Abstract
Over the course of infection, the human immunodeficiency virus type 1 (HIV-1) continuously adapts in part to evade the host’s neutralizing antibody response. Antibodies often target the HIV envelope proteins that mediate HIV fusion to its cellular targets. HIV virions pseudotyped with primary envelopes have often been used to explore the fusogenic properties of these envelopes. Unfortunately, these pseudotyped virions fuse with greatly reduced efficiency to primary cells. Here, we describe a relatively simple strategy to clone primary envelopes into a provirus and increase the sensitivity of the virion-based fusion assay.
Keywords: HIV fusion, primary envelopes, CD4 T-cells, monocyte-derived Dendritic Cells
Introduction
The human immunodeficiency virus (HIV) evolves rapidly, leading to an extensive genetic diversity within and between infected individuals. HIV diverged into several lineages or groups during multiple zoonotic infections occurring between nonhuman primates and humans (for review [1]). HIV type-1 (HIV-1) groups M, N, and O and HIV-2 appear to reflect different cross-species transmission events [2–5]. Group M has evolved into multiple subtypes due to adaptation and expansion in human hosts. These subtypes (or clades) are designated A through G and are joined in the pandemic by a few dozen additional strains that contain genetic segments derived from multiple subtypes, named circulating recombinant forms. HIV-1 subtypes share 70–90% sequence identity, groups share <70%, and HIV-1 and HIV-2 can differ by as much as 50% at the nucleotide level [6].
The genetic diversity of HIV-1 stems from the combination of point mutations and genetic recombinations [7]. Base substitutions are introduced principally by the error-prone reverse transcriptase [8] or by the mutagenic activities of host antiviral factors, such as the APOBEC3 (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like) family of cytidine deaminases [9, 10]. These processes introduce ~1 substitution per viral genome per generation. Genetic recombination results from the migration of reverse transcriptase between the two viral RNA templates encapsidated in the virion and happens several times in any given round of HIV-1 DNA synthesis. Both the incorporation of two heterologous genomic RNAs and recombination-prone replication machinery are critical for this high frequency of recombination. The vast sequence variability of the HIV-1 virus allows escape variants to arise upon selective pressure exerted by the immune system or antiviral drugs.
One of the most variable genes of HIV is the envelope (env). The env gene encodes a precursor, gp160, that is cleaved by the cellular protease furin into two subunits, gp120 and gp41. These proteins remain associated at the cell surface due to electrostatic interactions. Gp120 and gp41 heterodimers are anchored on the viral particle by the transmembrane domains of the gp41 subunit and assemble into trimeric structures to form the viral spike. Viral cell entry is initiated by the binding of gp120 to the CD4 receptor. Later binding of gp120 to the CCR5 or CXCR4 coreceptor [11] induces conformational changes that trigger the activation of gp41 [12–14] and leads to fusion of virions to the target cell [12, 15].
Gp120 consists of five highly variable regions (designated V1–V5) that are interspersed between five more conserved regions (C1–C5) [16]. The variable loops shield the more conserved regions that mediate binding to the receptors [17]. When gp120 binds to CD4, structural changes expose previously masked epitopes and surfaces [18, 19]. V1, V2, V4 and V5 are characterized by rapid changes in the length, number and localization of glycosylation sites [20, 21].
Because of the extreme genetic diversity of HIV-1 env, it is important that functional studies of HIV-1 envelopes be performed with primary rather than laboratory-adapted gene products. We previously described an HIV virion–based fusion assay that is able to measure the properties and kinetics of fusion of laboratory-adapted viruses to a wide variety of target cells, including biologically relevant primary cells. Unfortunately, this assay performs less efficiently when virions are pseudotyped with primary envelopes [22]. Thus, this shortcoming limited the type of target cells that could be studied as well as the analysis of fusion induced by various primary envelope proteins. Here, we describe the construction of viruses encoding primary envelopes in cis and their successful use to study the fusogenic properties of various primary HIV envelope proteins.
Materials and Methods
1. Proviral DNA and HIV envelopes
pCMV4-BlaM-Vpr is available upon request at Addgene (Cambridge, MA). pAdVAntage is a commercially available construct (Promega, Madison, WI). The proviral constructs pNL4-3ΔEnv and TN6-GFP are as previously described [23] and [24]. The pCR3.1 vectors encoding the primary envelopes 55FPB28a and 109FPB4 are as previously described [25]. The vectors expressing primary HIV envelope proteins (pSVIII-92RW020.5, pSVIII-92HT599.24, pSVIII-93MW965.26, pSVIII-92UG021.16) were obtained from the NIH AIDS Research & Reference Reagent Program [26].
2. Cloning the primary envelope into the TN6-GFP vector
To facilitate cloning of the primary envelopes into the proviral DNA, we selected the TN6-GFP proviral DNA expression vector, an NL4-3-based construct modified to contain a BstEII restriction site 15 nucleotides (nt) after the signal peptide of NL4-3 env and a NcoI site at the end of the envelope (for map see Fig. 1 [24]). Primary envelopes were amplified with the sense primer C6323+ as previously described [24] (ttgtgGGTCACCgtctattatgggg) and the antisense primer ASenvNcoI (ctgcatCCATGGtttattgtaaagctgcttc). The PCR amplification was performed in 50 µl of a solution containing 100–250 ng of purified vector encoding the envelopes, 20 pmol of each primer, 200 µM dNTPs, and 1X buffer containing 15 mM MgCl2, and 2.6 U of Taq DNA polymerase (Expand High Fidelity PCR System, Roche). The PCR parameters were 94°C for 2 min to achieve initial denaturation, followed by 30 cycles at 94°C for 30s, 58°C for 30s, 72°C for 3 min and a final elongation at 72°C for 30 min. The PCR products were analyzed on 1% agarose gels, purified using QIAquick kit (QIAGEN, Valencia, CA) and subcloned into the TOPO XL vector (Invitrogen, Carlsbad, CA). To release the insert, the TOPO clones were then digested by BstEII and NcoI (NEB, Ipswich, MA). After gel purification, these inserts were ligated using T4 ligase (NEB, Ipswich, MA) into TN6-GFP previously cut with BstEII and NcoI. Ligation was performed in 20 µl of a solution containing 50 mM Tris-HCl (pH7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, and 25 µg/ml bovine serum albumin, and 2,000 U of T4 DNA ligase (NEB). Use of approximately 3 inserts per 1 proviral vector yielded high levels of ligation. To further increase the ligation efficiency, temperatures were alternated between 16° and 37°C every 30 sec. Half of the ligation products (i.e., 10 µl of the ligation reaction) were used to transform MAX Efficiency Stbl2 competent cells (Invitrogen). The resulting TN6-GFP clones containing the primary envelopes were then amplified and purified using a QIAGEN plasmid mega kit. Sequences were confirmed by sequencing.
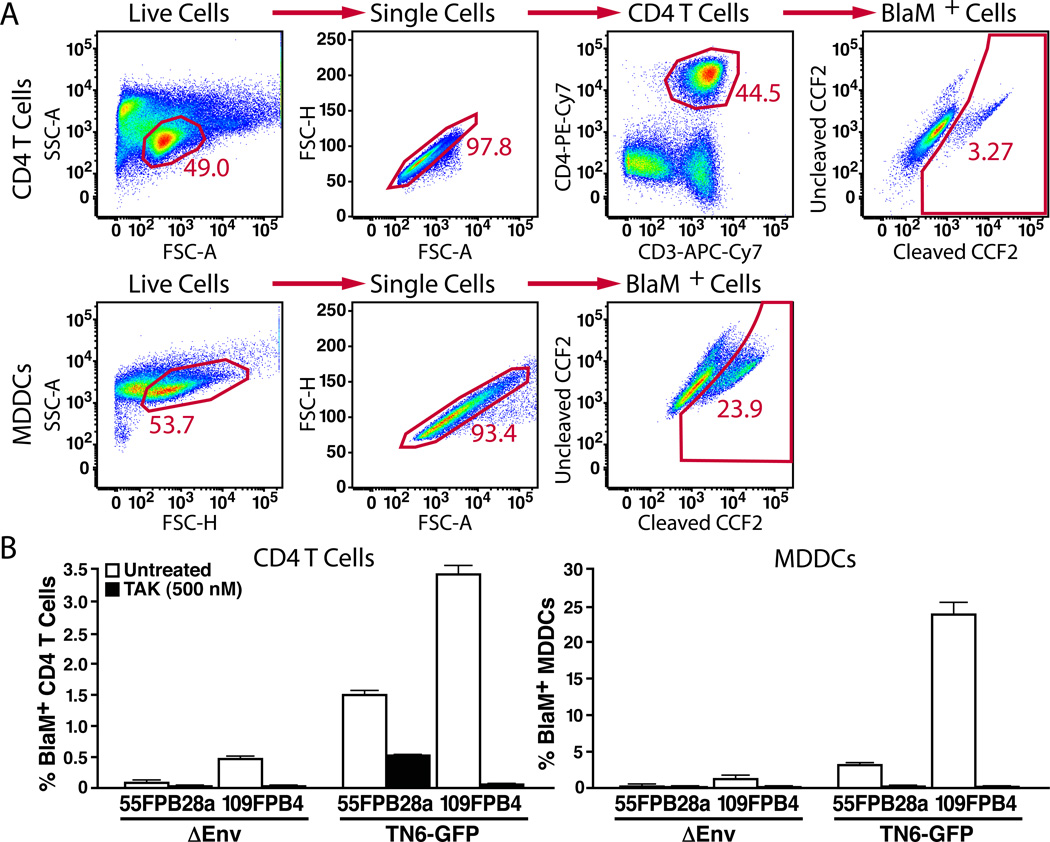
Figure 1. Comparison of fusion mediated by primary envelopes expressed in trans or in cis.
Virions containing BlaM-Vpr were produced by pseudotyping ΔEnv NL4-3 with an expression vector encoding the primary envelopes (pCR3.1-55FPB28a and pCR3.1-109FPB4) or by transfection of a proviral DNA encoding the same primary envelopes (TN6-55FPB28a and TN6-109FPB4). 2 × 106 PBLs or 5 × 105 MDDCs were infected with equal quantities of virions (equivalent of 400 ng of p24Gag) for 2 h at 37°C. (A) FACS plot representing the gating strategy for analysis of fusion of TN6-109FPB4 in CD4 T-cells (first row) or of MDDCs (second row). The BlaM+ cell gate was established based on uninfected samples. Percentages represent the fraction of cells displaying increased blue fluorescence indicative of virion fusion. (B) Histogram representing the level of fusion for experiment performed in triplicate with these envelopes. As a control, cells were treated with TAK-779 for 1 h before the addition of virus. Note the larger increase in fusion obtained when the envelopes were expressed in cis in the proviral construct (TN6-GFP) compared to pseudotyping virions (_Env) in trans. Data presented are representative of results obtained with two independent donors.
3. Generation and culture of PBLs and MDDCs
Peripheral blood mononuclear cells (PBMCs) were purified from fresh buffy coats on Ficoll gradients. CD14+ monocytes were positively selected using Miltenyi anti-CD14 magnetic beads, according to the manufacturer’s instructions. DCs were derived by culturing CD14+ monocytes (2 × 106 cells/ml) for 6 days with 25 ng/ml IL4 (R&D Systems, Minneapolis, MN) and 50 ng/ml GM-CSF (Biosource, Camarillo, CA) in RPMI medium supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 µg/ml streptomycin [27]. The medium was changed every 2 days. Autologous peripheral blood lymphocytes (PBLs) were maintained in RPMI medium until used in the fusion assay in parallel to the MDDCs.
4. Production of NL4-3 Virions Containing BlaM-Vpr
HIV virions containing BlaM-Vpr were produced by transfection of 293T cells with calcium phosphate induced DNA precipitates [28, 29]. 293T cells (1.5 × 107) were plated in 40 ml of DMEM culture medium in a T175 cm2 tissue-culture flask and cultured overnight at 37°C in a 5% CO2 humidified incubator. The medium was then exchanged with 40 ml of DMEM pre-warmed to 37°C. The DNA (60 µg of TN6-GFP proviral DNA, 20 µg of pCMV-BlaM-Vpr and 10 µg of pAdVAntage vectors) was diluted in 1.75 ml of H2O and precipitates formed by successively adding 2 ml of 2X HBS (280 mM NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 12 mM dextrose, 50 mM Hepes, pH 7.05), and 250 µl of 2 M CaCl2 drop by drop. After 30 min of incubation at room temperature, the precipitate was added dropwise to the 293T cells. Pseudotyped viruses were produced by transfection of 40 µg of NL4-3ΔEnv proviral DNA, 20 µg of envelope expressing constructs, 20 µg of pCMV-BlaM-Vpr and 10 µg of pAdVAntage vector DNA. After 16 h at 37°C, the medium was replaced with 20 ml of fresh DMEM complete medium prewarmed to 37°C. Viral production was allowed to continue for 24 h. Virion-containing supernatants were harvested and centrifuged at 800 × g at room temperature for 10 min to remove cellular debris. Virions were next pelleted by ultracentrifugation at 72,000 × g at 4°C for 90 min. The viral pellet was then resuspended in 400 µl of DMEM, aliquoted and stored at −80°C. The p24Gag content of the viral preparations was quantified with the Alliance HIV-I p24 ELISA Kit (Perkin Elmer), according to the manufacturer’s instructions. Virion yield after ultracentrifugation was 50–100 µg of p24Gag/ml.
5. HIV-1 virion fusion assay
The virion-based fusion assay has been previously described in considerable detail [28, 29]. This assay involves three steps: (1) incubation of target cells with NL4-3 virions containing BlaM-Vpr, (2) loading of target cells with the CCF2-AM substrate and development of the BlaM reaction, and (3) immunostaining and fixation. 5×105 MDDCs or 2 × 106 PBLs were infected with HIV virions containing BlaM-Vpr (equivalent of 500 ng of p24Gag) for 2 h at 37°C in a volume of 100 µl. For convenience, the infection was performed in a V-bottom 96-well plate. After infection, the cells were washed once in CO2-independent DMEM medium and loaded with the CCF2-AM substrate (Invitrogen) by incubating the cells for 1 h at room temperature in a solution of CO2-independent medium containing 1 µM CCF2-AM and a dilution 1/100 of the solution B provided by Invitrogen (100 mg/ml Pluronic-F127 and 0.1% acetic acid). The cells were next washed again in CO2-independent media containing 10% FBS and incubated overnight in 200 µl of CO2-independent media containing 10% FBS and 250 mM probenecid, an inhibitor of anion transport, at room temperature and in the dark. For the analysis of fusion to CD4 T cell PBLs, the fusion assay was combined with an immunostaining with anti-human CD3 conjugated to APC-Cy7, and anti-human CD4 conjugated to PE-Cy7. Briefly, the PBLs were collected by centrifugation, washed once with 200 µL of FACS staining buffer (PBS with 2% FBS) and incubated with 50 µl of FACS staining buffer containing 1/100 dilution of anti-CD3-APC-Cy7, 1/100 dilution of anti-CD4-PE-Cy7. This immunostaining step was omitted when HIV-1 fusion was analyzed in MDDCs. After 30 min at room temperature, the PBLs or MDDCs were washed and fixed in 1.2% paraformaldehyde overnight at 4°C.
6. Data collection by flow cytometry and analysis with FlowJo software
Virion fusion was monitored by flow cytometry using an LSRII equipped with four lasers [blue (488 nm–20 mW), red (633 nm–25 mW), violet (409 nm–50 mW) and green (531 nm–150 mW)]. Uncleaved CCF2-AM was detected using the violet laser on the “amcyan” detector harboring a 505LP splitter and a 515/20PB filters. Cleaved CCF2 was detected using the violet laser on the “pacific blue” detector containing 450/50PB filters. All fluorescence detection employed log amplification. The combination of forward scatter and side scatter from the first laser was used to delineate the cells of interest. Samples were acquired in the absence of fluorescence compensation. The data were subsequently compensated and analyzed using FlowJo software (Treestar, Ashland, OR).
Results
HIV-1 fusion to target cells can be studied in detail using the HIV-1 virion fusion assay. This assay relies on the incorporation of β-lactamase–Vpr chimeric (BlaM-Vpr) proteins into HIV-1 virions and their subsequent delivery into the cytoplasm of target cells after fusion. This transfer is monitored by the enzymatic cleavage of the CCF2-AM dye, a fluorescent substrate of β-lactamase (BlaM), loaded into the target cells. BlaM cleavage of the beta-lactam ring in CCF2-AM prevents fluorescence resonance energy transfer (FRET) between the coumarin and fluorescein moieties of the dye. This event changes fluorescence emission spectrum from green (520 nm) to blue (447 nm) and thus permits the detection of virion fusion by flow cytometry.
We previously reported the feasibility of studying fusion mediated by primary envelopes that were provided in trans [22, 23]. However, our data indicated that the levels of fusion were substantially lower with this approach than obtained when proviruses were constructed containing a laboratory-adapted env gene in cis [22]. To formally compare the fusion efficiency of virions containing envelopes introduced in cis versus in trans, we used primary subtype C envelopes 55FPB28a and 109FPB4 [25]. These envelopes were amplified by PCR with C6323+ and EnvASNco primers and then subcloned into the TN6-GFP proviral construct [24] harboring a unique BstEII site after the end of env signal peptide and a NcoI site at the end of the coding sequence of env. The resulting construct leads to the expression of an envelope protein from which 41 amino acids correspond to NL4-3 and the remaining amino acids are derived from the inserted primary env cDNA. Virions containing BlaM-Vpr were produced by transfection of 293T cells with either TN-55FPB28a- or TN-109FPB4-expressing envelope in cis or with _Env NL4-3 and envelope expressing constructs pCR3.1-55FPB28a or pCR3.1-109FPB4 expressing envelope in trans. After normalization for p24Gag, these viral preparations were used to infect PBLs or MDDCs for 2h at 37°C. CCF2-AM, the membrane-permeable ester form of the BlaM substrate, was then loaded into the target cells and enzymatic cleavage was allowed to proceed overnight at room temperature. To specifically measure fusion to the CD4 T cells in the PBL population, the fusion assay was combined with anti-CD3 and anti-CD4 immunostaining. The cells were then fixed in 1.2% paraformaldhehyde and analyzed by flow cytometry. Fusion levels were significantly higher for the replication competent virus (TN6-GFP with 55FPB28a or 109FPB4) than for the pseudotyped viruses (_Env and 55FPB28a or 109FPB4) (Figure 1), independent of the target cell tested. Thus, expression of the envelope in cis leads to the production of more infectious viruses for the 2 envelopes tested.
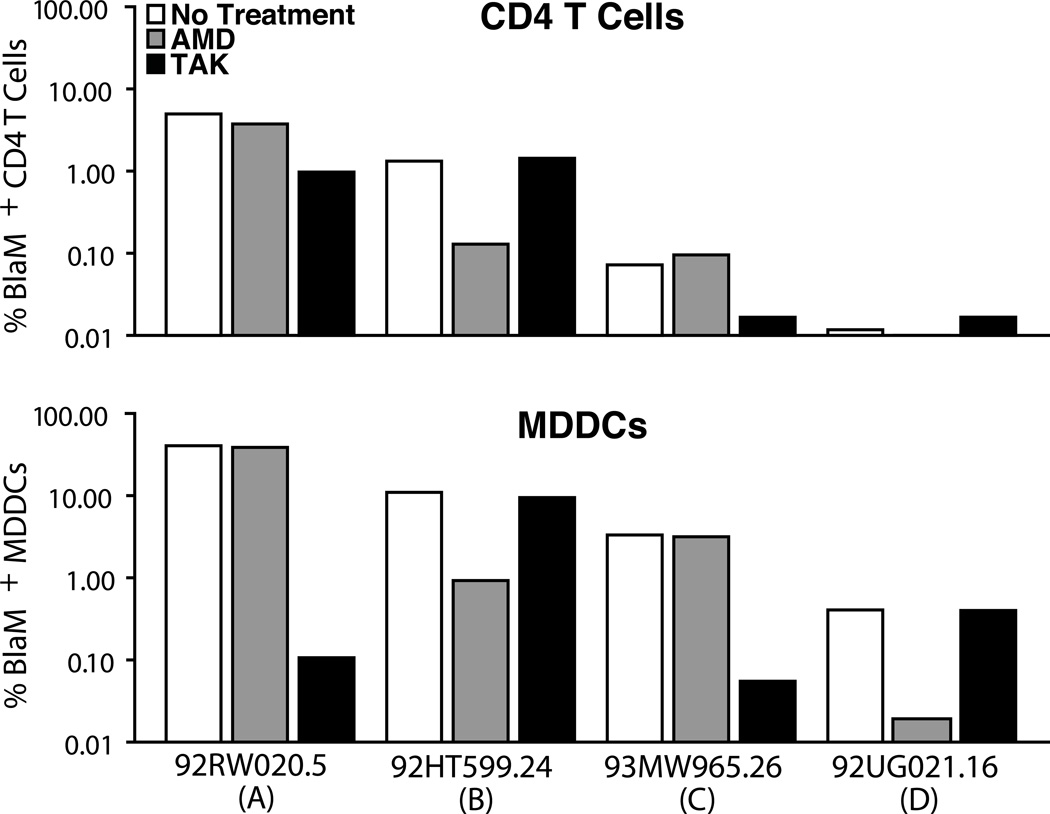
Next, we investigated whether this strategy could be extended to analyze fusion mediated by other HIV-1 subtypes. A panel of four primary envelopes, subtype A, B, C and D, were obtained from the AIDS Research & Reference Reagent Program [26] and subcloned into the TN6-GFP proviral vector. The resulting constructs, TN-92RW020.5 (subtype A), TN-92HT599.24 (subtype B), TN-93MW965.26 (subtype C) and TN-92UG021.16 (subtype D) were used to produce HIV-1 virions containing BlaM-Vpr. PBLs and MDDCs were then infected with equal quantities of virions based on p24Gag. The tropism of the envelope was determined by sensitivity to TAK-779 or AMD3100, which inhibit the entry of CCR5- and CXCR4- tropic viruses, respectively (Figure 2). As reported earlier, we observed a broad range of fusion levels and confirmed the tropism described [26]. Therefore, the cloning strategy can be extended to study fusion mediated by varied primary envelopes displaying different coreceptor tropisms.
Figure 2. Measurement of fusion mediated by envelopes from subtype A, B, C and D.
PBLs were infected for 2 h with HIV virions containing BlaM–Vpr in the presence or absence of TAK-779 (500 nM) or AMD3100 (500 nM). The inhibitors were added 1 h prior to infection and maintained throughout the experiment. For the PBLs, HIV-1 fusion was analyzed in CD3+CD4+ subset of cells. The histogram indicates the number of BlaM+ cells. Note the broad range of fusion observed despite the similar amount of virus used to infect the cells. Data are representative of results obtained with two independent donors.
Discussion
In this study, we investigated whether a relatively simple cloning strategy where primary envelope cDNAs are inserted into a provirus in cis would lead to an improved ability to study fusogenic properties of these Env proteins compared to pseudotyping of HIV virions. Several cloning strategies have been developed including methods including shuttle vectors passed though yeast or laborious multi step cloning [30, 31]. We took advantage of the molecular clone TN6-GFP developed by Neumman et al., that harbors two cloning sites on each side of env, to construct a set of six clones containing primary envelopes. When used to generate HIV-1 virions containing BlaM-Vpr, these proviral constructs result in the production of viruses exhibiting ~7–80-fold more fusogenicity to primary cells than the equivalent viruses produced by pseudotyping. This difference likely stems from improved efficiency of envelope incorporation and gp160 cleavage when env is expressed in cis.
The envelope glycoprotein is produced as a precursor polyprotein gp160, which is cleaved in a Golgi or post-Golgi compartment by a cellular furin-type protease yielding the gp120 and gp41 subunits [32–36]. Although Env cleavage during its transport in the secretory pathway depends on both the cell type and the viral isolate, this process is usually very inefficient. The majority of the Env glycoproteins remain uncleaved and are retained in the endoplasmic reticulum (ER) or in a cis-Golgi compartment [37]. Several studies have shown that matched virions produced by infectious molecular clones or produced by pseudotyping systems lead to physically different virions [38, 39]. Infectious molecular clones, in general, produced virions containing higher amounts of envelope that are more efficiently processed. These differences do not significantly alter the measurement of IC50 by neutralizing antibodies or entry inhibitors but these differences can greatly influence fusogenicity of these envelopes. Careful titration of the Env-encoding constructs used to pseudotype HIV virions can improve gp160 cleavage, however, such titration would make comparisons between multiple viruses problematic at best.
We further demonstrated that the in cis env cloning strategy can be used for multiple HIV-1 subtypes and for both X4 and R5 tropic env gene products. The sense primer (C6223+) used to amplify the HIV envelopes is located in a well-conserved region 9 nt after the end of the signal peptide (SP). The antisense primer (envASNcoI) was designed using the subtype C envelope 109FBP4. It encodes both the env stop codon and the GFP initiation codon. This antisense primer allows for envelope amplification of the different subtypes used in this study. However, since this region is less conserved among HIV subtypes, the sequence of this anti-sense primer might require optimization for better amplification of other envelopes. After cloning into TN6-GFP, the resulting envelope encodes a gp160 envelope protein of ~840 aa, including the 41 initial and five terminal amino acids from NL4-3 envelope. Of the 41 N-terminal aa, 30 aa correspond to the signal peptide targeting the gp160 to the endoplasmic reticulum and processing via the secretory pathway [40]. Therefore, this simple cloning strategy will not allow sampling of functional differences conferred by the signal peptides encoded by the various primary envelopes, which do exhibit a high degree of variability [41]. This experimental approach will also not transpose the regulatory sequences in the rev-vpu region, which were recently reported to regulate envelope expression [42]. For some studies, the optimal clones will remain the full-length primary infectious proviruses that are both difficult and require extensive effort to construct. For other studies, such as characterization of HIV-1 resistance to CCR5 antagonists, our strategy will allow studying most of the resistance mutations and will importantly allow the determination of the shift in IC50 in PBMCs rather than in cell lines, which can be misleading [43]. In conclusion, the assay that we describe here presents a valuable alternative to studying fusion mediated by primary envelope proteins in biologically relevant primary cell targets.
Acknowledgements
We thank Mario Santiago and Kara Lassen for discussions, Stephen Ordway and Gary Howard for editorial assistance, John C.W. Carroll for graphic arts, and Sue Cammack and Robin Givens for administrative assistance. We also thank the NIH AIDS Research & Reference Reagent Program, Division of AIDS, NIAID, NIH for the vectors expressing various primary envelopes. These studies were supported by funding from the National Institutes of Health (P01 AI083050). The authors also acknowledge the UCSF-GIVI CFAR for infrastructure support (P30 AI27763).
Abbreviations
- MDDCs
monocyte-derived dendritic cells
- PBMCs
peripheral blood mononuclear cells
- PBLs
peripheral blood lymphocytes
- BlaM
beta lactamase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, Hahn BH. The origins of acquired immune deficiency syndrome viruses: where and when? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:867–876. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 3.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, Loul S, Ngole EM, Bienvenue Y, Delaporte E, Brookfield JF, Sharp PM, Shaw GM, Peeters M, Hahn BH. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santiago ML, Rodenburg CM, Kamenya S, Bibollet-Ruche F, Gao F, Bailes E, Meleth S, Soong SJ, Kilby JM, Moldoveanu Z, Fahey B, Muller MN, Ayouba A, Nerrienet E, McClure HM, Heeney JL, Pusey AE, Collins DA, Boesch C, Wrangham RW, Goodall J, Sharp PM, Shaw GM, Hahn BH. SIVcpz in wild chimpanzees. Science. 2002;295:465. doi: 10.1126/science.295.5554.465. [DOI] [PubMed] [Google Scholar]
- 5.Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 6.Arien KK, Vanham G, Arts EJ. Is HIV-1 evolving to a less virulent form in humans? Nat. Rev. Microbiol. 2007;5:141–151. doi: 10.1038/nrmicro1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 2009;73:451–480. doi: 10.1128/MMBR.00012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Preston BD, Dougherty JP. Mechanisms of retroviral mutation. Trends Microbiol. 1996;4:16–21. doi: 10.1016/0966-842x(96)81500-9. [DOI] [PubMed] [Google Scholar]
- 9.Bourara K, Liegler TJ, Grant RM. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 2007;3:1477–1485. doi: 10.1371/journal.ppat.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 12.Blish CA, Nedellec R, Mandaliya K, Mosier DE, Overbaugh J. HIV-1 subtype A envelope variants from early in infection have variable sensitivity to neutralization and to inhibitors of viral entry. AIDS. 2007;21:693–702. doi: 10.1097/QAD.0b013e32805e8727. [DOI] [PubMed] [Google Scholar]
- 13.Hollier MJ, Dimmock NJ. The C-terminal tail of the gp41 transmembrane envelope glycoprotein of HIV-1 clades A, B, C, and D may exist in two conformations: an analysis of sequence, structure, and function. Virology. 2005;337:284–296. doi: 10.1016/j.virol.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sattentau QJ. HIV gp120: double lock strategy foils host defences. Structure. 1998;6:945–949. doi: 10.1016/s0969-2126(98)00096-3. [DOI] [PubMed] [Google Scholar]
- 15.Root MJ, Hamer DH. Targeting therapeutics to an exposed and conserved binding element of the HIV-1 fusion protein. Proc. Natl. Acad. Sci. USA. 2003;100:5016–5021. doi: 10.1073/pnas.0936926100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton DR, Stanfield RL, Wilson IA. Antibody vs. HIV in a clash of evolutionary titans. Proc. Natl. Acad. Sci. USA. 2005;102:14943–14948. doi: 10.1073/pnas.0505126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 18.Rits-Volloch S, Frey G, Harrison SC, Chen B. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 2006;25:5026–5035. doi: 10.1038/sj.emboj.7601358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Canziani G, Plugariu C, Wyatt R, Sodroski J, Sweet R, Kwong P, Hendrickson W, Chaiken I. Conformational changes of gp120 in epitopes near the CCR5 binding site are induced by CD4 and a CD4 miniprotein mimetic. Biochemistry. 1999;38:9405–9416. doi: 10.1021/bi990654o. [DOI] [PubMed] [Google Scholar]
- 20.Blay WM, Gnanakaran S, Foley B, Doria-Rose NA, Korber BT, Haigwood NL. Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 2006;80:999–1014. doi: 10.1128/JVI.80.2.999-1014.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology. 2004;14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 22.Cavrois M, Neidleman J, Kreisberg JF, Fenard D, Callebaut C, Greene WC. Human immunodeficiency virus fusion to dendritic cells declines as cells mature. J. Virol. 2006;80:1992–1999. doi: 10.1128/JVI.80.4.1992-1999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yonezawa A, Cavrois M, Greene WC. Studies of ebola virus glycoprotein-mediated entry and fusion by using pseudotyped human immunodeficiency virus type 1 virions: involvement of cytoskeletal proteins and enhancement by tumor necrosis factor alpha. J. Virol. 2005;79:918–926. doi: 10.1128/JVI.79.2.918-926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann T, Hagmann I, Lohrengel S, Heil ML, Derdeyn CA, Krausslich HG, Dittmar MT. T20-insensitive HIV-1 from naive patients exhibits high viral fitness in a novel dual-color competition assay on primary cells. Virology. 2005;333:251–262. doi: 10.1016/j.virol.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 26.Gao F, Morrison SG, Robertson DL, Thornton CL, Craig S, Karlsson G, Sodroski J, Morgado M, Galvao-Castro B, von Briesen H, Beddows S, Weber J, Sharp PM, Shaw GM, Hahn BH. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J. Virol. 1996;70:1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavrois M, De Noronha C, Greene WC. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 29.Cavrois M, Neidleman J, Bigos M, Greene WC. Fluorescence resonance energy transfer-based HIV-1 virion fusion assay. Methods Mol. Biol. 2004;263:333–344. doi: 10.1385/1-59259-773-4:333. [DOI] [PubMed] [Google Scholar]
- 30.Marozsan AJ, Arts EJ. Development of a yeast-based recombination cloning/system for the analysis of gene products from diverse human immunodeficiency virus type 1 isolates. J. Virol. Methods. 2003;111:111–120. doi: 10.1016/s0166-0934(03)00166-6. [DOI] [PubMed] [Google Scholar]
- 31.Tsibris AM, Sagar M, Gulick RM, Su Z, Hughes M, Greaves W, Subramanian M, Flexner C, Giguel F, Leopold KE, Coakley E, Kuritzkes DR. In vivo emergence of vicriviroc resistance in a human immunodeficiency virus type 1 subtype C-infected subject. J. Virol. 2008;82:8210–8214. doi: 10.1128/JVI.00444-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kantanen ML, Leinikki P, Kuismanen E. Endoproteolytic cleavage of HIV-1 gp160 envelope precursor occurs after exit from the trans-Golgi network (TGN) Arch. Virol. 1995;140:1441–1449. doi: 10.1007/BF01322670. [DOI] [PubMed] [Google Scholar]
- 33.Merkle RK, Helland DE, Welles JL, Shilatifard A, Haseltine WA, Cummings RD. gp160 of HIV-I synthesized by persistently infected Molt-3 cells is terminally glycosylated: evidence that cleavage of gp160 occurs subsequent to oligosaccharide processing. Arch. Biochem. Biophys. 1991;290:248–257. doi: 10.1016/0003-9861(91)90616-q. [DOI] [PubMed] [Google Scholar]
- 34.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. Transfer of endoplasmic reticulum and Golgi retention signals to human immunodeficiency virus type 1 gp160 inhibits intracellular transport and proteolytic processing of viral glycoprotein but does not influence the cellular site of virus particle budding. J. Gen. Virol. 1997;78(Pt 7):1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 35.Raja NU, Vincent MJ, Jabbar MA. Analysis of endoproteolytic cleavage and intracellular transport of human immunodeficiency virus type 1 envelope glycoproteins using mutant CD4 molecules bearing the transmembrane endoplasmic reticulum retention signal. J. Gen. Virol. 1993;74(Pt 10):2085–2097. doi: 10.1099/0022-1317-74-10-2085. [DOI] [PubMed] [Google Scholar]
- 36.Salzwedel K, West JT, Jr, Mulligan MJ, Hunter E. Retention of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum does not redirect virus assembly from the plasma membrane. J. Virol. 1998;72:7523–7531. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crise B, Buonocore L, Rose JK. CD4 is retained in the endoplasmic reticulum by the human immunodeficiency virus type 1 glycoprotein precursor. J. Virol. 1990;64:5585–5593. doi: 10.1128/jvi.64.11.5585-5593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Provine NM, Puryear WB, Wu X, Overbaugh J, Haigwood NL. The infectious molecular clone and pseudotyped virus models of human immunodeficiency virus type 1 exhibit significant differences in virion composition with only moderate differences in infectivity and inhibition sensitivity. J. Virol. 2009;83:9002–9007. doi: 10.1128/JVI.00423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrera C, Klasse PJ, Michael E, Kake S, Barnes K, Kibler CW, Campbell-Gardener L, Si Z, Sodroski J, Moore JP, Beddows S. The impact of envelope glycoprotein cleavage on the antigenicity, infectivity, and neutralization sensitivity of Env-pseudotyped human immunodeficiency virus type 1 particles. Virology. 2005;338:154–172. doi: 10.1016/j.virol.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Land A, Braakman I. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie. 2001;83:783–790. doi: 10.1016/s0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- 41.Louwagie J, Janssens W, Mascola J, Heyndrickx L, Hegerich P, van der Groen G, McCutchan FE, Burke DS. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 1995;69:263–271. doi: 10.1128/jvi.69.1.263-271.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus MH, Parrish NF, Shaw KS, Decker JM, Keele BF, Salazar-Gonzalez JF, Grayson T, McPherson DT, Ping LH, Anderson JA, Swanstrom R, Williamson C, Shaw GM, Hahn BH. A rev1-vpu polymorphism unique to HIV-1 subtype A and C strains impairs envelope glycoprotein expression from rev-vpu-env cassettes and reduces virion infectivity in pseudotyping assays. Virology. 2010;397:346–357. doi: 10.1016/j.virol.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pugach P, Marozsan AJ, Ketas TJ, Landes EL, Moore JP, Kuhmann SE. HIV-1 clones resistant to a small molecule CCR5 inhibitor use the inhibitor-bound form of CCR5 for entry. Virology. 2007;361:212–228. doi: 10.1016/j.virol.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]