SUMMARY
Previous work demonstrated recently the adaptation of the E. coli biotin ligase BirA - biotin acceptor sequence (BAS) labeling system to produce human immunodeficiency virus type 1 viruses with biotinylated integrase (NLXINB) and matrix (NLXMAB) proteins (Belshan et al., 2009). This report describes the construction of an HIV permissive cell line stably expressing BirA (SupT1.BirA). Consistent with the results in the previous report, NLXMAB replicated similar to wild-type levels and expressed biotinylated Gag and MA proteins in the SupT1.BirA cells, whereas the replication of NLXINB was reduced severely. Three additional HIV type 2 (HIV-2) viruses were constructed with the BAS inserted into the vpx and vpr accessory genes. Two BAS insertions were made into the C-terminal half of the Vpx, including one internal insertion, and one at the N-terminus of Vpr. All three viruses were replication competent in the SupT1.BirA cells and their target proteins biotinylated efficiently and incorporated into virions. These results demonstrate the potential utility of the biotinylation system to label and capture HIV protein complexes in the context of replicating virus.
Keywords: human immunodeficiency virus type 1 and type 2, biotinylation, matrix, integrase, vpx, vpr
Human immunodeficiency virus 1 (HIV-1), a member of the genus Lentivirus in the Retroviridae family interacts with a myriad of cellular proteins to complete a productive replication cycle. Identifying and characterizing these interactions may lead to an increased knowledge of HIV pathology and the eventual discovery of new targets for therapeutic inhibition of HIV replication. Three recent genome-wide siRNA screens individually identified >200 potential factors critical for the productive infection by the virus (Brass et al., 2008; Konig et al., 2008; Zhou et al., 2008). Unexpectedly the overlap between these screens was minimal (~16 genes). Moreover, the validation of all the potential targets will take years to complete. An alternative method to genome wide screening for viral factors is the characterization of HIV-host cell proteome. Whole cell, or so-called “shotgun” proteomics, has proven to be of limited utility given current mass spectrometry (MS) technologies. Historically the analyses of protein-protein interactions have been limited to screens performed with individual viral proteins. A common protein-based purification strategy that has been utilized to identify potential HIV-interacting cellular proteins is to capture protein complexes using single or tandem-affinity epitope tagged viral proteins to capture protein complexes. This approach is not ideal since the viral bait protein is usually expressed as a standalone expression construct, and the experiments are performed in cell lines that are transfected efficiently, but not necessarily permissive for HIV-1 infection. Genetic studies have established that HIV infection results in a rearrangement of the expression of a multitude of genes and gene pathways (van 't Wout et al., 2003). Therefore it would be advantageous to study HIV-cellular interactions in the context of productive HIV infection. This requires the insertion of an affinity tag into a HIV-1 protein that maintains protein function and virus replication. The effectiveness of this strategy was demonstrated by the identification of the interaction of HIV-1 Vif with Cul5 using a molecular clone containing HA-tagged Vif (Yu et al., 2003).
A previous study reported the adaptation of the E. coli biotin ligase BirA - biotin acceptor sequence (BAS) labeling system to biotinylate HIV-1 in vivo (Belshan et al., 2009). For this system the 20 amino acid BAS is inserted into the bait protein. Co-expression of BirA with a BAS-containing protein results in the covalent attachment of biotin to a central lysine residue in the BAS (Beckett et al., 1999; Schatz, 1993). Two HIV-1 molecular clones were characterized previously that contained BAS insertions into the C-terminus of the matrix (NLXMAB) and integrase (NLXINB) proteins (Belshan et al., 2009). Both viruses produced virions containing their respective biotinylated proteins from 293T cells expressing BirA. Replication assay in T-cells demonstrated that the MA insertion was well tolerated, but the IN insertion reduced virus viability. Single-round infectivity assays with biotinylated viruses confirmed that the MA virus was as fit as wild-type virus, but the biotinylated IN virus was not infectious. The construction of a 293T cell line that exhibited stable expression BirA for the consistent production of biotinylated virus was also reported. A recent report also described the production of biotinylated IN and MA using a lentiviral transduction system (Benkhelifa-Ziyyat et al., 2010); however that system does not permit productive replication of biotinylated proteins.
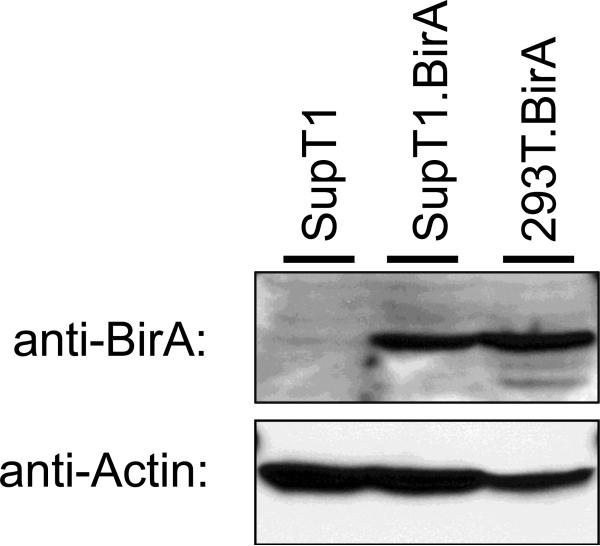
Belshan et al. (2009) demonstrated the feasibility and specificity of the system to biotinylate HIV proteins in vivo, however the labeling was limited to virus produced by 293T cells expressing BirA. The next goal of these studies was to label HIV proteins in the context of replicating virus. To accomplish this, a HIV-1 permissive cell line that expressed stable levels of BirA was produced. 5×107 SupT1 cells were electroporated with 20 μg of the pc6BirA expression plasmid in a 0.4 cm gap electoporation cuvette and electroporated using a Gene Pulser II electroporator (Bio-Rad Laboratories, Hercules, CA USA) set to 300 V and 975 μF. After electroporation the cells were expanded for two days and then passaged into media supplemented with 10 μg/ml Blasticidin S to select for pc6BirA plasmid maintenance. Cells were propagated, and media changed every 2-3 days, until visible cell growth was observed by media color change (approximately three weeks). Single cell clones were obtained by seeding the cells into 96-well plates at 0.1 cells/well. Cells were grown until colonies were visible and only wells with single colonies were expanded. Once expanded sufficiently the cell clones were examined for BirA expression by Western blot. Several SupT1.BirA cell lines were identified to express BirA at high levels and a single high expressing cell line was chosen for future experiments. The expression of BirA in the SupT1.BirA cell line as well as the 293T.BirA cell line described previously is demonstrated in Figure 1.
Fig. 1.
Cell lines that stably express BirA. Western blot analysis of BirA expression in SupT1, SupT1.BirA, and the previously described 293T.BirA cell lines (top panel). Cells were lysed with M-PER solution (Pierce Biotechnology, Rockford, IL USA), the lysates clarified by centrifugation and separated by SDS-PAGE. BirA was detected by Western blot using an anti-BirA antibody (Genway Biotech, San Diego, CA USA) followed by an anti-chicken IgY-HRP conjugated secondary antibody. A control Western blot for Actin (Santa Cruz Biotechnology, Santa Cruz CA USA) was also performed as a normalization control (lower panel).
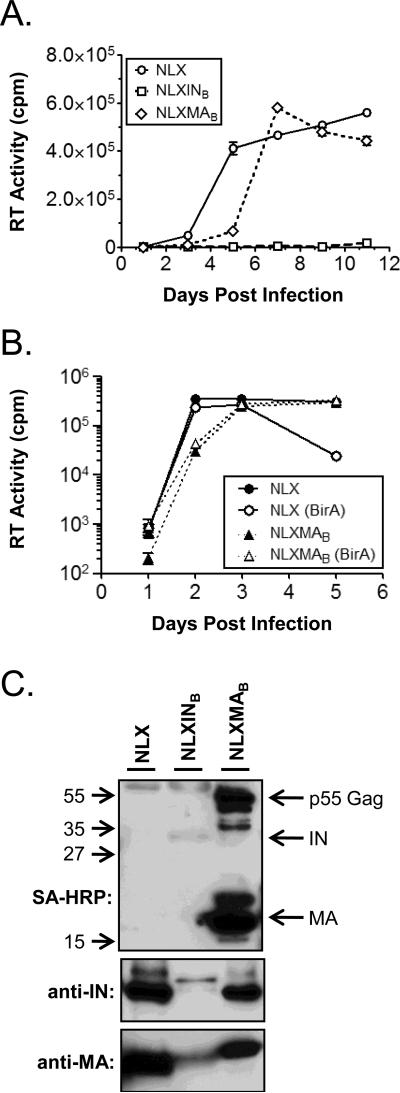
To establish that the SupT1.BirA cells retained the ability to support HIV replication and test their ability to biotinylate HIV proteins in the context of ongoing replication, the cells were infected with wild-type HIV-1 NLX and the NLXMAB and NLXINB viruses and supernatants were collected (Fig. 2A). SupT1.BirA cells were inoculated overnight with normalized amounts of each virus, the virus removed the next day, and the cells propagated for 11 days. The supernatants were sampled in triplicate at various days post infection and virus replication measured by an in vitro [32P]TTP incorporation reverse transcriptase (RT) assay (Belshan et al., 2009). The parental NLX virus replicated well in the SupT1.BirA cells indicating that the expression of BirA was not deleterious for HIV replication. The replication of the NLXMAB virus was similar to wild-type NLX virus, consistent with the previous findings in parental SupT1 cells ((Belshan et al., 2009) and summarized in Table 1). In contrast, NLXINB failed to substantially replicate in the SupT1.BirA cells. These results differed with the previous results for SupT1 cells, in which NLXINB replicated at a delayed rate compared to NLX, but this finding was consistent with the loss of infectivity of biotinylated NLXINB in single-round infectivity assays (Belshan et al., 2009). Next it was assessed whether the SupT1.BirA cells supported HIV-1 infection similar to parental SupT1 cells. SupT1 and SupT1.BirA cells were infected in parallel with normalized amounts of either NLX or NLXMAB and replication again monitored by RT assay of supernatants (Fig. 2B). The replication of both NLX and NLXMAB was similar in the SupT1 and SupT1.BirA cells. This data demonstrated that the SupT1.BirA cells support HIV-1 replication levels comparable to the parental SupT1 cells and confirmed that the biotinylation of MA was not deleterious to the replication of NLXMAB.
Fig. 2.
HIV-1 replication and biotinylation in SupT1.BirA cells. (A) SupT1.BirA cells were inoculated overnight with normalized levels of the indicated viruses, the cells washed, and propagated. Supernatants were collected in triplicate and clarified by centrifugation on the days indicated. At the end of the experiment the samples were analyzed for exogenous RT activity using a [32P]TTP incorporation assay. (B) SupT1 (black symbols) and SupT1.BirA cells (open symbols, (BirA)) were infected in parallel with normalized amounts of the indicated virus and monitored for replication as described in (A). Error bars in both (A) and (B) denote the standard deviation for the triplicate samples in one experiment. (C) At the end of replication experiments 12 ml of supernatant was collected, clarified, and concentrated by ultracentrifugation through a 20% sucrose cushion (w/v in PBS). Samples were resuspended in 1x Sample Buffer, separated by SDS-PAGE and the indicated proteins detected by Western blot. The location of viral proteins in the streptavidin-horseradish peroxidase (SA-HRP) blot is shown at the right of the blot and the approximate location of molecular weight markers is shown on the left. The data shown in (A) and (C) is representative of three independent experiments and (B) is representative of duplicate experiments.
Table 1.
Summary of HIV-BAS mutants
| Virus | Protein | Site of Insertion1 | Biotinylated?2 | Replication Competence2 | Notes2 |
|---|---|---|---|---|---|
| HIV-1 | MA | 120 | Yes | ++++ | |
| IN | 5 | No | - | Unstable IN | |
| 176 | No | - | No infectious virus | ||
| 212 | ND | - | No infectious virus | ||
| 247 | ND | - | No infectious virus | ||
| 2893 | Yes | +/- | |||
| HIV-2 | Vpr | 1 | Yes | ++++ | |
| Vpx | 71 | Yes | ++++ | ||
| 112 | Yes | ++++ |
Amino acid immediately prior to insertion
Data not shown for IN-5 and -176; IN-212 & -247 previously reported in (Belshan et al., 2009)
Clone referred to as NLXINB in text
At the termination of each experiment supernatants were collected and concentrated to confirm the expression and biotinylation of the target proteins. Supernatants were clarified and virus particles concentrated by ultracentrifugation through a 20% sucrose (w/v) cushion. Virus samples were resuspended in 1x Sample Buffer, separated by SDS-PAGE, and Western blots performed to detect MA, IN, and biotinylated proteins (Fig. 2C). Consistent with the overall levels of replication of each virus, there were substantially lower levels of MA detected in the NLXINB sample compared to either NLX or NLXMAB samples (lower panel). As expected, a slightly larger band was observed in the NLXMAB lane compared to NLX and NLXINB, corresponding to the larger MA-BAS fusion protein. No biotinylation of either MA or IN was detected by Western blot with streptavidin-horseradish peroxidase (SA-HRP) in the NLX sample demonstrating the specificity of the system (top panel). Biotinylated proteins were detected at approximately 17 and 55 kDa in the NLXMAB sample, corresponding to the MW of MA and full-length p55 Gag. Surprisingly, and despite the low level of NLXINB replication, a faint biotinlyated band was discernable in the NLXINB sample at ~32 kDa, demonstrating that the low level of NLXINB replication produced biotinylated IN. Western blot with an anti-IN antibody confirmed the presence of low levels of the slightly larger IN-BAS fusion protein (middle panel), but also detected a small amount of wild-type size IN protein, suggesting that the BAS insertion in NLXINB was unstable and had partially reverted to wild-type virus. Supporting this supposition, NLXINB replication was observed in one replicate experiment, but subsequent analysis of the virion proteins by Western blot detected no biotinylated IN using SA-HRP and only a single band at ~32 kDa, the size of wild-type IN, was detected by anti-IN. This data suggested the virus mutated to remove expression of the BAS in this experiment.
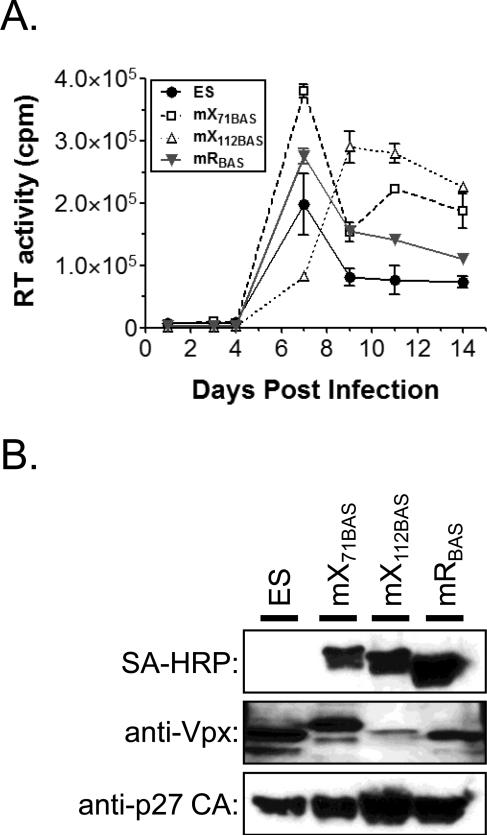
The in vivo biotinylation system represents a powerful tool to dissect viral protein complexes and identify novel cellular factors that interact with individual viral proteins. To expand the repertoire of biotinylatable viruses, several additional HIV-2 molecular clones were constructed with the BAS inserted into the vpx and vpr open reading frames. HIV-2 Vpr (2Vpr) and Vpx split the functions of HIV-1 Vpr. 2Vpr mediates cell cycle arrest and Vpx is required for efficient infection of non-dividing cells and the dissemination of virus in vivo (Di Marzio et al., 1995; Fletcher et al., 1996). The insertions were made in the HIV-2 ES molecular clone by PCR-overlap mutagenesis as previously described (Belshan et al., 2009; Hu et al., 1989). The BAS was inserted after the initiator methionine of 2Vpr (mRB) and into two positions in Vpx, after amino acid 71 (mX71B) and at the C-terminus of the protein (mX112B), shown previously to be amenable to insertions by transposon mutagenesis (Mahnke et al., 2006). To test the replication competence of the viruses, equivalent amounts of viruses were inoculated overnight onto SupT1.BirA cells and their replication monitored over 14 days by RT assay (Fig. 3A). The replication of the mRB and mX71B viruses was comparable to the parental ES virus, reaching peak levels by day 7 post-infection, and the peak level of mX112B virus was slightly delayed (10 days post infection). At the end of the time-course, the supernatants were collected and virus particles concentrated by ultracentrifugation. The expression and labeling of the target proteins was assessed by SDS-PAGE and Western blot (Fig. 3B). Western blot with a p27 CA antibody demonstrated that wild-type levels of virus particles were produced by the BAS-tagged viruses (bottom panel). The target proteins of all three viruses was detected by Western blot with SA-HRP (top panel), indicating each protein was biotinylated and incorporated into virus particles. The presence of Vpx in each sample was confirmed using a monoclonal anti-Vpx antibody (middle panel). Despite equivalent detection of mX71B and mX112B by SA-HRP, there was a reduced level of mX112B detected by the anti-Vpx antibody. This suggests that the insertion of the BAS at the C-terminus of Vpx resulted in a loss of the epitope recognized by the anti-Vpx antibody. Combined, these data indicate that the insertion of BAS into N-terminus of 2Vpr, Vpx position 71, or the Vpx C-terminus was tolerated by the virus for replication in SupT1 cells. Since Vpx and 2Vpr are dispensable for replication in T-cells, additional studies will need to be performed to investigate whether the insertion-containing proteins maintain their respective functions.
Fig. 3.
Replication and biotinylation of HIV-2 viruses with the BAS inserted into Vpx and Vpr. (A) Replication assays of wild-type (ES) and BAS containing viruses. The assays were performed as described in Fig. 2 legend. Error bars denote the standard deviation of the triplicate samples harvested at each timepoint. (B) Detection of biotinylated (SA-HRP) and viral proteins (indicated at left) from virus particles concentrated by ultracentrifugation. Supernatants were collected at the end of the infection and virus particles concentrated as described in the Fig. 2 legend. Data shown is representative of two independent experiments.
These studies further demonstrate the utility of the biotinylation system to study HIV protein complexes. The SupT1.BirA cells that stably express the E. coli biotin ligase BirA and are permissive for HIV-1 and HIV-2 replication are a powerful tool to tag HIV proteins in the context of replicating virus and study HIV protein complexes in vivo. Three additional HIV-2 viruses with the BAS inserted into vpr and vpx were also constructed and characterized. A summary of the molecular clones that have been constructed and tested is given in Table 1. Thus far HIV-1 MA and IN, and HIV-2 Vpr and Vpx were tagged with the BAS and observed to produce virus containing biotinylated proteins. All of these viruses except NLXINB also replicates productively in SupT1.BirA cells. The IN protein has proven to be refractory for any insertion except the C-terminus. A recent study confirmed the amenability of the C-terminus of IN for BAS insertion and biotinylation using lentiviral vector transduction, but concluded that the C-terminal tag did not permit efficient capture of IN from cellular lysates (Benkhelifa-Ziyyat et al., 2010). Summarized in Table 1 are several additional clones with the BAS inserted into different locations throughout the HIV-1 IN ORF that were constructed and tested, however additional IN-BAS viruses that produce infectious virions have not yet been identified. Given the potential to capture RTCs and PICs with a tagged IN protein, studies continue to construct and test additional IN-BAS clones.
Acknowledgements
This study was supported by NIH grants P20RR01635 and 1R01AI080348. The following reagent was obtained from the NIH AIDS Research and Reference Reagent Program: Anti-Vpx monoclonal antibody from John C. Kappes, and monoclonal antibody to HIV-1 p24 from Jonathan Allan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Beckett D, Kovaleva E, Schatz P. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshan M, Schweitzer CJ, Donnellan MD, Lu R, Engelman A. In vivo biotinylation and capture of HIV-1 matrix and integrase proteins. J Virol Methods. 2009;159:178–184. doi: 10.1016/j.jviromet.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkhelifa-Ziyyat S, Bucher S, Zanta-Boussif M-A, Pasquet J, Danos O. Changes in the accessibility of the HIV-1 Integrase C-terminus in the presence of cellular proteins. Retrovirology. 2010;7:27. doi: 10.1186/1742-4690-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau NR. Mutational analysis of cell cycle arrest, nuclear localization and virion packaging of human immunodeficiency virus type 1 Vpr. J. Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TM, 3rd, Brichacek B, Sharova N, Newman MA, Stivahtis G, Sharp PM, Emerman M, Hahn BH, Stevenson M. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). Embo J. 1996;15:6155–65. [PMC free article] [PubMed] [Google Scholar]
- Hu W, Vander Heyden N, Ratner L. Analysis of the function of viral protein X (VPX) of HIV-2. Virology. 1989;173:624–30. doi: 10.1016/0042-6822(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke LA, Belshan M, Ratner L. Analysis of HIV-2 Vpx by modeling and insertional mutagenesis. Virology. 2006;348:165–74. doi: 10.1016/j.virol.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Schatz PJ. Use of Peptide Libraries to Map the Substrate Specificity of a Peptide-Modifying Enzyme: A 13 Residue Consensus Peptide Specifies Biotinylation in Escherichia coli. Nat Biotech. 1993;11:1138. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- van 't Wout AB, Lehrman GK, Mikheeva SA, O'Keeffe GC, Katze MG, Bumgarner RE, Geiss GK, Mullins JI. Cellular Gene Expression upon Human Immunodeficiency Virus Type 1 Infection of CD4+-T-Cell Lines. J. Virol. 2003;77:1392–1402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu X-F. Induction of APOBEC3G Ubiquitination and Degradation by an HIV-1 Vif-Cul5-SCF Complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, Espeseth AS. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]