Abstract
1,2-Benzisothiazol-3(2H)-ones and 1,3,4-oxadiazoles individually have recently attracted considerable interest in drug discovery, including as antibacterial and antifungal agents. In this study, a series of functionalized 1,2-benzisothiazol-3(2H)-one - 1,3,4-oxadiazole hybrid derivatives were synthesized and subsequently screened against Dengue and West Nile virus proteases. Ten out of twenty-four compounds showed greater than 50% inhibition against DENV2 and WNV proteases ([I] = 10 μM). The IC50 values of compound 7n against DENV2 and WNV NS2B/NS3 were found to be 3.75 ± 0.06 and 4.22 ± 0.07 μM, respectively. The kinetics data support a competitive mode of inhibition by compound 7n. Molecular modeling studies were performed to delineate the putative binding mode of this series of compounds. This study reveals that the hybrid series arising from the linking of the two scaffolds provides a suitable platform for conducting a hit-to-lead optimization campaign via iterative structure-activity relationship studies, in vitro screening and X-ray crystallography.
Introduction
Dengue virus (DENV) and West Nile virus (WNV) are the most prevalent mosquito-borne viruses affecting humans.1 DENV and WNV belong to the family Flaviviridae (genus Flavivirus) which comprises several human and animal pathogens. Other noteworthy members in this family include Yellow fever virus (YFV), Japanese encephalitis virus (JEV) and Hepatitis C virus (HCV).2 Four distinct Dengue serotypes exist based on their antigenic properties and/or sequence similarities. Each serotype can cause Dengue disease. Only one serotype exists for WNV. Dengue fever is a relatively mild illness with rash; however, infection by Dengue virus can potentially progress to a more severe illness, called Dengue hemorrhagic fever. Approximately 2.5 billion people worldwide are at risk for Dengue infection alone. Indeed, Dengue virus is estimated to infect 50-100 million individuals every year and causes approximately 1.5 million clinical cases of Dengue disease annually.3-4 Currently, there is no effective chemotherapeutic agent or vaccine for the management and treatment of Dengue infection.5
Both Dengue and West Nile viruses contain a single-strand sense RNA (ssRNA) genome with positive polarity, approximately 11 kb in length flanked by a 5’ cap but no poly(A) tail at the 3’ end non-translated regions. After endocytosis and release of the viral nucleocapsid from the viral RNA, the single long open reading frame (ORF) of the viral RNA encodes a polyprotein, which is processed by both a cellular signal peptidase and a viral serine protease at the surface of the endoplasmic reticulum (ER), resulting in the formation of three structural proteins, capsid (C), precursor membrane (prM) and envelope (E), and seven non-structural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5). 6-7 NS3 is a multi-functional protein containing a serine protease with a prototypical catalytic triad (H51, D75, S135) within the N-terminal 185 amino acid residues, an NTPase/RNA helicase, and a 5’-RNA triphosphatase within the C-terminal (161-618 amino acid residues) region of NS3.6 However, NS3 alone is not active. NS2B is an ER-resident integral membrane protein consisting of three hydrophobic domains and a conserved hydrophilic domain. The hydrophobic sequence of NS2B is essential for co-translational insertion of the protease cofactor into ER membranes for efficient cleavage of the NS2B/NS3 junction. The hydrophilic region of 49-92 amino acids of NS2B serves as an activator and is sufficient for interaction with and activation of the NS3 protease domain (hereafter referred to as “NS2B/NS3pro”) in vitro.7-8
There are many Dengue and West Nile viral proteins that have been targeted for drug discovery, including helicase,9-10 methyl transferase,11-13 serine protease14-18 and viral RNA.19-20 In general, the blockage of NS2B/NS3pro activity provides an effective means for designing DENV and WNV small-molecule therapeutics.21-23 Small-molecule inhibitors of DENV2 and WNV proteases are of interest both as therapeutic agents and as chemical probes for investigating biological functions related to virus replication and reproduction. 24-25 To date only a small number of non-peptidyl inhibitor scaffolds for DENV and WNV proteases have been described and the available structure-activity relationship data are rather limited.5,23,25
1,2-Benzisothiazol-3(2H)-one derivatives play a significant role in various pharmaceutical applications.26-28 As an important class of heterocyclic compounds, 1,2-benzisothiazol-3(2H)-ones show broad spectrum bioactivity.29-32 Among these, suitably-substituted 1,2-benzisothiazol-3(2H)-ones have been shown to exhibit potent antifungal33 and antivirus activity.17 Compounds containing a 1,3,4-oxadiazole template have also received significant attention in medicinal and pharmaceutical research as this structural motif has found use in a variety of applications,34 including as an essential component of the pharmacophore and as a bioisosteric replacement for carbonyl-containing compounds, such as esters, amides and others. Recently, sulfonamide-1,3,4-oxadiazole derivatives have been shown to exhibit noteworthy antibacterial and antifungal activity.35
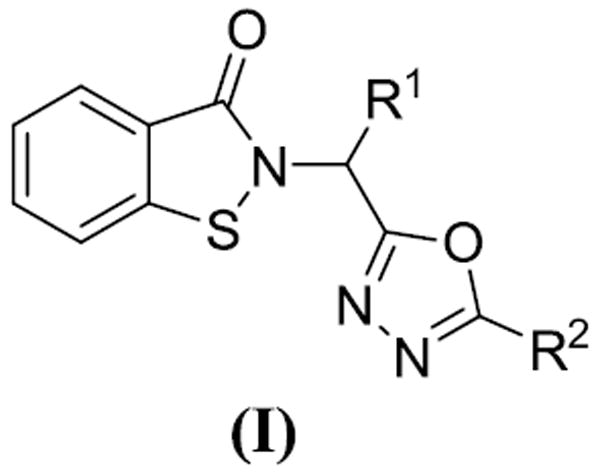
In this study, we describe the design, synthesis, and structure-activity relationship (SAR) studies of a series of novel 1,2-benzisothiazol-3(2H)-one and 1,3,4-oxadiazole hybrid derivatives, represented by general structure (I) (Figure 1) as potential inhibitors of DENV2 and WNV NS2B/NS3 proteases. Inhibition percentage and steady state kinetics against DENV2 and WNV NS2B/NS3 proteases were also carried out, leading to the identification of a low micromolar inhibitor.
Figure 1. General structure of inhibitor (I).
Results and Discussion
Chemistry
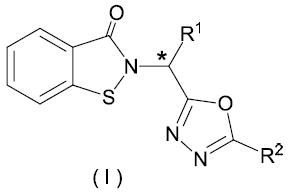
In this study, twenty four 1,2-benzisothiazol-3(2H)-one - 1,3,4-oxadiazole hybrid derivatives were synthesized. The synthetic routes leading to compounds 7a-x (Table 1) are illustrated in the synthetic scheme. Compounds 7a-d were synthesized from compound 2, which was in turn synthesized33 by the sequential treatment of 1,2-benzisothiazolin-3[2H]-one with NaH and t-butyl bromoacetate in trifluoroethanol, followed by hydrolysis of the resulting ester with trifluoroacetic acid (Scheme). Coupling of 2 with a series of hydrazides36 followed by p-toluenesulfonyl chloride-mediated cyclodehydration of the corresponding products, yielded the desired compounds. Compounds 7e-l and 7w-x were prepared using 2,2’-dithiodibenzoic acid as the starting material (Scheme) via treatment with 1,1’-carbonyldiimidazole (CDI) and an appropriate amino acid methyl ester hydrochloride to afford the corresponding ester 3. Intermediate 4 was obtained by reacting 3 with bromine, followed by the addition of triethylamine. The corresponding acids 5 were obtained following hydrolysis with LiOH in aqueous THF. Substituted hydrazide derivatives 6 were obtained by reacting 5 with 1,1’-carbonyldiimidazole in THF and an array of substituted hydrazides. Finally, treatment of hydrazides 6 with p-toluenesulfonyl chloride in the presence of triethylamine in CH2Cl2 under ambient temperature gave cyclodehydration products 7. Compounds 7m-v were conveniently prepared using a one-pot synthesis, as described in the literature37 (Scheme). The synthesized compounds are listed in Table 1.
Table 1.
Inhibition of DENV2 and WNV NS2B/NS3pro by compounds 7a-7x at 10 and 25 μM.
| ||||||
|---|---|---|---|---|---|---|
| Inhibitor | R1 | R2 | DENV2 | WNV | ||
|
| ||||||
| % inhibition at 10 μM | % inhibition at 25 μM | % inhibition at 10 μM | % inhibition at 25 μM | |||
| 7a | H | m-fluorophenyl | 3.24 ± 0.93 | 9.11 ± 1.76 | 7.68 ± 0.98 | 10.07 ± 0.87 |
| 7b | H | m-chlorophenyl | 5.21 ± 2.17 | 16.74 ± 1.97 | 6.15 ± 1.11 | 9.15 ± 0.79 |
| 7c | H | benzyl | 3.56 ± 1.09 | 16.43 ± 1.84 | 8.27 ± 1.37 | 14.73 ± 1.04 |
| 7d | H | p-methoxybenzyl | 5.27 ± 1.46 | 7.90 ± 1.94 | 5.96 ± 1.07 | 8.68 ± 0.95 |
| 7e | methyl | p-methoxybenzyl | 8.08 ± 1.56 | 17.82 ± 1.99 | 4.17 ± 0.63 | 9.79 ± 1.02 |
| 7f | benzyl | benzyl | 22.38 ± 0.87 | 31.23 ± 1.23 | 20.54 ± 0.91 | 30.12 ± 1.02 |
| 7g | benzyl | p-methoxybenzyl | 28.49 ± 1.82 | 38.64 ± 1.69 | 24.23 ± 0.61 | 34.86 ± 0.45 |
| 7h | benzyl | m-fluorophenyl | 39.03 ± 1.06 | 50.87 ± 1.55 | 30.36 ± 0.84 | 38.91 ± 0.61 |
| 7i | benzyl | m-chlorophenyl | 34.27 ± 2.07 | 42.12 ± 1.89 | 29.01 ± 0.75 | 43.11 ± 0.71 |
| 7j | benzyl | p-fluorophenyl | 60.34 ± 1.12 | 75.74 ± 1.95 | 51.12 ± 1.47 | 59.09 ± 1.35 |
| 7k | benzyl | p-chlorophenyl | 53.25 ± 1.64 | 70.52 ± 2.37 | 53.46 ± 1.27 | 63.24 ± 1.24 |
| 7l | benzyl | p-cyanophenyl | 63.86 ± 1.71 | 72.94 ± 1.24 | 63.57 ± 1.59 | 70.87 ± 1.32 |
| 7m | benzyl | phenyl | 56.73 ± 0.95 | 69.24 ± 1.42 | 51.71 ± 1.16 | 64.09 ± 0.91 |
| 7n | benzyl | p-methoxyphenyl | 65.49 ± 1.18 | 80.01 ± 1.73 | 68.58 ± 1.38 | 73.42 ± 1.22 |
| 7o | benzyl | p-ethoxyphenyl | 40.65 ± 1.79 | 61.27 ± 1.71 | 56.09 ± 1.18 | 71.84 ± 2.67 |
| 7p | benzyl | p-pyridyl | 62.43 ± 1.82 | 70.87 ± 1.27 | 63.13 ± 1.49 | 72.68 ± 0.88 |
| 7q | benzyl | m-pyridyl | 58.06 ± 1.55 | 69.97 ± 1.02 | 55.83 ± 1.32 | 62.52 ± 0.98 |
| 7r | benzyl | 2-furyl | 52.28 ± 1.74 | 64.74 ± 1.51 | 44.32 ± 1.53 | 57.67 ± 1.02 |
| 7s | benzyl | 2-thiophenyl | 52.34 ± 2.23 | 69.81 ± 1.64 | 43.68 ± 1.36 | 59.66 ± 1.46 |
| 7t | benzyl | methyl | 19.98 ± 1.45 | 31.59 ± 1.37 | 22.32 ± 0.82 | 40.39 ± 2.19 |
| 7u | benzyl | ethyl | 22.54 ± 1.55 | 34.22 ± 2.43 | 25.07 ± 1.75 | 44.26 ± 1.69 |
| 7v | benzyl | propyl | 25.58 ± 1.07 | 36.68 ± 1.91 | 34.94 ± 1.83 | 46.19 ± 2.25 |
| 7w | m-fluorobenzyl | m-chlorophenyl | 15.97 ± 1.72 | 31.61 ± 2.06 | 27.44 ± 0.59 | 35.65 ± 0.22 |
| 7x | phenethyl | m-chlorophenyl | 26.06 ± 1.15 | 35.01 ± 2.21 | 23.86 ± 0.84 | 33.85 ± 0.54 |
[WNV protease] = 28 nM; [DENV2 protease] = 25 nM *(DL) methyl, (L) benzyl, (DL) m-fluorobenzyl, (DL) phenethyl
Scheme. Synthesis of compounds 3a-d, 4a-d, 5a-d, 6a-x and 7a-x.
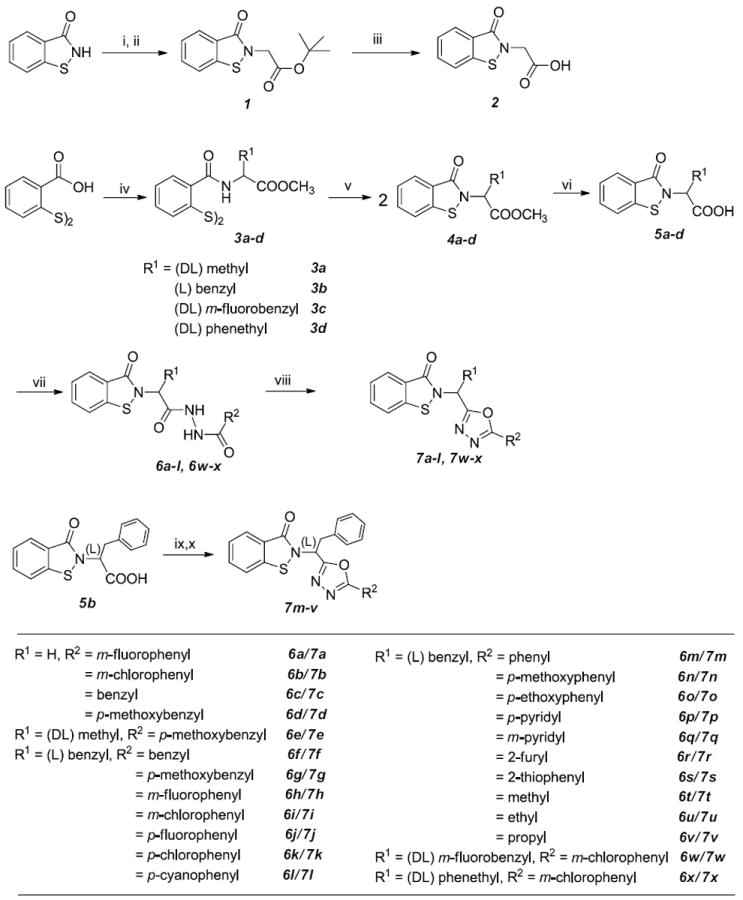
Reagents and conditions: i) NaH/CH3CN; ii) BrCH2COOC(CH3)3 /CF3CH2OH; iii) TFA; iv) CDl/THF, followed by amino acid and TEA; v) Br2/CH2Cl2, TEA; vi) LiOH/H2O, THF; vii) CDl/THF, R2CONHNH2; viii)p-TsCl/CH2Cl2, TEA; ix) R2CONHNH2, DlPEA, TBTU, dry CH3CN; x) DlPEA, p-TsCl
X-ray crystallography
The structure of a representative member of this series (compound 7p) was determined (Figure 2), and the crystal and structure refinement data are listed in Table 2.
Figure 2. ORTEP drawing of compound 7p showing the 30% thermal ellipsoids.
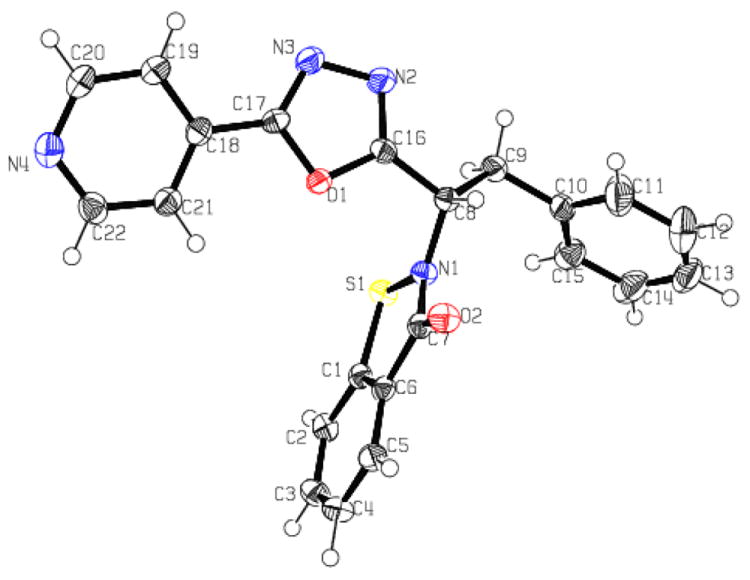
Hydrogen atoms have been omitted for clarity.
Table 2.
X-ray data collection and processing parameters for compound 7p.
| Molecular formula | C22H16N4O2S |
| Formula weight | 400.45 |
| Diffractometer | Bruker Kappa Apex II |
| Radiation/λ (Å) | Mo Kα/0.71073 |
| Temperature (K) | 150 |
| Color/habit | colorless/ |
| Crystal size (mm3) | 0.288 × 0.302 × 0.318 |
| Crystal system | Orthorhombic |
| Space group | P212121 |
| a (Å) | 10.397(3) |
| b (Å) | 11.313(3) |
| c (Å) | 16.606(5) |
| V (Å3) | 1953.1(10) |
| Z-value | 4 |
| Dcalc (g cm-3) | 1.362 |
| Octants collected | ±h, ±k, ±l |
| Max. h, k, l | 12, 13, 20 |
| Θ range (°) | 3.60 to 25.99 |
| Absorption coefficient (mm-1) | 0.192 |
| F(000) | 832 |
| Reflections/unique (Rint) | 35207/3812 (0.0405) |
| Observed (>2σ)/parameters | 3812 /262 |
| Robs/Rall | 0.0266/0.0307 |
| Goodness-of-fit | 1.030 |
| Absolute structure parameter | 0.02(5) |
| Largest diff. peak and hole (e.Å-3) | 0.159-0.178 |
Biochemistry
The expression and purification of DENV2 NS2B/NS3pro and WNV NS2B/NS3pro have been previously described.38 Enzyme assays and inhibition studies were carried out as previously described39-40 and the results are summarized in Tables 1, 3 and Figures 3-5.
Table 3.
Kinetic parameters for the tetra-peptide substrate and compound 7n against DENV2 NS2B/NS3pro at 37 °C.
| 7n (μM) | Km, app (μM) | kcat (s-1) | kcat/Km (M-1·s-1) |
|---|---|---|---|
| 0 | 11.69 ± 0.41 | 0.064 ± 0.0008 | 5751 ± 275 |
| 1 | 17.59 ± 0.54 | 0.065 ± 0.0009 | 3879 ± 169 |
| 3 | 20.98 ± 1.05 | 0.0623 ± 0.0014 | 3176 ± 225 |
| 5 | 31.47 ± 3.25 | 0.062 ± 0.0034 | 2327 ± 349 |
Figure 3. Inhibition of DENV2 and WNV NS2B/NS3pro by selected compounds at 25 μM.
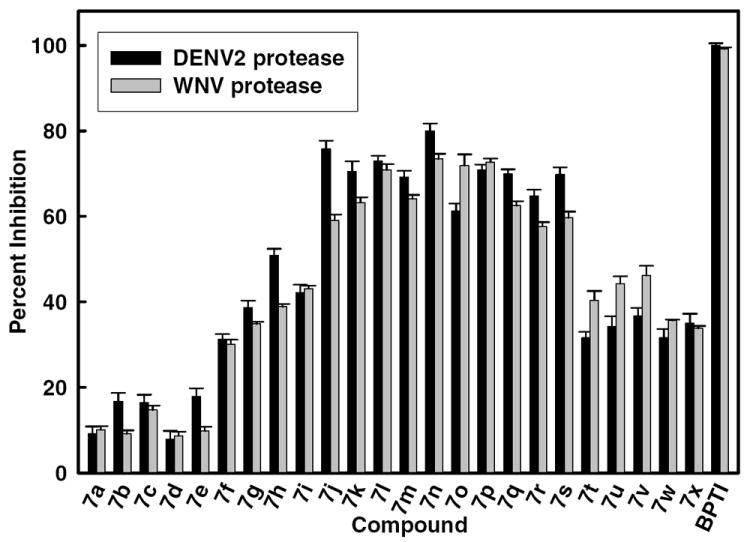
The concentrations of DENV2 and WNV proteases were 25 nM and 28 nM, respectively. Substrate concentration was 5.0 μM. The buffer used contained 200 mM Tris·HCl, 6.0 mM NaCl, 30 % glycerol, and 0.1% CHAPS, pH 9.5 and the reactions were run at 37°C. The percent values were calculated from the relative fluorescence units obtained in the presence and absence of tested compound. BPTI (Aprotinin) and DMSO were used as a positive and negative control, respectively. All assays were performed in triplicate and the average values are shown.
Figure 5. Inhibition of DENV2 NS2B/NS3 protease activity by compound 7n.
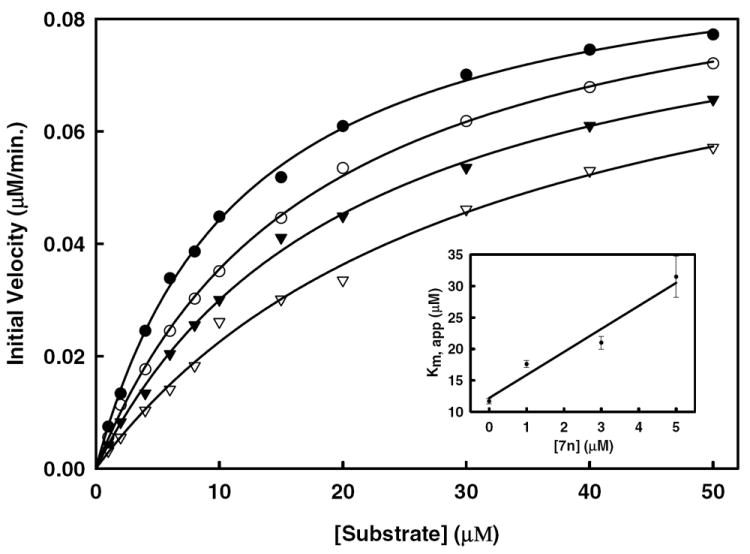
Initial reaction rates of the substrate (Bz-Nle-Lys-Arg-Arg-AMC) cleavage catalyzed by DENV2 NS2B/NS3 protease (25 nM) in 200 mM Tris·HCl (pH 9.5), 6.0 mM NaCl, 30% glycerol and 0.1% CHAPS at 37 °C were determined by varying the substrate concentrations in the range of 0, 1, 2, 4, 6, 8, 10, 15, 20, 30, 40 and 50 μM at each concentration of inhibitor fixed at 0 (solid circle), 1.0 μM (open circle), 3.0 μM (solid triangle) and 5.0 μM (open triangle). The reactions were initiated by the addition of DENV2 NS2B/NS3 protease and the fluorescence intensity at 460 nm was monitored with an excitation at 380 nm. Reactions were less than 5% complete in all cases to maintain valid steady-state measurements. The solid lines are fitted lines using the Michaelis-Menten equation. Inset: Secondary plot of Km, app against the concentration of selected compound 7n. Kinetics studies were carried out as described utilizing substrate concentrations of 0-50 μM Bz-Nle-Lys-Arg-Arg-AMC. Each experiment was performed in duplicate and repeated three times. Data were analyzed using SigmaPlot 2001 v7.0 software to determine values for apparent Km and kcat.
The armamentarium of antiviral therapeutics for the treatment of Dengue and West Nile virus infections is currently very limited; consequently, small-molecule anti-Dengue and anti-West Nile therapeutics are urgently needed. A series of structurally-diverse 1,2-benzisothiazol-3(2H)-one - 1,3,4-oxadiazole hybrid derivatives represented by general structure (I) (Figure 1) were synthesized and their activity against DENV2 and WNV NS2B/NS3 proteases investigated. The design of this series of inhibitors was based on the following considerations: (a) it was envisaged that an entity comprised of two heterocyclic scaffolds would provide multiple loci for hydrogen bond donor and/or acceptor interactions between the protease and inhibitors, leading to enhanced pharmacological activity. This approach was anticipated to be advantageous in the case of DENV2 NS2B/NS3pro because of its rather shallow active site; (b) molecular modeling using the known X-ray crystal structures of DENV2 NS2B/NS3pro23,41-42 suggested that derivatives of (I) can be accommodated at the protease active site and, (c) linking the two heterocyclic scaffolds could potentially result in the creation of a novel entity that displays superior drug-like characteristics. Thus, the 1,2-benzisothiazoli-2(3H)-one-1,3,4-oxadiazole hybrid platform was chosen for generating a focused library of compounds and for conducting exploratory studies.
The desired compounds were readily obtained as shown in the synthetic scheme and the structure and stereochemistry of a representative member was established using X-ray crystallography (Figure 2). The compounds were subsequently screened against DENV2 NS2B/NS3pro and WNV NS2B/NS3pro. It is evident from Figure 3 and Table 1 that several derivatives of (I) are active against both proteases, with nine out of twenty-four compounds exhibiting more than 50% inhibitory activity ([Inhibitor] = 10 μM). It can be generally inferred from the SAR studies that the nature of both R1 and R2 influences activity. Furthermore, R1 exerts a more dominant effect on activity than R2. Variation of R1 while keeping R2 constant reveals that inhibitory activity follows the order for R1 benzyl > H, corresponding to 7i, and 7b, respectively. This hierarchy is further confirmed when R2 is substituted with a p-methoxybenzyl group: benzyl is better than H (7g > 7d). Similarly, when R2 is a benzyl group, compound 7f (R1 = benzyl) is a better inhibitor than 7c (R1 = H). The inhibition studies indicate that highest inhibitory activity is manifested where R1 is a benzyl group, while lowest inhibition is observed where R1 is hydrogen. A direct comparison between 7w and 7x versus 7i cannot be made because the former compounds were screened as racemic mixtures while the latter compound was screened as the pure (L) enantiomer.
One of the most potent compounds (compound 7n), was chosen for further kinetics studies. The percent inhibition of compound 7n (R1 = benzyl and R2 = p-methoxyphenyl) against both DENV2 and WNV proteases is higher than the rest of the compounds (Figure 3). The apparent IC50 values of compound 7n (Figure 4) against DENV2 and WNV proteases were determined to be 3.75 ± 0.06 and 4.22 ± 0.07 μM, respectively. We performed kinetics analysis to determine the Km, kcat and Vmax values in the presence and absence of compound 7n at four different concentrations (Figure 5 and Table 3). The apparent Michaelis-Menten constants (Km, app) increased and kcat/Km decreased proportionally with increasing concentration of compound 7n. These results are supportive of a competitive mode of inhibition. The inhibition constant (Ki) of 7n against DENV2 NS2B/NS3pro was determined to be 3.61 μM. Next, we employed molecular modeling to identify a plausible binding mode for a series of compounds, including 7n and analogs thereof, with the goal of identifying the binding domains of 1,2-benzisothiazol-3[2H]-one and 1,3,4-oxadiazole moieties as a basis for pharmacophore perception. In the case of DENV2 NS2B/NS3pro, we compared the distribution of docked conformers for compounds 7b, 7i, 7g, 7n, 7w and 7x within models produced from the 3U1I23 and 2FOM41 crystal structures to determine whether a consistent ligand binding mode was evident for either case. For the 3U1I receptor model, all compounds except 7b exhibited at least one predicted binding mode within the five top-scoring poses that satisfied both of the following two pharmacophore features: H-bond acceptance of the Gly 153 backbone amide proton by the 1,2-benzisothiazol-3(2H)-one O, and H-bond acceptance of the Ser 135 side chain hydroxyl proton by one or two nitrogens on the 1,3,4-oxadiazole ring. A similar binding mode was predicted for these compounds in docking to WNV NS2B/NS3pro with additional conservation of ligand hydrophobic interactions with His 51 and Tyr 161, however, no consistent binding mode was resolved within the manifold of top five poses of these ligands for the DENV2 NS2B/NS3pro model based on the 2FOM41 crystal structure. For consistency of interpretation, we have thus chosen to focus on the docking poses generated for the 3U1I (DENV)23, 3E90 (WNV)43 and 2FP7 (WNV)42 structure models.
Figure 4. Determination of IC50 values of inhibitor 7n against DENV2 and WNV proteases.
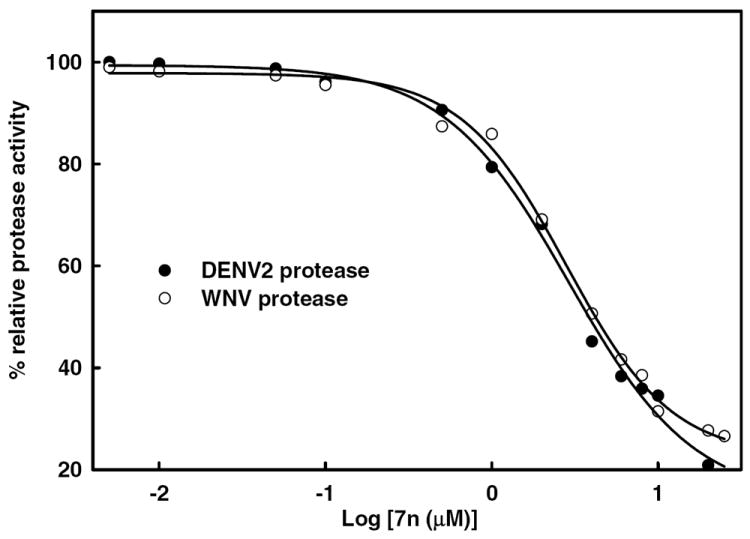
Inhibitor 7n was incubated with DENV2 NS2B/NS3pro (25 nM) or WNV NS2B/NS3pro (28 nM) in buffer (200 mM Tris·HCl, 6 mM NaCl and 30% glycerol, pH 9.5) for 15 min at 37°C. Bz-Nle-Lys-Arg-Arg-AMC (5.0 μM) was added to the mixture in a final volume of 100 μL. The fluorescence intensity was measured at 460 nm with excitation at 380 nm and converted to the percentage of protease activity in the absence and presence of inhibitors. The solid line is the theoretical fitting curve based on the Sigmoidal Equation. The apparent IC50 values for compounds 7n were 3.75 ± 0.06 and 4.22 ± 0.07 μM against DENV2 (solid circle) and WNV (open circle), respectively.
In a preliminary sense, the docking results corroborate some general activity trends observed in the tested compounds. Variation of R1 with constant R2 (Figure 6A) suggests that the significant penalty incurred by absence of a hydrophobic R1 group (compound 7b) likely arises from lost hydrophobic interactions which our model suggests as arising for DENV2 NS2B/NS3pro from contact with His 51, resulting in a failure to dock to the receptor in a pose analogous to those of other more potent inhibitors. A benzyl R1 group as per compound 7i, appears to fit well in this hydrophobic pocket, whereas the larger fluorobenzyl (7w) and phenethyl (7x) groups, while not sterically forbidden, may incur a marginal receptor desolvation penalty by protruding into a more polar region of the cavity. Variation of R2 (Figure 6B) suggests that, relative to benzyl analogs, the substituted phenyl groups in our library achieve a marginally better steric fit within the cavity around Tyr 161 and Val/Ile 155 for DENV2/WNV, respectively, which could explain a binding preference for the latter compounds. The p-methoxy group located on the phenyl group of 7n seems to possess an excellent steric fit for this cavity, which may help to explain the strong potency of this compound. Furthermore, the 7n pose solved for the 3U1I23 crystal structure (Figure 6B) suggests the possibility that the methoxy group may serve as a H-acceptor for the receptor hydroxy proton on Tyr 150, although this is not corroborated by the other crystal structures.
Figure 6. Putative binding mode of selected compounds interacting with DENV2 (A and B) and WNV (C) proteases, as predicted by molecular modeling.
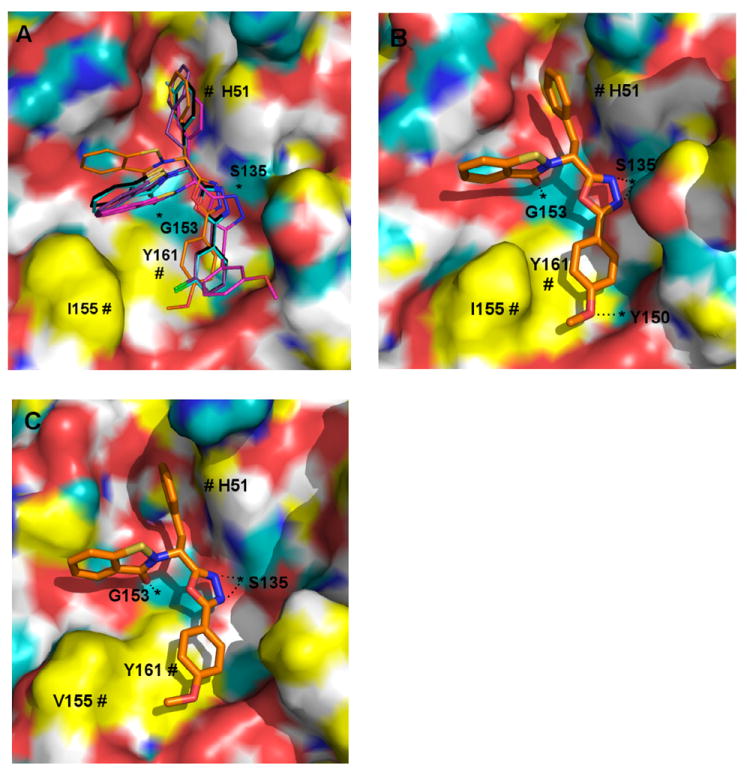
All compounds have CPK-colored heteroatoms, and are distinguished according to the color of their carbon atoms as follows: black = 7i, cyan = 7w, purple = 7x, magenta = 7g and orange = 7n. The protease receptor surfaces are colored as follows: yellow = lipophilic, white = weakly polar, cyan = polar H, blue = polar N and red = polar O. Apparent H-bonding features of the receptor are marked with “*” and hydrophobic interactions with “#”
Conclusion
In summary, we have synthesized a focused library of 1,2-benzisothiazol-3(2H)-one – 1,3,4-oxadiazole hybrid derivatives and have investigated their inhibitory activity against DENV2 NS2B/NS3pro and WNV NS2B/NS3pro. Several members of this series of compounds were found to inhibit the two enzymes, and a low micromolar inhibitor capable of inhibiting both enzymes has been identified. Importantly, biochemical studies indicate that the compounds act as competitive inhibitors. Furthermore, the interaction of the compounds with the two enzymes was probed using molecular graphics and modeling and a mode of binding that is congruent with the results of SAR studies is proposed. The findings reported herein have laid the groundwork for conducting hit-to-lead optimization studies44-46 encompassing iterative medicinal chemistry, in vitro screening, and X-ray crystallography.
Experimental Section
General
The 1H NMR spectra were recorded on a Varian XL-300 or XL-400 NMR spectrometer. Melting points were determined on a Mel-Temp apparatus and are uncorrected. High resolution mass spectrometry (HRMS) was performed using an LCT Premier mass spectrometer (Waters, Milford MA) equipped with a time of flight mass analyzer and an electrospray ion source, at the University of Kansas Mass Spectrometry Lab. Reagents and solvents were purchased from various chemical suppliers (Aldrich, Acros Organics, TCI America, and Bachem). Silica gel (230-450 mesh) used for flash chromatography was purchased from Sorbent Technologies (Atlanta, GA). Thin layer chromatography was performed using Analtech silica gel plates. The TLC plates for the final compounds were eluted using two different solvent systems and were visualized using iodine and/or UV light. Each individual compound was identified as a single spot on TLC plate. DENV2 NS2B/NS3pro (or WNV NS2B/NS3pro) substrate Bz-Nle-Lys-Arg-Arg-AMC was purchased from Bachem, Torrance, CA or custom synthesized by NeoBioScience, Cambridge, MA.
Representative syntheses
Compounds 1 and 2 were synthesized using a previously published procedure33.
t-Butyl (3-oxo-1,2-benzisothiazol-3(2H)-2-yl)-acetate (1)
Yellowish oil (20.5 g; 77% yield) which was used in the next step without further purification. 1H NMR (CDCl3): δ 1.51 (s, 9H), 4.57 (s, 2H), 7.20-8.29 (m, 4H).
(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-acetic acid (2)
White solid (11.2 g; 69% yield), mp 229-230 °C. 1H NMR (CDCl3): δ 4.57 (s, 2H), 7.40-8.12 (m, 4H).
2,2’-Dithiodibenzoyl bis-alanine methyl ester (3a)
Compound 3a was prepared as described previously.33 Briefly, a solution of 2,2’-dithiodibenzoic acid (1.53 g; 5 mmol) in 10 mL THF and 6 mL DMF was added dropwise a suspension of 1,1’-carbonyl diimidazole (1.70 g; 10 mmol) in 10 mL THF. The reaction mixture was stirred at room temperature for 20 min and refluxed for 20 min. The reaction mixture was allowed to cool to room temperature and (DL) alanine methyl ester hydrochloride (1.40 g; 10 mmol) and triethylamine (1.5 mL; 12 mmol) were added. The reaction mixture was stirred at room temperature overnight and concentrated. The residue was taken up in ethyl acetate (40 mL) and water (30 mL). The organic layer was washed with 5% HCl (3 × 30 mL), saturated NaHCO3 (3 × 30 mL) and brine (30 mL). The organic layer was dried over anhydrous sodium sulfate, filtered, and concentrated, leaving behind 3a as an yellow solid (1.8 g; 76% yield), mp 139-141°C. 1H NMR (DMSO-d6): δ 1.44 (d, J = 7.3 Hz, 6H), 3.68 (s, 6H), 4.46-4.59 (m, 2H), 7.25-7.33 (m, 2H), 7.43-7.53 (m, 2H), 7.65-7.79 (m, 4H), 8.98 (d, J = 6.3 Hz, 2H).
2,2’-Dithiodibenzoyl bis-phenylalanine methyl ester (3b)
White solid (68% yield), mp 165-167 °C. 1H NMR (CDCl3) δ 3.24 (dd, J = 13.9, 5.3 Hz, 2H), 3.36 (dd, J = 13.7, 5.8 Hz, 2H), 3.79 (s, 6H), 5.11 (dd, J = 10, 5.8 Hz, 2H), 6.58 (d, J = 7.6 Hz, 2H), 7.13-7.22 (m, 8H), 7.25-7.43 (m, 8H), 7.73-7.78 (m, 2H).
2,2’-Dithiodibenzoyl bis-(m-fluoro)phenylalanine methyl ester (3c)
Light brown solid (75% yield). mp 141-143 °C. 1H NMR (CDCl3): δ. 3.25 (dd, J = 14.1, 5.1 Hz, 2H), 3.38 (dd, J = 15.8, 5.9 Hz, 2H), 3.80 (s, 6H), 5.14 (q, J = 5.4 Hz, 2H), 6.61 (d, J = 7.6 Hz, 2H), 6.82-7.00 (m, 6H), 7.17-7.28(m, 4H), 7.33-7.44 (m, 4H), 7.76 (d, J = 8.1 Hz, 2H).
2,2’-Dithiodibenzoyl bis-benzylalanine methyl ester (3d)
White solid (55% yield), mp 190-192 °C. 1H NMR (DMSO-d6): δ 2.09 (q, J = 14.7 Hz, 4H), 2.62-2.82 (m, 4H), 3.65 (s, 6H), 4.38 (q, J = 7.3 Hz, 2H), 7.14-7.37 (m, 12H), 7.43-7.51 (m, 2H), 7.65-7.74 (m, 4H), 9.07 (d, J = 7.4 Hz, 2H).
Methyl 2-(3-oxo-1,2-benzisothiazol-3(2H)-2-yl)-propanoic acetate (4a)
Compound 4a was prepared as described previously.33 Briefly, a chilled solution of 3a (1.8 g; 3.78 mmol) in 25 mL CH2Cl2 was added bromine (0.60 g; 3.78 mmol) and the reaction mixture was stirred for 30 min. Triethylamine (0.77 g; 7.5 mmol) was then added and the resulting mixture was stirred for 5 min. The reaction mixture was washed with water (2 × 15 mL) and the organic layer was dried over anhydrous sodium sulfate, dried and concentrated, leaving a pure compound 4a as yellowish oil (1.8 g; 100% yield). 1H NMR (CDCl3): δ 1.70 (d, J = 7.8 Hz, 3H), 3.77 (s, 3H), 5.50 (q, J = 8.2 Hz, 1H), 7.42 (t, J = 7.2 Hz, 1H), 7.56-7.66 (m, 2H), 8.09 (d, J = 8.8 Hz, 1H).
Methyl 2-(3-oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acetate (4b)
Yellow oil (14.92g; 70% yield). 1H NMR (CDCl3): δ 3.24 (dd, J = 13.69, 9.83 Hz, 1H), 3.51 (dd, J = 14.52, 5.79 Hz, 1H), 3.74 (s, 3H), 5.68 (dd, J = 10.09, 5.80 Hz, 1H), 7.17-7.25 (m, 5H), 7.33-7.40 (m, 1H), 7.50-7.63 (2H), 7.95-7.99 (m, 1H).
Methyl 2-(3-oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-(3-fluorophenyl)propanoic acetate (4c)
Reddish oil (2.3g; 92% yield). 1H NMR (CDCl3): δ 3.23 (dd, J = 11.2, 8.9 Hz, 1H), 3.49 (dd, J = 11.2, 8.9 Hz, 1H), 3.76 (s, 3H), 5.62-5.69 (m, 1H), 6.82-7.00 (m, 3H), 7.10-8.00 (m, 5H).
Methyl 2-(3-oxo-1,2-benzisothiazol-3(2H)-2-yl)-4-phenylbutanoic acetate (4d)
Yellow oil (1.61g; 90% yield). 1H NMR (CDCl3): δ 2.23-2.35 (m, 1H), 2.43-2.55 (m, 1H), 2.64 (t, J = 8.3 Hz, 2H), 3.75 (s, 3H), 5.40-5.46 (m, 1H), 7.16-7.68 (m, 5H), 8.03-8.08 (m, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-propanoic acid (5a)
Compound 5a was prepared as described previously.33 Briefly, a solution of compound 4a (2.37g; 10 mmol) was dissolved in 15 mL 1,4-dioxane and treated with 20 mL 1 M LiOH at 0 °C for 1 h. The solution was neutralized and the solvent was removed. The residue was adjusted to pH 1, forming a precipitate which was collected with vacuum filtration and washed with diethyl ether to give pure compound 5a as a gray solid (1.73 g; 77% yield), mp 201-202 °C. 1H NMR (DMSO-d6): δ 1.57 (d, J = 6.0 Hz, 3H), 5.14 (q, J = 6.5 Hz, 1H), 7.45 (t, J = 6.2 Hz, 1H), 7.70 (t, J = 6.0 Hz, 1H), 7.88 (d, J = 6.5 Hz, 1H), 7.97 (d, J = 6.5 Hz, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid (5b)
White solid (2.43g; 44% yield), mp 180-182 °C. 1H NMR (DMSO-d6): δ 3.23 (dd, J = 15.1, 11.0 Hz, 1H), 3.45 (dd, J = 15.1, 4.7 Hz, 1H), 5.42 (dd, J = 10.4, 4.7 Hz, 1H), 7.12-7.32 (m, 5H), 7.37-7.44 (m, 1H), 7.63-7.70 (m, 1H), 7.77-7.82 (m, 1H), 7.90-7.95 (m, 1H), 13.40 (br s, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-(3-fluorophenyl)propanoic acid (5c)
Yellow oil (1.87g; 98% yield). 1H NMR (CDCl3): δ 3.29-3.39 (m, 1H), 3.52-3.60 (m, 1H), 5.52-5.58 (m, 1H), 6.80-7.72 (m, 7H), 7.98-8.03 (m, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-4-phenylbutanoic acid (5d)
Yellow solid (1.36g; 80% yield), mp 149-150 °C. 1H NMR (DMSO-d6): δ 2.20-2.48 (m, 2H), 2.61 (t, J = 6.7 Hz, 2H), 5.05-5.11 (m, 1H), 7.18-7.36 (m, 5H), 7.46 (t, J = 6.7 Hz, 1H), 7.71 (t, J = 6.7 Hz, 1H), 7.90 (d, J = 8.3 Hz, 1H), 8.00 (d, J = 8.3 Hz, 1H).
(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-acetic acid, 2’-(3-fluorobenzoyl)hydrazide (6a)
To a stirred solution of compound 2 (0.58 g; 2mmol) in dry DMF (6 mL) was added EDCI (0.46 g; 2.4 mmol), followed by m-fluorobenzhydrazide (0.31 g; 2 mmol), and the reaction mixture was stirred at room temperature overnight. The solvent was removed and the residue was taken up in ethyl acetate (50 mL). The organic layer was washed with brine (15 mL), dried over anhydrous sodium sulfate, filtered, and concentrated. The crude product was purified using flash chromatography (silica gel/ethyl acetate/hexanes) to give pure compound 6a, as a white solid (0.27 g; 32% yield), mp 207-209 °C. 1H NMR (DMSO-d6): δ 4.63 (s, 2H), 7.40-8.01 (m, 8H), 10.41 (s, 1H), 10.60 (s, 1H).
(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-acetic acid, 2’-(3-chlorobenzoyl)hydrazide (6b)
Yellow solid (42% yield), mp 88-90 °C. 1H NMR (DMSO-d6): δ 4.66 (s, 2H), 7.35-7.97 (m, 8H), 10.40 (s, 1H), 10.58 (s, 1H).
(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-acetic acid, 2-(2-phenylacetyl)hydrazide (6c)
White solid (28% yield), mp 230-232 °C. 1H NMR (DMSO-d6): δ 3.47 (s, 2H), 4.56 (s, 2H), 7.20-7.35 (m, 5H), 7.43 (t, J = 5.3 Hz, 1H), 7.69 (t, J = 5.3 Hz, 1H), 7.86 (d, J = 6.7 Hz, 1H), 7.98 (d, J = 6.7 Hz, 1H), 10.21 (s, 1H), 10.29 (s, 1H).
(3-Oxo-1,2-benzisothiazol-(3(2H)-2-yl)-acetic acid, 2-[2-(4-methoxyphenyl)acetyl]hydrazide (6d)
Gray solid (47% yield), mp 203-205 °C. 1H NMR (DMSO-d6): δ 3.40 (s, 2H), 3.74 (s, 3H), 4.56 (s, 2H), 6.88 (d, J =8.0 Hz, 2H), 7.20 (d, J = 8.0 Hz, 2H), 7.45 (t, J = 7.3 Hz, 1H), 7.71 (t, J = 7.3 Hz, 1H), 7.89 (d, J = 8.0 Hz, 1H), 7.98 (d, J = 8.0 Hz, 1H), 10.18 (s, 1H), 10.28 (s, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-propanoic acid, 2-[2-(4-methoxyphenyl) acetyl]hydrazide (6e)
White solid (21% yield), mp 135-137 °C. 1H NMR (DMSO-d6): δ 1.46 (d, J = 4.7 Hz, 3H), 3.40 (s, 2H), 3.72 (s, 3H), 5.30-5.40 (m, 1H), 6.82 (d, J = 9.3 Hz, 2H), 7.19 (d, J = 9.3 Hz, 2H), 7.40 (t, J = 6.7 Hz, 1H), 7.65 (t, J = 6.7 Hz, 1H), 7.86 (d, J = 7.3 Hz, 1H), 7.95 (d, J = 7.3 Hz, 1H), 10.11 (s, 1H), 10.30 (s, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-(2-phenylacetyl)hydrazide (6f)
Yellow oil (52% yield). 1H NMR (DMSO-d6): δ 3.18-3.42 (m, 2H), 3.56 (s, 2H), 5.81-5.87 (m, 1H), 7.00-7.60 (m, 14H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-[2-(4-methoxyphenyl)acetyl]hydrazide (6g)
Yellow oil (53% yield). 1H NMR (DMSO-d6): δ 3.16-3.45 (m, 2H), 3.51 (s, 2H), 3.75 (s, 3H), 5.76-5.81 (m, 1H), 6.78-7.82 (m, 13H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-(3-fluorobenzoyl)hydrazide (6h)
Brown oil (53% yield). 1H NMR (DMSO-d6): δ 3.10-3.50 (m, 2H), 5.82-5.91 (m, 1H), 6.97-7.65 (m, 13H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-(3-chlorobenzoyl)hydrazide (6i)
Brown solid (50% yield), mp 96-97 °C. 1H NMR (DMSO-d6): δ 3.20-3.60 (m, 2H), 5.64-5.75 (m, 1H), 7.20-7.98 (m, 13H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-(4-fluorobenzoyl)hydrazide (6j)
White solid (77% yield), mp 189-191 °C. 1H NMR (CDCl3): δ 3.55 (dd, J = 41.8, 13.5 Hz, 2H), 5.85-5.90 (m, 1H), 7.00-7.47 (m, 10H), 7.71-7.83 (m, 2H), 8.05-8.13 (m, 1H), 9.08 (s, 1H), 9.50 (s, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-(4-chlorobenzoyl)hydrazide (6k)
Yellow-green solid (92% yield), mp 110-112 °C. 1H NMR (CDCl3): δ 3.29 (dd, J = 13.9, 9.1 Hz, 1H), 3.51 (dd, J = 14.3, 6.3 Hz, 1H), 5.82 (dd, J = 9.0, 6.6 Hz, 1H), 7.09-7.19 (m, 4H), 7.27-7.37 (m, 4H), 7.49-7.59 (m, 2H), 7.68-7.74 (m, 2H), 7.86-7.91 (m, 2H), 9.10 (s, 1H), 10.06 (s, 1H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-phenylpropanoic acid, 2-(4-cyanobenzoyl)hydrazide (6l)
White solid (42% yield), mp 168-170 °C. 1H NMR (DMSO-d6): δ 3.55 (dd, J = 26.0, 14.3 Hz, 2H), 5.75 (dd, J = 9.6, 4.7 Hz, 1H), 7.20-7.45 (m, 6H), 7.49-7.55 (m, 1H), 7.63-7.69 (m, 1H), 7.87-8.03 (m, 4H), 8.73 (br s, 1H), 10.51 (br s, 1H), 10.75-10.87 (m, 1H)
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-3-(3-fluorophenyl)propanoic acid, 2-(3-fluorobenzoyl)hydrazide (6w)
White solid (63% yield), mp 93-95 °C. 1H NMR (CDCl3): δ 3.15-3.40 (m, 2H), 5.39 (t, J = 7.1 Hz, 1H), 6.78-7.99 (m, 12H).
2-(3-Oxo-1,2-benzisothiazol-3(2H)-2-yl)-4-phenylbutanoic acid, 2-(3-fluorobenzoyl)hydrazide (6x)
White solid (47% yield), mp 105-106 °C. 1H NMR (CDCl3): δ 2.45-2.72 (m, 4H), 5.11-5.17 (m, 1H), 7.00-8.02 (m, 13H).
2-[(2-m-Fluorophenyl-1,3,4-oxadiazol-5-yl)methyl]-1,2-benzisothiazol-3(2H)-3-one (7a)
To a stirred solution of compound 6a (0.27 g; 0.63 mmol) in CH2Cl2 (10 mL) was added p-toluenesulfonyl chloride (0.12 g; 0.63 mmol) followed by triethylamine (0.20 mL; 1.5 mmol) and the resulting reaction mixture was stirred at room temperature overnight. CH2Cl2 (30 mL) was added and the solution was washed with water (30 mL), and brine (30 mL). The organic layer was dried over anhydrous sodium sulfate, filtered and concentrated. The crude product was purified by flash chromatography (silica gel/ethyl acetate/hexanes) to give 7a as a white solid (0.1g; 39% yield), mp 149-151 °C. 1H NMR (CDCl3): δ 5.40 (s, 2H), 7.20-7.82 (m, 7H), 8.09-8.12 (m, 1H). 13C NMR (CDCl3): δ 165.4, 164.0, 161.9, 161.5, 140.5, 132.7, 131.0, 127.2, 126.0, 125.2, 123.2, 122.9, 120.6, 119.3, 119.1, 114.3, 37.7. HRMS (ESI): Calcd for C16H10FN3O2SNa, m/z [M+Na]+: 350.0375, found 350.0366.
2-[(2-m-Chlorophenyl-1,3,4-oxadiazol-5-yl)methyl]-1,2-benzisothiazol-3(2H)-3-one (7b)
White solid (57% yield), mp 136-137 °C. 1H NMR (CDCl3): δ 5.40 (s, 2H), 7.40-7.68 (m, 5H), 7.90-8.12 (m, 3H). 13C NMR (CDCl3): δ 165.4, 164.8, 161.9, 140.5, 135.3, 132.7, 132.2, 130.4, 129.1, 127.2, 127.2, 126.0, 125.2, 125.0, 123.3, 120.6, 37.7. HRMS (ESI): Calcd for C16H10ClN3O2SNa, m/z [M+Na]+: 366.0080, found 366.0068.
2-[(2-Benzyl-1,3,4-oxadiazol-5-yl)methyl]-1,2-benzisothiazol-3(2H)-3-one (7c)
Yellow solid (0.10 g; 35% yield), mp 93-95 °C. 1H NMR (CDCl3): δ 4.16 (s, 2H), 5.24 (s, 2H), 7.23-7.35 (m, 5H), 7.43 (t, J = 5.1 Hz, 1H), 7.53 (d, J = 5.5 Hz, 1H), 7.64 (t, J = 5.1 Hz, 1H), 8.05 (d, J = 5.5 Hz, 1H). 13C NMR (CDCl3): δ 166.7, 165.4, 162.2, 140.5, 133.3, 132.6, 128.9, 128.8, 127.6, 127.1, 126.0, 123.3, 120.6, 37.7, 31.7. HRMS (ESI): Calcd for C17H13N3O2SNa, m/z [M+Na]+: 346.0626, found 346.0635.
2-[(2-p-Methoxybenzyl-1,3,4-oxadiazol-5-yl)methyl]-1,2-benzisothiazol-3(2H)-3-one (7d)
Yellow solid (52% yield), mp 92-93 °C. 1H NMR (CDCl3): δ 3.79 (s, 3H), 4.11 (s, 2H), 5.26 (s, 2H), 6.83 (d, J = 8.8 Hz, 2H), 7.20 (d, J = 8.8 Hz, 2H), 7.45 (d, J = 7.7 Hz, 1H), 7.53 (d, J = 8.9 Hz, 1H), 7.65 (t, J = 7.7 Hz, 1H), 8.08 (d, J = 8.9 Hz, 1H). 13C NMR (CDCl3): δ 167.1, 165.4, 162.1, 159.0, 140.5, 132.6, 129.9, 127.1, 125.9, 125.2, 123.2, 120.6, 114.3, 55.3, 37.7, 30.9. HRMS (ESI): Calcd for C18H15N3O3SNa, m/z [M+Na]+: 376.0732, found 376.0735.
2-[(2-p-Methoxybenzyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-3-one (7e)
White solid (48% yield), mp 139-141 °C. 1H NMR (CDCl3): δ 1.86 (d, J = 7.7 Hz, 3H), 3.76 (s, 3H), 4.09 (dd, J = 19.6, 13.4 Hz, 2H), 6.20 (q, J = 7.2 Hz, 1H), 6.79 (d, J = 8.8 Hz, 2H), 7.16 (d, J = 8.8 Hz, 2H), 7.40 (t, J = 6.7 Hz, 1H), 7.52 (d, J = 8.3 Hz, 1H), 7.61 (t, J = 6.7 Hz, 1H), 8.03 (d, J = 8.3 Hz, 1H). 13C NMR (CDCl3): δ 166.8, 165.4, 165.1, 158.9, 140.4, 132.4, 129.8, 127.0, 125.8, 125.3, 124.0, 120.6, 114.2, 55.2, 44.6, 30.8, 17.9. HRMS (ESI): Calcd for C19H17N3O3SNa, m/z [M+Na]+: 390.0888, found 390.0869.
2-[2-Phenyl-1-(2-benzyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-3-one (7f)
Colorless oil (56% yield). 1H NMR (CDCl3): δ 3.47 (dd, J = 13.9, 8.6 Hz, 1H), 3.67 (dd, J = 13.9, 8.6 Hz, 1H), 4.12 (d, J = 12.2 Hz, 2H), 6.38 (dd, J = 8.9, 6.9 Hz, 1H), 7.12-7.62 (m, 13H), 7.96 (d, J = 9.1 Hz, 1H). 13C NMR (CDCl3): δ 166.3, 165.3, 164.6, 140.4, 134.8, 133.3, 132.4, 129.1, 129.0, 128.8, 128.7, 127.5, 127.3, 127.0, 125.7, 123.6, 120.5, 49.8, 38.2, 31.7. HRMS (ESI): Calcd for C24H19N3O2SNa, m/z [M+Na]+: 436.1096, found 436.1077.
2-[2-Phenyl-1-(2-p-mehoxybenzyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-3-one (7g)
Colorless oil (64% yield). 1H NMR (CDCl3): δ 3.48 (dd, J = 13.6, 9.9 Hz, 1H), 3.68 (dd, J = 13.6, 9.9 Hz, 1H), 3.77 (s, 3H), 4.06 (d, J = 3.1 Hz, 2H), 6.37 (dd, J = 9.1, 7.0 Hz, 1H), 6.77-7.62 (m, 12H), 7.92-7.98 (m, 1H). 13C NMR (CDCl3): δ 152.3, 150.8, 150.1, 126.0, 120.5, 117.9, 117.6, 115.4, 114.7, 114.2, 112.9, 112.6, 111.3, 110.9, 109.3, 106.1, 99.9, 40.9, 35.5, 23.8, 16.4. HRMS (ESI): Calcd for C25H21N3O3SNa, m/z [M+Na]+: 466.1201, found 466.1178.
2-[2-Phenyl-1-(2-m-fluorophenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7h)
Colorless oil (33% yield). 1H NMR (CDCl3): δ 3.54 (dd, J = 12.9, 9.2 Hz, 1H), 3.78 (dd, J = 12.9, 9.2 Hz, 1H), 6.52 (dd, J = 8.8, 7.0 Hz, 1H), 7.19-8.03 (m, 13H). 13C NMR (CDCl3): δ 165.3, 164.4, 164.2, 161.1, 134.9, 132.5, 131.0, 129.2, 128.8, 127.4, 127.1, 125.8, 122.9, 120.6, 119.3, 119.0, 114.3, 114.0, 49.9, 38.2. HRMS (ESI): Calcd for C23H17N3O2SF, m/z [M + Na]+: 440.0845, found 440.0840.
2-[2-Phenyl-1-(2-m-chlorophenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7i)
Colorless oil (26% yield). 1H NMR (CDCl3): δ 3.55 (dd, J = 13.1, 8.2 Hz, 1H), 3.81 (dd, J = 13.1, 8.2 Hz, 1H), 6.53 (dd, J = 8.8, 7.0 Hz, 1H), 7.20-7.63 (m, 11H), 7.97-8.02 (m, 2H). 13C NMR (CDCl3): δ 165.6, 165.4, 164.2, 164.0, 135.1, 132.4, 132.0, 129.4, 129.2, 129.0, 128.7, 127.4, 127.2, 125.8, 125.1, 123.8, 123.4, 120.6, 50.0, 38.3. HRMS (ESI): Calcd for C23H17N3O2SCl, m/z [M + Na]+: 456.0550, found 456.0559.
2-[2-Phenyl-1-(2-p-fluorophenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7j)
Light yellow solid (23% yield), mp 161-163 °C. 1H NMR (CDCl3): δ 3.56 (dd, J = 14.3, 8.9 Hz, 1H), 3.8 (dd, J = 14.2, 7.0 Hz, 1H), 6.51 (dd, J = 8.7, 7.0 Hz, 1H), 7.12-7.44 (m, 9H), 7.51-7.66 (m, 2H), 7.96-8.04 (m, 2H). 13C NMR (CDCl3): δ 166.2, 165.2, 164.8, 140.4, 134.9, 132.4, 129.5, 129.2, 128.7, 127.4, 127.1, 125.8, 123.7, 120.6, 119.6, 116.5, 116.3, 49.8, 38.2. HRMS (ESI): Calcd for C23H16FN3O2SNa, m/z [M+Na]+: 440.0845; found 440.0824.
2-[2-Phenyl-1-(2-p-chlorophenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7k)
White solid (24% yield), mp 164-166 °C. 1H NMR (CDCl3): δ 3.55 (dd, J = 12.8, 8.8 Hz, 1H), 3.80 (dd, J = 14.7, 7.0 Hz, 1H), 6.51 (dd, J = 8.7, 7.0 Hz, 1H), 7.18-7.35 (m, 5H), 7.37-7.48 (m, 3H), 7.52-7.57 (m, 1H), 7.59-7.66 (m, 1H), 7.89-7.96 (m, 2H), 7.99-8.04 (m, 1H). 13C NMR (CDCl3): δ 165.2, 164.8, 164.1, 140.4, 138.4, 134.9, 132.5, 129.4, 129.2, 128.7, 128.4, 127.4, 127.1, 125.8, 123.6, 121.7, 120.6, 49.8, 38.2. HRMS (ESI): Calcd for C23H16 ClN3O2SNa, m/z [M+Na]+: 456.0549; found 456.0554.
2-[2-Phenyl-1-(2-p-cyanophenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7l)
White solid (20% yield), mp 182-184 °C. 1H NMR (CDCl3): δ 3.57 (dd, J = 14.4, 8.7 Hz, 1H), 3.81 (dd, J = 14.3, 7.1 Hz, 1H), 6.52 (dd, J = 8.7, 7.1 Hz, 1H), 7.18-7.35 (m, 5H), 7.38-7.44 (m, 1H), 7.53-7.57 (m, 1H), 7.60-7.66 (m, 1H), 7.74-7.80 (m, 2H), 7.98-8.04 (m, 1H), 8.08-8.14 (m, 2H). 13C NMR (CDCl3): δ 165.3, 164.8, 164.1, 140.3, 134.7, 132.8, 132.6, 130.2, 129.4, 129.1, 128.8, 127.6, 127.1, 125.9, 123.5, 120.6, 117.8, 115.5, 49.8, 38.2. HRMS (ESI): Calcd for C24H16N4O2SNa, m/z [M+Na]+: 447.0892; found 447.0883.
2-[2-Phenyl-1-(2-phenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7m)
To a stirred solution of compound 5b (1.20 g; 4mmol) in dry CH3CN (40 mL) was added phenyl hydrazide (0.55 g; 4 mmol) followed by diisopropylethylamine (1.55 g; 12 mmol), and then TBTU (1.42 g; 4.4 mmol). The resulting reaction mixture was stirred at room temperature for 24 h. TLC of the reaction mixture showed formation of the intermediate. Diisopropylethyl amine (1.03 g; 8 mmol) and p-toluenesulfonyl chloride (2.29 g; 12 mmol) were added and the reaction mixture was stirred at room temperature overnight. Removal of the solvent left a crude residue which was adsorbed on silica gel and purified using flash chromatography (silica gel/ethyl acetate/ hexanes) to give compound 7m as an yellow oil (0.37g; 23% yield). 1H NMR (CDCl3): δ 3.55 (dd, J = 14.3, 8.7 Hz, 1H), 3.81 (dd, J = 14.8, 6.9 Hz, 1H), 6.53 (dd, J = 8.8, 6.9 Hz, 1H), 7.18-7.29 (m, 3H), 7.31-7.35 (m, 2H), 7.38-7.42 (m, 1H), 7.44-7.55 (m, 4H), 7.59-7.65 (m, 1H), 7.97-8.04 (m, 3H). 13C NMR (CDCl3): δ 165.6, 165.2, 163.9, 140.4, 135.0, 132.4, 132.0, 129.2, 129.0, 128.7, 127.4, 127.1, 127.0, 125.7, 123.7, 123.3, 120.6, 49.9, 38.2. HRMS (ESI): Calcd for C23H17N3O2SNa, m/z [M+Na]+: 422.0939; found 422.0923.
2-[2-Phenyl-1-(2-p-methoxyphenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7n)
White solid (20% yield), mp 187-189 °C. 1H NMR (CDCl3): δ 3.54 (dd, J = 13.3, 8.7 Hz, 1H), 3.79 (dd, J = 14.3, 7.0 Hz, 1H), 3.85 (s, 3H), 6.49 (dd, J = 8.9, 7.0 Hz, 1H), 6.93-6.98 (m, 2H), 7.17-7.35 (m, 5H), 7.37-7.43 (1H), 7.51-7.55 (m, 1H), 7.59-7.64 (m, 1H), 7.90-7.95 (m, 2H), 7.99-8.03 (m, 1H). 13C NMR (CDCl3): δ 165.6, 165.2, 163.4, 162.5, 140.4, 135.1, 132.4, 129.2, 128.9, 128.7, 127.3, 127.0, 125.7, 123.7, 120.6, 115.7, 114.4, 55.5, 49.8, 38.2. HRMS (ESI): Calcd for C24H19N3O3SNa, m/z [M+Na]+: 452.1045, found 452.1042.
2-[2-Phenyl-1-(2-p-ethoxyphenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7o)
White solid (28% yield), mp 98-100 °C. 1H NMR (CDCl3): δ 1.43 (t, J = 7.0 Hz, 3H), 3.54 (dd, J = 14.0, 8.9 Hz, 1H), 3.79 (dd, J = 14.7, 6.9 Hz, 1H), 4.07 (q, J = 7.0, 2H), 6.49 (dd, J = 8.5, 6.9 Hz, 1H), 6.91-6.96 (m, 2H), 7.17-7.28 (m, 3H), 7.30-7.35 (m, 2H), 7.37-7.42 (m, 1H), 7.51-7.55 (m, 1H), 7.59-7.64 (m, 1H), 7.89-7.93 (m, 2H), 7.99-8.03 (m, 1H). 13C NMR (CDCl3): δ 165.6, 165.2, 163.3, 162.0, 140.4, 135.1, 132.4, 129.2, 128.9, 128.7, 127.3, 127.0, 125.7, 123.7, 120.6, 115.5, 114.8, 63.7, 49.8, 38.2, 14.6. HRMS (ESI): Calcd for C25H21N3O3SNa, m/z [M+Na]+: 466.1201; found 466.1196.
2-[2-Phenyl-1-(2-p-pyridyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3-one (7p)
White solid (20% yield), mp 176-177 °C. 1H NMR (CDCl3): δ 3.57 (dd, J = 14.2, 8.7, 1H), 3.81 (dd, J = 14.3, 6.9 Hz, 1H), 6.53 (dd, J = 8.6, 7.1 Hz, 1H),7.19-7.35 (m, 5H), 7.39-7.44 (m, 1H), 7.53-7.57 (m, 1H), 7.61-7.66 (m, 1H), 7.82-7.87 (m, 2H), 8.00-8.05 (m, 1H), 8.75-8.82 (m, 2H). 13C NMR (CDCl3): δ 165.3, 165.0, 163.8, 150.9, 140.3, 134.7, 132.6, 130.4, 129.2, 128.8, 127.5, 127.1, 125.9, 123.5, 120.6, 120.5, 49.9, 38.2. HRMS (ESI): Calcd for C22H16N4O2SNa, m/z [M+Na]+: 423.0892; found 423.0880.
2-[2-Phenyl-1-(2-m-pyridyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7q)
White solid (19% yield), mp 148-149 °C. 1H NMR (CDCl3): δ 3.57 (dd, J = 14.1, 8.7 Hz, 1H), 3.81 (dd, J = 13.8, 7.1 Hz, 1H), 6.53 (dd, J = 8.9, 7.1 Hz, 1H), 7.18-7.35 (m, 5H), 7.38-7.45 (m, 2H), 7.53-7.56 (m, 1H), 7.61-7.66 (m, 1H), 8.00-8.04 (m, 1H), 8.25-8.30 (m, 1H), 8.73-8.78 (m, 1H), 9.19-9.23 (m, 1H). 13C NMR (CDCl3): δ 165.3, 164.5,163.5, 152.7, 148.0, 140.4, 134.8, 134.4, 132.5, 129.2, 128.8, 127.5, 127.1, 125.8, 123.8, 123.6, 120.6, 49.9, 38.3 HRMS (ESI): Calcd for C22H16N4O2SNa, m/z [M+Na]+: 423.0892; found 423.0884.
2-[2-Phenyl-1-(2-(2-furyl)-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7r)
White solid (32% yield), mp 142-144 °C. 1H NMR (DMSO-d6): δ 3.56 (dd, J = 14.0, 10.8 Hz, 1H), 3.73 (dd, J = 14.3, 5.1 Hz, 1H), 6.42 (dd, J = 10.6, 5.1 Hz, 1H), 6.77-6.80 (m, 1H), 7.14-7.27 (m, 3H), 7.28-7.33 (m, 2H), 7.36-7.45 (m, 2H), 7.65-7.72 (m, 1H), 7.80-7.84 (m, 1H), 7.94-7.99 (m, 1H), 8.04-8.07 (m, 1H). 13C NMR (CDCl3): δ 165.4, 163.4, 158.6, 146.2, 140.5, 139.0, 135.1, 132.6, 129.4, 128.9, 127.6, 127.3, 126.0, 123.8, 120.8, 115.1, 112.4, 49.9, 38.4. HRMS (ESI): Calcd for C21H15N3O3SNa, m/z [M+Na]+:412.0732; found 412.0746.
2-[2-Phenyl-1-(2-(2-thiophenyl)-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7s)
White solid (25% yield), mp 128-130 °C. 1H NMR (CDCl3): δ 3.51 (dd, J = 14.6, 8.8 Hz, 1H), 3.76 (dd, J = 14.2, 7.0 Hz, 1H), 6.46 (dd, J = 9.3, 7.0 Hz, 1H), 7.08-7.12 (m, 1H), 7.15-7.32 (m, 5H), 7.35-7.40 (m, 1H), 7.49-7.54 (m, 2H), 7.57-7.63 (m, 1H), 7.65-7.69 (m, 1H), 7.96-8.00 (m, 1H). 13C NMR (CDCl3): δ 165.2, 163.3, 161.8, 140.4, 134.9, 132.4, 130.7, 130.5, 129.2, 128.7, 128.2, 127.4, 127.0, 125.8, 124.5, 123.7, 120.6, 49.8, 38.3. HRMS (ESI): Calcd for C21H15N3O2S2Na, m/z [M+Na]+: 428.0503; found 428.0496.
2-[2-Phenyl-1-(2-methyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7t)
White solid (25% yield), mp 98-99 °C. 1H NMR (CDCl3): δ 2.48 (s, 3H), 3.49 (dd, J = 14.1, 9.1 Hz, 1H), 3.75 (dd, J = 14.3, 6.6 Hz, 1H), 6.39 (dd, J = 9.1, 6.6 Hz, 1H), 7.17-7.31 (m, 5H), 7.37-7.42 (m, 1H), 7.52-7.55 (m, 1H), 7.59-7.64 (m, 1H), 7.98-8.01 (m, 1H). 13C NMR (CDCl3): δ 165.2, 164.8, 164.3, 140.3, 135.0, 132.4, 129.1, 128.7, 127.3, 127.0, 125.7, 123.7, 120.6, 49.6, 37.9, 11.0. HRMS (ESI): Calcd for C18H15N3O2SNa, m/z [M+Na]+:360.0783; found 360.0779.
2-[2-Phenyl-1-(2-ethyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7u)
White solid (21% yield), mp 78-80 °C. 1H NMR (CDCl3): δ 1.32 (t, J = 7.6 Hz, 3H), 2.84 (q, J = 7.6 Hz, 2H), 3.49 (dd, J = 14.1, 9.0 Hz, 1H), 3.73 (dd, J = 14.2, 6.8 Hz, 1H), 6.39 (dd, J = 8.9, 6.9 Hz, 1H), 7.16-7.32 (m, 5H), 7.36-7.43 (m, 1H), 7.50-7.66 (m, 2H), 7.97-8.02 (m, 1H). 13C NMR (CDCl3): δ 168.9, 165.2, 164.1, 140.4, 135.0, 132.4, 129.2, 128.7, 127.3, 127.0, 125.7, 123.7, 120.6, 49.7, 38.1, 19.1, 10.6. HRMS (ESI): Calcd for C19H17N3O2SNa, m/z [M+Na]+: 374.0939; found 374.0932.
2-[2-Phenyl-1-(2-propyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7v)
Light yellow solid (25% yield), mp 83-84 °C. 1H NMR (CDCl3): δ 0.95 (t, J = 7.4 Hz, 3H), 1.69-1.80 (m, 2H), 2.74-2.81 (m, 2H), 3.49 (dd, J = 14.1, 9.0 Hz, 1H), 3.73 (dd, J = 14.4, 6.8 Hz, 1H), 6.40 (dd, J = 8.7, 6.8 Hz, 1H), 7.17-7.30 (m, 5H), 7.37-7.42 (m, 1H), 7.52-7.56 (m, 1H), 7.59-7.64 (m, 1H), 7.97-8.02 (m, 1H). 13C NMR (CDCl3): δ 168.0, 165.2, 164.1, 140.4, 135.0, 132.4, 129.1, 128.7, 127.3, 127.0, 125.7, 123.7, 120.6, 49.7, 38.2, 27.1, 19.9, 13.5. HRMS (ESI): Calcd for C20H19N3O2SNa, m/z [M+Na]+:388.1096; found 388.1111.
2-[2-m-Fluorophenyl-1-(2-m-chlorophenyl-1,3,4-oxadiazol-5-yl)ethyl]-1,2-benzisothiazol-3(2H)-one (7w)
Yellow oil (29% yield). 1H NMR (CDCl3): δ 3.56 (dd, J = 12.4, 9.1 Hz, 1H), 3.80 (dd, J = 12.4, 9.1 Hz, 1H), 6.51 (dd, J = 8.9, 6.8 Hz, 1H), 6.85-8.05 (m, 12H). 13C NMR (CDCl3): δ 165.3, 164.6, 164.4, 164.0, 140.3, 137.3, 135.2, 132.2, 130.8, 130.4, 130.0, 127.1, 125.9, 125.2, 124.8, 123.5, 120.7, 116.4, 116.1, 114.6, 114.4, 49.6, 37.8. HRMS (ESI): Calcd for C23H16N3O2SClF, m/z [M+H]+: 452.0636, found 452.0637.
2-[3-Phenyl-1-(2-m-chlorophenyl-1,3,4-oxadiazol-5-yl)propyl]-1,2-benzisothiazol-3(2H)-one (7x)
Yellow oil (31% yield). 1H NMR (CDCl3): δ 2.60-2.82 (m, 4H), 6.21-6.27 (m, 1H), 7.18-8.11 (m, 13H). 13C NMR (CDCl3): δ 165.7, 164.7, 164.6, 140.7, 139.7, 135.3, 132.8, 132.3, 130.6, 128.8, 128.7, 127.3, 127.2, 126.7, 126.1, 125.4, 125.1, 123.9, 120.9, 48.4, 34.1, 32.0. HRMS (ESI): Calcd for C24H19N3O2SCl, m/z [M+H]+: 448.0887, found 448.0875.
X-Ray crystal analysis
Compound 7p crystallizes as colorless needles. The crystal was affixed to a nylon cryoloop using oil (Paratone-n, Exxon) and mounted in the cold stream of a Bruker Kappa-Apex-II area-detector diffractometer. The temperature of the crystal was maintained at 150 K using a Cryostream 700EX Cooler (Oxford Cryosystems). Data were measured with a redundancy using a CCD detector at a distance of 50 mm from the crystal with a combination of phi and omega scans. A scan width of 0.5 degrees and scan time of 10 seconds was employed using graphite monochromated Molybdenum Kα radiation (λ= 0.71073 Å) that was collimated to a 0.6 mm diameter. Data collection, reduction, structure solution, and refinement were performed using the Bruker Apex2 suite (v2.0-2).47 All available reflections to 2θmax = 52° were harvested (35207 reflections, 3812 unique) and corrected for Lorentz and polarization factors with Bruker SAINT (v6.45).48 Reflections were then corrected for absorption (numerical correction, μ=0.222 mm-1), interframe scaling, and other systematic errors with SADABS 2004/1.49 The structure was solved (direct methods) and refined (full-matrix least-squares against F2) with the Bruker SHELXTL package (v6.14-1).50 All non-hydrogen atoms were refined using anisotropic thermal parameters. All hydrogen atoms were included at idealized positions; hydrogen atoms were not refined. Absolute stereochemistry was assigned based on the synthetic protocol and confirmed by a Flack parameter of 0.02. Pertinent crystal, data collection, and refinement parameters are given in Table 2.
Biochemistry
DENV2 NS2B/NS3pro expression and purification
The construction of the pQE30-NS2BH(QR)NS3pro expression plasmid has been described previously.38 The protease was expressed from E. coli strain Top 10 F’ (Invitrogen) transformed by pQE30-NS2BH(QR)-NS3pro plasmid and purified as described.39 DENV2 NS2B/NS3pro contains the hydrophilic NS2B cofactor peptide (NS2BH) linked to the N-terminal NS3 protease domain via Q-R (P2 and P1 residues at the NS2B/S3 junction site).
WNV NS2B/NS3pro expression and purification
The expression and purification of the WNV NS2B/NS3pro containing the 5 amino acid spacer between the NS2B and NS3pro domains used in these experiments were previously described.40
In vitro DENV2 and WNV NS2B/NS3pro assays and inhibition studies
Selected compounds 7a-7x were analyzed by in vitro protease assays performed in opaque 96-well plates. Standard reaction mixtures (100 μL) containing 200 mM Tris·HCl (pH 9.5), 6 mM NaCl, 30 % glycerol, 25 nM DENV2 NS2B/NS3 protease, and 10 μM or 25 μM inhibitor (dissolved in DMSO) in each assay were incubated 15 min at 37 °C. Reactions were started by the addition of 5.0 μM tetra-peptide substrate Bz-Nle-Lys-Arg-Arg-AMC. Conditions were the same for WNV protease except 28 nM WNV NS2B/NS3pro was used in the assay.
Release of free AMC was measured using a spectrofluorometer (Molecular Devices) at excitation and emission wavelengths of 380 and 460 nm, respectively. Control fluorescence values obtained in the absence of inhibitor were taken as 100%, and those in the presence of inhibitors were calculated as the percentage of inhibition of the control using Microsoft Excel and plotted using SigmaPlot 2001 v7.1 (Systat Software Inc.). The background of AMC in the absence of protease was subtracted before the data analysis. All assays were performed in triplicate. Compounds with more than 50% percent inhibition at 25 μM were chosen to determine the IC50 values. Twelve data points obtained from the range of 10 nM, 50 nM, 0.1, 0.5, 1, 2, 4, 6, 8, 10, 20, and 25 μM inhibitor concentrations of selected compounds were used. IC50 values were calculated using the SigmaPlot 2001 v7.1 software.
Steady-state kinetics analysis
The tetra-peptide Bz-Nle-Lys-Arg-Arg-AMC was used as the substrate for analyzing the solution kinetics of protease inhibition. Reaction progress was monitored by release of free AMC every 1.5 min for at least 10 min in 200 mM Tris·HCl (pH 9.5), 6 mM NaCl, 30 % glycerol containing 0.1% CHAPS at 37 °C unless otherwise specified. The tetra-peptide with an AMC tag was incubated with and without the inhibitor in the above buffer in the presence of varying concentrations of substrate. The reactions were initiated by the addition of NS2B/NS3 protease. The protease activity was monitored by the initial rates of the increase of fluorescence intensity at 460 nm with an excitation wavelength of 380 nm. All reactions were carried out to less than 5% completion. The results were analyzed using Sigma-plot 2001 v7.1 and the Michaelis-Menten equation to obtain the apparent Michaelis-Menten constants and maximal velocities. All assays were performed two times in duplicate. For each tested compound, four different concentrations of inhibitor (0, 1.0, 3.0 and 5.0 μM) were used at varying concentrations (0-50 μM) of substrate. The mechanism of inhibition was determined to be competitive by observing the deviation of the apparent Km while a relative constant kcat was maintained.51 The kinetics analysis methods have been previously described.40Ki values were calculated from these data using a secondary plot of Km, app against the concentrations of selected compound 7n using SigmaPlot 2001 v7.0 software. The results are summarized in Figures 4 and 5, and Table 4.
Molecular Modeling
Molecular docking simulations were performed with the Vina program.52 To describe the DENV2 NS2B/NS3 enzyme, modeling was performed on both the active form (PDB: 3U1I)23 and on the structure with a co-crystallized substrate-based inhibitor (PDB: 2FOM)41. WNV NS2B/NS3pro was modeled via a structure (PDB: 2FP7)41 analogous to the latter DENV model and via a more recent structure (PDB: 3E90) in complex with the Naph-KKR-H inhibitor43. In each case, the receptor model was prepared by extracting all ligands and waters, and protonating the receptor according to assumption of anionic aspartate and glutamate groups, and cationic arginines and lysines. The co-crystallized ligand from each crystal structure was used to define center of the grid space into which the ligand was docked. In each case, a cubic grid was defined with a buffer that extended 8.0 Å beyond the extent of the cocrystallized ligand. Ligand docking was then pursued by requesting a level-10 search, so as to achieve a reasonably comprehensive conformational sampling. All other Vina parameters were left at default values.
Acknowledgments
The generous financial support of this work by the National Institutes of Health (AI082068) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindenbach BD, Thiel H-J, Rice CM. In: Fields Virology. 5. Knipe DM, Howley PM, editors. Vol. 1. Lippincott, Williams, and Wilkins; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- 2.Muñoz-Jordάn JL, Sάnchez-Burgos GG, Laurent-Rolle M, García-Sastre A. Proc Natl Acad Sci USA. 2003;100:14333. doi: 10.1073/pnas.2335168100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morens DM, Fauci AS. J Am Med Assoc. 2008;299:214. [Google Scholar]
- 4.Gulati S, Maheshwari A. Trop Med Int Health. 2007;12:1087. doi: 10.1111/j.1365-3156.2007.01891.x. [DOI] [PubMed] [Google Scholar]
- 5.Sampath A, Padmanabhan R. Antiviral Res. 2009;81:6. doi: 10.1016/j.antiviral.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lescar J, Luo D, Xu T, Sampath A, Lim SP, Canard B, Vasudevan SG. Antiviral Res. 2008;80:94. doi: 10.1016/j.antiviral.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. J Biol Chem. 2000;275:9963. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 8.Natarajan S. Gen Mol Biol. 2010;33:214. doi: 10.1590/S1415-47572010000200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampath A, Xu T, Chao A, Luo D, Lescar J, Vasudevan SG. J Virol. 2006;80:6686. doi: 10.1128/JVI.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwong AD, Rao BG, Jeang K–T. Nat Rev Drug Discovery. 2005;4:845. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim SP, Sonntag LS, Noble C, Nilar SH, Ng RH, Zou G, Monaghan P, Chung KY, Dong H, Liu B, Bodenreider C, Lee G, Ding M, Chan WL, Wang G, Jian YL, Chao AT, Lescar J, Yin Z, Vedananda TR, Keller TH, Shi PY. J Biol Chem. 2011;286:6233. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podvinec M, Lim SP, Schmidt T, Scarsi M, Wen D, Sonntag LS, Sanschagrin P, Shenkin PS, Schwede T. J Med Chem. 2010;53:1483. doi: 10.1021/jm900776m. [DOI] [PubMed] [Google Scholar]
- 13.Puig-Basagoiti F, Qing M, Dong H, Zhang B, Zou G, Yuan Z, Shi PY. Antiviral Res. 2009;83:71. doi: 10.1016/j.antiviral.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Antimicrob Agents Chemother. 2008;52:3385. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Int J Biochem Cell Biol. 2007;39:606. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 16.Dou D, Viswanathan P, Li Y, He G, Alliston KR, Lushington GH, Brown-Clay JD, Padmanabhan R, Groutas WC. J Comb Chem. 2010;12:836. doi: 10.1021/cc100091h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tiew KC, Dou D, Teramoto T, Lai H, Alliston KR, Lushington GH, Padmanabhan R, Groutas WC. Bioorg Med Chem. 2012;20:1213. doi: 10.1016/j.bmc.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravapalli S, Lai H, Teramoto T, Alliston KR, Ferguson EL, Padmanabhan R, Groutas WC. Bioorg Med Chem. 2012;20:4140. doi: 10.1016/j.bmc.2012.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y-L, Yin Z, Duraiswamy J, Schul W, Lim CC, Liu B, Xu HY, Qing M, Yip A, Wang G, Chan WL, Tan HP, Lo M, Liung S, Kondreddi RR, Rao R, Gu H, He H, Keller TH, Shi P-Y. Antimicrob Agents Chemother. 2010;54:2932. doi: 10.1128/AAC.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torrence PF, Gupta N, Whitney C, Morrey JD. Antiviral Res. 2006;70:60. doi: 10.1016/j.antiviral.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Yang CC, Hsieh YC, Lee SJ, Wu SH, Liao CL, Tsao CH, Chao YS, Chern JH, Wu CP, Yueh A. Antimicrob Agents Chemother. 2011;55:229. doi: 10.1128/AAC.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomlinson SM, Malmstrom RD, Russo A, Mueller N, Pang Y-P, Watowich SJ. Antiviral Res. 2009;82:110. doi: 10.1016/j.antiviral.2009.02.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble CG, She CC, Chao AT, Shi P-Y. J Virol. 2012;86:438. doi: 10.1128/JVI.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Julander JG, Perry ST, Shresta S. Antiviral Chem Chemother. 2011;21:105. doi: 10.3851/IMP1690. [DOI] [PubMed] [Google Scholar]
- 25.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Curr Med Chem. 2008;15:2771. doi: 10.2174/092986708786242804. [DOI] [PubMed] [Google Scholar]
- 26.Mor M, Zani F, Mazza P, Silva C, Bordi F, Morini G, Plazzi PV. Il Pharmaco. 1996;51:493. [PubMed] [Google Scholar]
- 27.Jorgensen WL, Trofimov A, Du X, Hare AA, Leng L, Bucala R. Bioorg Med Chem Lett. 2011;21:4545. doi: 10.1016/j.bmcl.2011.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matuszak N, Saadi BE, Labar G, Marchand-Brynaert J, Lambert DM. Bioorg Med Chem Lett. 2011;21:7321. doi: 10.1016/j.bmcl.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Morini G, Poli E, Comini M, Menozzi A, Possoli C. Arch Pharm Res. 2005;28:1317. doi: 10.1007/BF02977894. [DOI] [PubMed] [Google Scholar]
- 30.Mahmoud AH, Abouzid KAM, El Ella DA, El RA, Ismail MAH. Bioinformation. 2011;7:328. doi: 10.6026/97320630007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsunaga H, Matsui T, Ohya K, Okino K, Hayashida K, Maebayashi K, Kiriike N, Stein DJ. Int J Psychiatry Clin Pract. 2006;10:142. doi: 10.1080/13651500500487586. [DOI] [PubMed] [Google Scholar]
- 32.Wright SW, Petraitis JJ, Abelman MM, Bostrom LL, Corbett RL, Green AM, Kindt RM, Sherk SR, Magolda RL. Bioorg Med Chem Lett. 1993;3:2875. [Google Scholar]
- 33.Dou D, Alex D, Du B, Tiew K-C, Aravapalli S, Mandadapu SR, Calderone R, Groutas WC. Bioorg Med Chem. 2011;19:5782. doi: 10.1016/j.bmc.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 34.Bostrom J, Hogner A, Llinas A, Wellner E, Plowright AT. J Med Chem. 2012;55:1817. doi: 10.1021/jm2013248. [DOI] [PubMed] [Google Scholar]
- 35.Zoumpoulakis P, Camoutsis Ch, Pairas G, Soković M, Glamočlija J, Potamitis C, Pitsas A. Bioorg Med Chem. 2012;20:1569. doi: 10.1016/j.bmc.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 36.Pouliot M-F, Angers L, Hamel J-D, Paquin J-F. Org Biomol Chem. 2012;10:988. doi: 10.1039/c1ob06512b. [DOI] [PubMed] [Google Scholar]
- 37.Stabile P, Lamonica A, Ribecai A, Castoldi D, Guercio G, Curcuruto O. Tetrahedron Lett. 2010;51:4801. [Google Scholar]
- 38.Yon C, Teramoto T, Mueller N, Phelan J, Ganesh VK, Murthy KH, Padmanabhan R. J Biol Chem. 2005;280:27412. doi: 10.1074/jbc.M501393200. [DOI] [PubMed] [Google Scholar]
- 39.Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Antimicrob Agents Chemother. 2008;52:3385. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ezgimen M, Lai H, Mueller NH, Lee K, Cuny G, Ostrov DA, Padmanabhan R. Antiviral Res. 2012;94:18. doi: 10.1016/j.antiviral.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Nat Struct Mol Biol. 2006;13:372. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 42.De la Cruz L, Nguyen TH, Ozawa K, Shin J, Graham B, Huber T, Ottig G. J Am Chem Soc. 2011;133:19205. doi: 10.1021/ja208435s. [DOI] [PubMed] [Google Scholar]
- 43.Robin G, Chapell K, Stoermer MJ, Hu S-H, Young PR, Fairlie DP, Martin JL. J Mol Biol. 2009;385:1568. doi: 10.1016/j.jmb.2008.11.026. [DOI] [PubMed] [Google Scholar]
- 44.Gillespie P, Goodnow R. Ann Rep Med Chem. 2004;39:293. [Google Scholar]
- 45.Perola E. J Med Chem. 2010;53:2986. doi: 10.1021/jm100118x. [DOI] [PubMed] [Google Scholar]
- 46.Meanwell NA. Chem Res Toxicol. 2011;24:1420. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 47.APEX2 User Manual. Bruker AXS; Madison (USA): 2005. [Google Scholar]
- 48.SAINT software reference manual, Version 4. Bruker AXS; Madison (USA): 1994-1996. [Google Scholar]
- 49.Sheldrick G. SADABS (Version 2.03) University of Göttingen; Germany: 2002. [Google Scholar]
- 50.SHELXTL reference manual, Version 5.1. Bruker AXS; Madison (USA): 1997. [Google Scholar]
- 51.Segel IH. Enzyme Kinetics. John Wiley & Sons; NY: 1993. pp. 104–107. [Google Scholar]
- 52.Trott O, Olson J. J Comput Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]


