2H), 7.88 (d, J = 8.75 Hz, 2H), 8.04 (s, 1H), 9.12 (s, 1H), s (10.06, 1H). HRMS (ESI): Calculated for C24H21FN6O2Na (M+Na) 467.1608; found 467.1594.
Compound 7r
White solid (99% yield), mp 189-191 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.60 Hz, 2H), 4.43 (d, J = 5.65 Hz, 2H), 5.56 (s, 2H), 6.24-6.25 (m, 1H), 6.37-6.39 (m, 1H), 6.79-6.86 (m, 1H), 7.02 (s, 1H), 7.16-7.24 (m, 2H), 7.34-7.50 (m, 4H), 7.72-7.83 (m, 2H), 8.03 (s, 1H), 8.76 (t, J = 5.70Hz, 1H), 9.00 (s, 1H). HRMS (ESI): Calculated for C23H21FN6O3Na (M+Na) 471.1557; found 471.1555.
Compound 7s
(90% yield), mp 163-166 °C. 1H NMR (DMSO-d6) δ 2.35-2.47 (m, 8H), 3.51- 3.63 (m, 4H), 4.32 (d, J = 5.56 Hz, 2H), 5.56 (s, 2H), 6.83 (t, J = 5.62 Hz, 1H), 7.01 (s, 1H), 7.16-7.24 (m, 2H), 7.36-7.47 (m, 4H), 7.69-7.73 (m, 2H), 8.02 (s, 1H), 8.21 (t, J = 5.64 Hz, 1H), 9.01 (s, 1H). HRMS (ESI): Calculated for C24H29FN7O3 (M+Na) 482.2316; found 482.2297.
Compound 7t
Off white solid (16% yield), mp 211-212 °C. 1H NMR (DMSO-d6): δ 4.34 (d, J = 5.50 Hz, 2H), 5.56 (s, 2H), 6.69-6.80 (m, 2H), 7.00-7.09 (m, 2H), 7.10-7.27 (m, 3H), 7.29-7.46 (m, 5H), 7.47-7.62 (m, 4H), 7.84 (d, J = 8.78 Hz, 2H), 8.03 (s, 1H), 8.95 (s, 1H), 10.10 (s, 1H). 13C NMR (DMSO-d6): δ 164.96, 162.97, 160.54, 156.75, 156.43, 154.61, 145.61, 143.56, 140.89, 132.34, 130.24, 129.95, 128.66, 126.71, 123.39, 122.63, 118.68, 116.46, 115.58, 115.37, 114.89, 109.95, 51.79, 34.74. HRMS (ESI): Calculated for C30H25FN6O3Na (M+Na) 559.1870; found 559.1862.
Compound 8a
White solid (21% yield), mp 156-157 °C. 1H NMR (DMSO-d6): δ 4.30 (d, J = 5.64 Hz, 2H), 4.45 (d, J = 5.99 Hz, 2H), 5.93 (s, 2H), 6.65 (t, J = 5.67 Hz, 1H), 7.22-7.44 (m, 12H), 7.56-7.61(m, 1H), 7.85 (t, J =1.79 Hz, 1H), 7.91 (s, 1H), 8.75 (s, 1H), 8.97 (t, J = 6.07 Hz, 1H). HRMS (ESI): Calculated for C25H24N6O2SNa (M+Na) 495.1579; found 495.1567.
Compound 8b
White solid (57% yield), mp 155-156 °C. 1H NMR (DMSO-d6): δ 2.83 (t, J = 7.46 Hz, 2H), 3.45 (q, J = 6.30 Hz, 2H), 4.30 (s, 2H), 5.93 (s, 2H), 6.72 (br s, 1H), 7.19-7.42 (m, 12H), 7.55-7.59 (m, 1H), 7.79 (t, J = 1.80 Hz, 1H), 7.91 (s, 1H), 8.50 (t, J = 5.55 Hz, 1H), 8.83 (s, 1H). HRMS (ESI): Calculated for C26H26N6O2SNa (M+Na) 509.1736; found 509.1757.
Compound 8c
White solid (21% yield), mp 179-181 °C. 1H NMR (DMSO-d6): δ 4.30 (d, J = 5.65 Hz, 2H), 5.93 (s, 2H), 6.72-6.82 (m, 2H), 7.02-7.08 (m, 2H), 7.12-7.20 (m, 1H), 7.22-7.47 (m, 10H), 7.50-7.65 (m, 3H), 7.87 (t, J = 1.76 Hz, 1H), 7.91 (s, 1H), 8.90 (s, 1H), 10.28 (s, 1H). 13C NMR (DMSO-d6): δ 165.79, 156.79, 156.37, 154.89, 145.89, 140.70, 140.42, 135.42, 132.50, 130.25, 129.95, 129.84, 129.14, 128.54, 127.47, 123.43, 122.37, 120.63, 120.09, 118.71, 116.98, 114.87, 113.52, 109.93, 51.36, 34.69. HRMS (ESI): Calculated for C30H26N6O3SNa (M+Na) 573.1685; found 573.1693.
Compound 8d
White solid (80% yield), mp 162-164 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.58 Hz, 2H), 4.45 (d, J = 5.97 Hz, 2H), 5.57 (s, 2H), 6.67 (t, J = 5.61 Hz, 1H), 7.2-7.42 (m, 12H), 7.57-7.60 (m, 1H), 7.84 (t, J = 1.64 Hz, 1H), 8.01 (s, 1H), 8.75 (s, 1H), 8.96 (t, J = 5.96 Hz, 1H). HRMS (ESI): Calculated for C25H24N6O2Na (M+Na) 509.1736; found 509.1757.
Compound 8e
White solid (83% yield), mp 197-198 °C. 1H NMR (DMSO-d6): δ 2.83 (t, J = 7.45 Hz, 2H), 3.41-3.49 (m, 2H), 4.33 (d, J = 5.56 Hz, 2H), 5.57 (s, 2H), 6.68 (t, J = 5.65 Hz, 1H), 7.17-7.40 (m, 12H), 7.55-7.58 (m, 1H), 7.78 (t, J = 1.65 Hz, 1H), 8.01 (s, 1H), 8.48 (t, J = 5.60 Hz, 1H), 8.74 (s, 1H). HRMS (ESI): Calculated for C26H26N6O2Na (M+Na) 477.2015; found 477.2032.
Compound 8f
White solid (64% yield), mp 201-202 °C. 1H NMR (DMSO-d6): δ 4.33 (d, J = 5.53 Hz, 2H), 5.57 (s, 2H), 6.69-6.79 (m, 2H),7.02-7.08 (m, 2H), 7.13-7.19 (m, 1H), 7.29-7.46 (m, 10H), 7.51-7.64 (m, 3H), 7.86 (s, 1H), 8.01 (s, 1H), 8.83 (s, 1H), 10.28 (s, 1H). HRMS (ESI): Calculated for C30H26N6O3Na (M+Na) 541.1945; found 541.1945.
Biochemistry
DENV2 NS2B/NS3pro expression and purification
The construction of the pQE30-NS2BH(QR)NS3pro expression plasmid has been described previously.27 The protease was expressed from E. coli strain Top 10 F’ (Invitrogen) transformed by pQE30-NS2BH(QR)-NS3pro plasmid and purified as described.29 DENV2 NS2B/NS3pro contains the hydrophilic NS2B cofactor peptide (NS2BH) linked to the N-terminal NS3 protease domain via Q-R (P2 and P1 residues at the NS2B-NS3 junction site).
WNV NS2B/NS3pro expression and purification
The expression and purification of the WNV NS2BH-NS3pro containing the 5 amino acid spacer between the NS2B and NS3pro domains was previously described.29
In vitro DENV2 and WNV NS2B/NS3pro assays and inhibition studies
The compounds were dissolved in dimethyl sulfoxide (DMSO) to make 50 mM stock solutions. The compounds were screened at 25 μM in 1% v/v DMSO in the final reaction mixture. Protease assays were performed in triplicates in Greiner Black 96 well plates. Each assay consisted of the reaction mixture of 100 μL containing 200 mM Tris·HCl buffer, pH 9.5, 30% glycerol, 25 nM DENV2 NS2B-NS3 pro (or 28 nM WNV NS2B/NS3pro) and the compound. The enzyme and the compound were pre-incubated at room temperature for 15 min prior to addition of the substrate (5 μM), Bz-Nle-Lys-Arg-Arg-AMC. The time course of the reaction at 37 °C was followed at every 90 sec intervals for up to 30 min in a monochrometer-based spectrofluorometer (Molecular Devices, Sunnyvale, CA) at excitation and emission wavelengths of 380 and 460 nm, respectively. The percent inhibition for each compound at 25 μM was first determined. For determining IC50 values, twelve data points obtained from the range of 10 nM, 50 nM, 0.1, 0.5, 1, 2, 4, 6, 8, 10, 20, and 25 μM inhibitor concentrations of selected compounds were used. IC50 values were calculated using the SigmaPlot 2001 v7.0 software.41
Kinetics Analysis
To determine the Km and Vmax values of compound 7n, four different concentrations of inhibitor (0, 1.0, 3.0 and 5.0 μM) were assayed at varying concentrations (0-50 μM) of substrate. The kinetics analysis methods have been previously described.42 Ki values were calculated from these data using a secondary plot of Km, app against the concentrations of selected compound 7n using SigmaPlot 2001 v7.0 software.41
Molecular Modeling
Molecular docking simulations were performed with the Surflex program.43 The DENV NS3/NS2B receptor was modeled from the active form crystal structure of Noble et al. (PDB ID: 3U1I)12 by extracting all ligands and waters, and protonating the receptor according to assumption of anionic aspartate and glutamate groups, and cationic arginines and lysines). The WNV NS2B/NS3pro receptor model was prepared in an analogous manner using the crystal structure of Erbel et al. (PDB ID 2FP7)13. In both cases, the co-crystallized ligand was used to define the Surflex protomol structure (which guides the original ligand binding site prediction), and 5 distinct randomized starting conformations and 100 final conformations were specified for the ligand. Ligands were constructed and refined according to default molecular mechanics constraints, force fields and optimization settings via the SYBYL 8.1 program (Tripos Associates, St. Louis, MO, 2009).
Acknowledgments
The generous financial support of this work by the National Institutes of Health (AI082068) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindenbach BD, Thiel H-J, Rice CM. In: Fields Virology. 5. Knipe DM, Howley PM, editors. one. Lippincott, Williams, and Wilkins; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- 2.Stevens AJ, Gahan ME, Mahalingam S, Keller PA. J Med Chem. 2009;52:7911. doi: 10.1021/jm900652e. [DOI] [PubMed] [Google Scholar]
- 3.Cregar-Hernandez L, Jiao GS, Johnson AT, Lehrer AT, Wong TAS, Margosiak SA. Antiviral Chem Chemother. 2011;21:209. doi: 10.3851/IMP1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble CG, Chen Y-L, Dong H, Gu F, Lim SP, Schull W, Wang Q-Y, Shi P-Y. Antiviral Res. 2010;85:450. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Sampath A, Padmanabhan R. Antiviral Res. 2009;81:6. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Antiviral Res. 2008;80:94. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. J Biol Chem. 2000;275:9963. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 8.Natarajan S. Gen Mol Biol. 2010;33:214. doi: 10.1590/S1415-47572010000200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schechter I, Berger A. Biochem Biophys Res Comm. 1967;27:157. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 10.Niyomrattanakit P, Winoyanuwattikun P, Chanprapaph S, Angsuthanasombat C, Panyim S, Katzenmeier G. J Virol. 2004;78:13708. doi: 10.1128/JVI.78.24.13708-13716.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wichapong K, Pianwanit S, Sippl W, Kokpol S. J Mol Recognit. 2010;23:283. doi: 10.1002/jmr.977. [DOI] [PubMed] [Google Scholar]
- 12.Noble CG, She CC, Chao AT, Shi PY. J Virol. 2012;86:438. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH. Nat Struct Mol Biol. 2006;13:372. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 14.de la Cruz L, Nguyen TH, Ozawa K, Shin J, Graham B, Huber T, Otting G. J Am Chem Soc. 2011;133:19205. doi: 10.1021/ja208435s. [DOI] [PubMed] [Google Scholar]
- 15.Schuller A, Yin Z, Chia BCS, Doan DNP, Kim H-K, Shang L, Loh TP, Hill J, Vasudevan SG. Antiviral Res. 2011;92:96. doi: 10.1016/j.antiviral.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Yin Z, Patel SJ, Wang WL, Wang G, Chan WL, Rao KR, Alam J, Jeyaraj DA, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. Bioorg Med Chem Lett. 2006;16:36. doi: 10.1016/j.bmcl.2005.09.062. [DOI] [PubMed] [Google Scholar]
- 17.Yin Z, Patel SJ, Wang WL, Chan WL, Ranga Rao KR, Ngew X, Patel V, Beer D, Knox JE, Ma NL, Ehrhardt C, Lim SP, Vasudevan SG, Keller TH. Bioorg Med Chem Lett. 2006;16:40. doi: 10.1016/j.bmcl.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 18.Knox JE, Ma NL, Yin Z, Patel SJ, Wang WL, Wang G, Chan WL, Vanga Rao KR, Wang G, Ngew X, Patel V, Beer D, Lim SP, Vasudevan SG, Keller TH. J Med Chem. 2006;49:6585. doi: 10.1021/jm0607606. [DOI] [PubMed] [Google Scholar]
- 19.Steuer C, Gege C, Fischl W, Heinonen KH, Bartenschlager R, Klein CD. Bioorg Med Chem. 2011;19:4067. doi: 10.1016/j.bmc.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Bodenreider C, Beer D, Keller TH, Sonntag S, Wen D, Yap L, Yau YH, Sochat SG, Huang D, Zhou T, Caflisch A, Su X-C, Ozawa K, Otting G, Vasudevan SG, Lescar J, Lim SP. Anal Biochem. 2009;395:195. doi: 10.1016/j.ab.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Nitsche C, Steuer C, Klein CD. Bioorg Med Chem. 2011;19:7318. doi: 10.1016/j.bmc.2011.10.061. [DOI] [PubMed] [Google Scholar]
- 22.Nitsche C, Behnam MAM, Steuer C, Klein CD. Antiviral Res. 2012;94:72. doi: 10.1016/j.antiviral.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Tiew KC, Dou D, Teramoto T, Lai H, Alliston KR, Lushington GH, Padmanabhan R, Groutas WC. Bioorg Med Chem. 2012;20:1213. doi: 10.1016/j.bmc.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens AJ, Gahan ME, Mahalingam S, Keller PA. J Med Chem. 2009;52:7911. doi: 10.1021/jm900652e. [DOI] [PubMed] [Google Scholar]
- 25.Parkinson T, Pryde DC. Future Med Chem. 2010;2:1181. doi: 10.4155/fmc.10.195. [DOI] [PubMed] [Google Scholar]
- 26.(a) Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; (b) Shen J, Woodward R, Kedenburg JP, Liu X, Chen M, Fang L, Sun D, Wang PG. J Med Chem. 2008;51:7417. doi: 10.1021/jm8005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KH, Padmanabhan R. J Biol Chem. 2005;280:27412. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 28.Yusof RS, Clum M, Wetzel H, Murthy HM, Padmanabhan R. J Biol Chem. 2000;265:9963. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 29.Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Int J Biochem Cell Biol. 2007;39:606. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Dou D, Viwanathan P, Li Y, He G, Alliston KR, Lushington GH, Brown-Clay JD, Padmanabhan R, Groutas WC. J Comb Chem. 2010;12:836. doi: 10.1021/cc100091h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Lim P, Beer D, Patel V, Wen D, Tumanut C, Tully DC, Williams JA, Jiricek J, Priestle JP, Harris JL, Vasudevan SG. J Biol Chem. 2005;280:28766. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- 32.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Antimicrob Agents Chemother. 2008;52:3385. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gleeson M. J Med Chem. 2008;51:817. doi: 10.1021/jm701122q. [DOI] [PubMed] [Google Scholar]
- 34.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Delivery Rev. 1997;23:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 35.Keller TH, Pichota A, Yin Z. Curr Opin Chem Biol. 2006;10:357. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Johnson TW, Dress KR, Edwards M. Bioorg Med Chem Lett. 2009;19:5560. doi: 10.1016/j.bmcl.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 37.Perola E. J Med Chem. 2010;53:2986. doi: 10.1021/jm100118x. [DOI] [PubMed] [Google Scholar]
- 38.Leeson PD, Empfield JR. Ann Rep Med Chem. 2010;45:393. [Google Scholar]
- 39.Walters WP, Green J, Weiss JR, Murcko MA. J Med Chem. 2011;54:6405. doi: 10.1021/jm200504p. [DOI] [PubMed] [Google Scholar]
- 40.Waring MJ. Expert Opin Drug Discov. 2010;5:235. doi: 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- 41.Systat Software Inc.; San Jose, CA: [Google Scholar]
- 42.Ezgimen M, Lai H, Mueller NH, Lee K, Cuny G, Ostrov DA, Padmanabhan R. Antiviral Res. 2012;94:18. doi: 10.1016/j.antiviral.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jain AN. J Med Chem. 2003;46:499. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]


