Directional neuronal migration is mediated by a dynamic SDF1a source generated through localized SDF1a expression followed by regulated mRNA and protein turnover.
Abstract
During animal development, SDF1 simultaneously guides various cell types to different targets. As many targets are in close proximity to one another, it is unclear how the system avoids mistargeting. Zebrafish trigeminal sensory neurons express the SDF1 receptor Cxcr4b and encounter multiple SDF1 sources during migration, but ignore all but the correct one. We show that miR-430 and Cxcr7b regulation of SDF1a are required for precise guidance. In the absence of miR-430 or Cxcr7b, neurons responded to ectopic SDF1a sources along their route and did not reach their target. This was due to a failure to clear SDF1a transcript and protein from sites of expression that the migrating neurons had already passed. Our findings suggest an “attractive path” model in which migrating cells closely follow a dynamic SDF1a source that is refined on a transcript and protein level by miR-430 and Cxcr7b, respectively.
Introduction
During development the vertebrate embryo organizes cells into tissues and organs. To achieve this organization, thousands of cells must migrate from their birthplaces to their final destinations. During their journey, cells follow guidance cues that lead them to their different destinations. In contrast to the large number of migrating cells, there are relatively few guidance cues. How a small number of cues can guide a large number of cells to different destinations at the same time without creating confusion is unclear.
The chemokine SDF1 (also known as CXCL12) is one of these cues. SDF1 signaling through the G protein–coupled receptor CXCR4 (Bleul et al., 1996; Oberlin et al., 1996) attracts many classes of cells (Tiveron and Cremer, 2008) that migrate concurrently and in close proximity to each other to different destinations. Classical models of chemotaxis in Dictyostelium discoideum have suggested that chemoattractants form long-range concentration gradients that guide migrating cells (Van Haastert and Devreotes, 2004). Consistent with such models, misexpression studies have demonstrated that SDF1 can act as a long-range attractant (Blaser et al., 2005; Li et al., 2008). However, it is difficult to imagine how migrating cells that rely on long-range attraction could distinguish between different SDF1 sources.
To address this question, we analyzed the migration of trigeminal sensory neurons (TgSNs). TgSNs are born from neural crest and placodal cells and assemble into two bilateral ganglia (Baker and Bronner-Fraser, 2001). Neurons within the ganglia extend peripheral axons underneath the skin to detect mechanical, thermal, and chemical stimuli on the head, and they relay this information through central axons to the hindbrain (Davies, 1988). In zebrafish, TgSNs are initially distributed as single cells or small clusters of cells along the anterior–posterior axis lateral to the midbrain–hindbrain boundary (MHB). This stripe-like arrangement changes as neurons move posteriorly past the MHB to the ganglion assembly site, against anteriorly directed general tissue movements. The migration distance varies with more anterior-born neurons migrating ∼120 µm and more posterior-born neurons moving ∼20 µm. Correct trigeminal sensory ganglion assembly requires the chemokine SDF1a and its receptor Cxcr4b. The TgSNs express Cxcr4b and migrate toward a local SDF1a source at the ganglion assembly site. In the absence of SDF1a signaling, the TgSNs form one or more displaced clusters rather than a single compact cluster, whereas artificial SDF1a sources attract neurons to ectopic sites (Knaut et al., 2005). TgSNs are born close to two different SDF1a sources, one source at the trigeminal sensory ganglion assembly site and one source at the olfactory placode, but respond only to the source at the ganglion assembly site. Thus, these neurons efficiently distinguish between sources of the same attractant, but the mechanisms that allow them to do so are unclear.
Our results suggest that TgSNs follow the correct source of SDF1a and ignore other nearby sources because they closely associate with a dynamic SDF1a expression domain. Initially, this expression domain delineates the migratory route. Over time it gradually shifts toward the neuron assembly site. The neurons track the shifting expression domain and, thus, are led to their destination. We find that two processes facilitate the rapid shift of the SDF1a expression domain. First, miR-430 is required to clear SDF1a transcripts from sites of expression that the neurons have already passed. Second, Cxcr7b is required to clear SDF1a protein from tissues the neurons have already traversed. In the absence of either process, TgSNs respond to ectopic sources of SDF1a and fail to reach their destination. These results define synergistic pathways that are required for precise cell guidance by a dynamic, attractive path of SDF1a. We propose that this “attractive path” model may explain how other shared guidance cues can direct different cells to different destinations without causing erroneous migration.
Results
TgSNs follow a shifting SDF1a expression domain
To understand why TgSNs are attracted to the SDF1a expression domain at the future site of the trigeminal sensory ganglion rather than to other SDF1a expression domains, we used HuC as a neuronal marker (Kim et al., 1996) to analyze the relationship of migrating TgSNs to SDF1a mRNA-expressing tissues. At the beginning of their migration, TgSNs are distributed bilaterally along the anterior–posterior axis between the future sites of the olfactory placode (anterior to the neurons) and the trigeminal sensory ganglion (posterior to the neurons; Fig. 1 A). Coincident with the initiation of SDF1a mRNA expression at the future site of the olfactory placode (Fig. 1, E–H, arrowheads), migrating TgSNs are closely associated with a stripe of SDF1a expression that extends from the TgSNs to the ganglion assembly site (Fig. 1 B), prefiguring the migratory route of the neurons (Fig. 1 E and Fig. S1 A). As the neurons migrate, the anterior border of this SDF1a expression stripe shifts posteriorly, such that by 14–15 h postfertilization (hpf; 10–12 somite stage) the border has shifted past the MHB to the ganglion assembly site (Fig. 1, C, D, G, and H; and Fig. S1 B). Thus, the TgSNs are closely associated with a shifting SDF1a expression domain throughout their migration.
Figure 1.
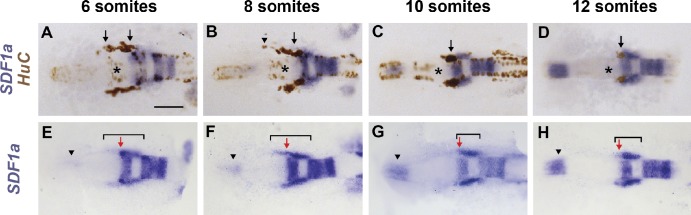
TgSNs follow a dynamic SDF1a expression domain. (A–D) SDF1a (blue) and HuC (brown, TgSNs indicated by black arrows) mRNA distribution in 6-, 8-, 10-, and 12-somite-stage embryos. TgSNs are initially distributed along the anterior–posterior axis (A). They gradually move posterior (B) until they assemble in tight bilateral clusters lateral and posterior to the MHB (asterisks in A–D) by the 10-somite stage (C) and cease migration (D). SDF1a mRNA-expressing cells were often found in close association with migrating TgSNs (arrow in B), but SDF1a mRNA in this region was never detected further anterior than the anterior-most TgSN (arrowhead in B). (D–G) 6-, 8-, 10-, and 12-somite-stage embryos stained for SDF1a mRNA only so that TgSNs do not obscure the SDF1a expression domain. Shown is the dynamic SDF1a expression domain (bracket in E–H) that delineates the TgSN migration route. In relation to the anterior border of rhombomere 2 (red arrow in E–H), the anterior border of the SDF1a expression domain gradually shifted posteriorly (F and G) to come to lie posterior to the ganglion assembly site by the 10- and 12-somite stages (G and H). Arrowheads in E–H denote SDF1a mRNA expression at the site of the future olfactory placode, a location to which trigeminal sensory neurons are not normally attracted. Dorsal view, anterior to the left. See also Fig. S1. Bar, 100 µm.
miRNAs clear SDF1a mRNA and are required for neuron migration
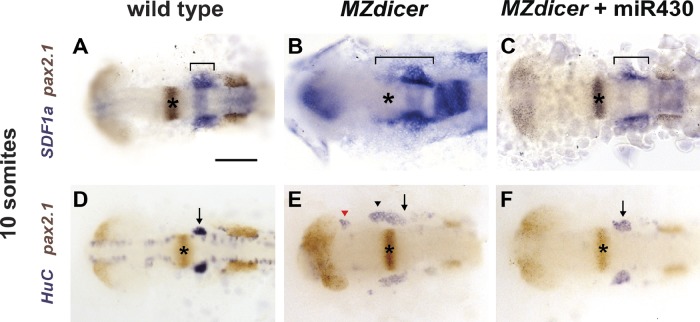
The anterior border of the SDF1a mRNA expression domain that the TgSNs follow shifts posteriorly by 50–100 µm within 90 min (Fig. 1, F–H; and Fig. S1, A and B). Such a rapid change in expression likely requires efficient clearance of SDF1a transcripts from tissues that no longer actively transcribe SDF1a. One possible clearance mechanism is miRNA-mediated mRNA degradation (Bartel, 2004). To test this idea, we investigated SDF1a expression in relation to the MHB, stained with pax2.1 (Krauss et al., 1991b; a), in embryos that lack the maternal and zygotic contributions of Dicer (MZdicer), an RNase required for the maturation of most miRNAs (Bernstein et al., 2001; Grishok et al., 2001; Hutvágner et al., 2001; Cheloufi et al., 2010; Cifuentes et al., 2010). In contrast to 10-somite-stage wild-type embryos (Fig. 2 A), the SDF1a expression domain in stage-matched MZdicer embryos fails to shift posteriorly. Instead, it extends beyond the MHB (Fig. 2 B), resembling the SDF1a expression pattern observed during neuron migration in wild-type embryos (Fig. 1, B and F). We then analyzed the migration of TgSNs in wild-type and MZdicer embryos at the 10-somite stage. Using HuC to mark the neurons and the pax2.1 expression domain at the MHB as a positional landmark for assessing correct migration, we found that TgSNs in MZdicer embryos form one or more clusters displaced anteriorly toward the eyes (Fig. 2 E) rather than single bilateral clusters as seen in wild-type embryos (Fig. 2 D). Together, these data indicate that miRNA-mediated clearance of SDF1a transcripts from sites that the neurons have passed is required for correct migration.
Figure 2.
miR-430 refines SDF1a mRNA expression. 10-somite-stage embryos were stained for SDF1a (blue, A–C) or HuC (blue, D–F) and pax2.1 (brown) mRNA to visualize the SDF1a mRNA expression domain (A–C) or TgSNs (D–F) in relation to the MHB (asterisks), respectively. The bracket marks the anterior–posterior extent of SDF1a mRNA expression in A–C. Arrows and arrowheads denote correctly positioned and mispositioned TgSNs, respectively, in D–F. The red arrowhead in E denotes mispositioned neurons located close to the eye. Dorsal view, anterior to the left. (A and D) Uninjected wild-type embryos. (B and E) Uninjected MZdicer mutant embryos. (C and F) MZdicer mutant embryos injected with miR-430 RNA. Bar, 100 µm.
miR-430 refines SDF1a mRNA expression
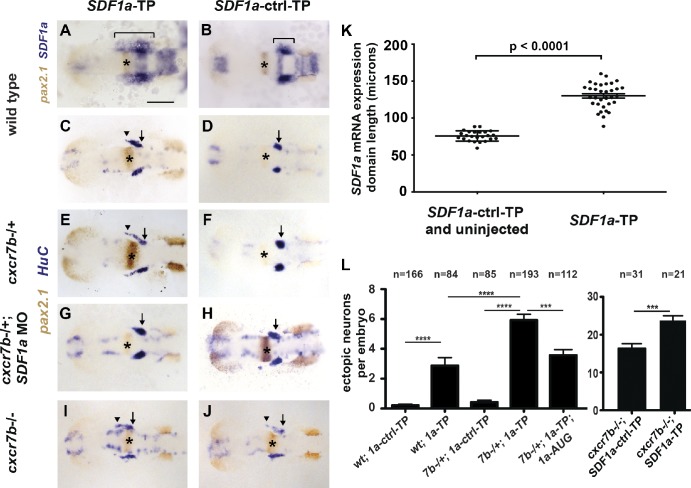
The 3′ UTR of an SDF1a transcript contains binding sites for the miRNA miR-430 (Staton et al., 2011), which suggests that miR-430 might mediate SDF1a refinement. To test this notion, we resupplied miR-430 to MZdicer embryos and found that it is sufficient to restore both the refinement of the SDF1a expression domain (Fig. 2 C) and TgSN migration (Fig. 2 F). Although these observations are consistent with the idea that lack of miR-430 regulation of SDF1a transcripts underlies the defects observed in MZdicer embryos, miR-430 regulates many other mRNAs (Giraldez et al., 2006) that could contribute to these defects. To interfere specifically with miR-430–mediated regulation of SDF1a transcripts, we used target protectors. Target protectors are antisense morpholinos that are complementary to the miRNA binding site in the transcript of interest (Choi et al., 2007) and should disrupt interaction of an miRNA with a single target. In embryos injected with a target protector against an miR-430 binding site in the SDF1a 3′ UTR (SDF1a-TP), we found that the SDF1a mRNA expression domain fails to refine posteriorly (Fig. 3 A) and has an anterior–posterior length of 130 ± 3.0 µm (Fig. 3 K) at the 10-somite stage. In embryos injected with a control target protector morpholino (SDF1a-ctrl-TP), this domain refines to a length of 76 ± 1.4 µm (Fig. 3, B and K). Consequently, TgSNs are often mislocalized along the unrefined expression domain in SDF1a-TP embryos as compared with SDF1a-ctrl-TP embryos (Fig. 3, C, D, and L; and Table 1). First, these observations suggest that miR-430 clears SDF1a transcripts from cells that no longer actively transcribe the SDF1a locus, preventing migrating neurons from being retained at past sites of expression by protein translated from remaining transcripts. Second, the stronger neuronal migration defect in MZdicer mutants than in SDF1a-TP embryos indicates that other miRNAs are required for correct neuron migration, probably indirectly because morphogenesis is also affected in MZdicer embryos (Giraldez et al., 2005).
Figure 3.
miR-430 is required for TgSN migration. 10-somite-stage embryos in A and B were stained for with SDF1a (blue) and pax2.1 (brown, MHB marked with asterisks) mRNA. Embryos in C–J were stained for HuC (blue, neurons) and pax2.1 (brown, MHB with asterisks) mRNA. Arrows and arrowheads in C–J denote correctly and incorrectly positioned TgSNs, respectively. Dorsal view, anterior to the left. (A and C) Wild-type embryos injected with SDF1a-TP. (B and D) Wild-type embryos injected with SDF1a-ctrl-TP. (E and F) cxcr7b−/+ embryos injected with SDF1a-TP or SDF1a-ctrl-TP. (G and H) cxcr7b−/+ embryos injected with SDF1a-TP or SDF1a-ctrl-TP and a suboptimal dose of a translation blocking SDF1a morpholino. (I and J) cxcr7b−/− embryos injected with SDF1a-TP or SDF1a-ctrl-TP. (K) Quantification of the SDF1a expression domain in SDF1a-TP embryos (n = 33) compared with controls (n = 8 for SDF1a-ctrl-TP, n = 16 for uninjected wild type). Anterior–posterior lengths in microns correspond to the bracketed region in A and B. Student’s t test: P < 0.0001. (L) Quantification of TgSN positioning defects. Neurons adjacent or anterior to the MHB were defined as ectopic. Student’s t test: ***, P = 0.001; ****, P < 0.0001. See also Table 1. Bar, 100 µm. Data are shown as mean ± SEM (error bars).
Table 1.
Quantification of neuron positioning defects in SDF1a-TP– and SDF1a-ctrl-TP–injected embryos
| Genotype | Embryos with mispositioned neurons | Total embryos | Total mispositioned neurons | Severity | Penetrance |
| % | |||||
| Wild-type; SDF1a-ctrl-TP | 12 | 166 | 36 | 0.22 | 7.23 |
| Mean | 0.18 | ||||
| SD | 0.9 | ||||
| SEM | 0.06 | ||||
| Wild-type; SDF1a-TP | 41 | 84 | 242 | 2.88 | 48.81 |
| Mean | 2.88 | ||||
| SD | 4.83 | ||||
| SEM | 0.53 | ||||
| Wild-type; SDF1a-TP; SDF1a-AUG | 28 | 74 | 198 | 2.68 | 37.84 |
| Mean | 2.68 | ||||
| SD | 4.72 | ||||
| SEM | 0.55 | ||||
| cxcr7b−/+; SDF1a-ctrl-TP | 11 | 85 | 36 | 0.42 | 12.94 |
| Mean | 0.42 | ||||
| SD | 1.23 | ||||
| SEM | 0.13 | ||||
| cxcr7b−/+; SDF1a-TP | 130 | 193 | 1143 | 5.92 | 67.36 |
| Mean | 5.92 | ||||
| SD | 5.5 | ||||
| SEM | 0.4 | ||||
| cxcr7b−/+; SDF1a-TP; SDF1a-AUG | 71 | 112 | 400 | 3.57 | 63.39 |
| Mean | 3.57 | ||||
| SD | 4.05 | ||||
| SEM | 0.38 | ||||
| cxcr7b−/−; SDF1a-ctrl-TP | 31 | 31 | 506 | 16.32 | 100 |
| Mean | 16.32 | ||||
| SD | 7.05 | ||||
| SEM | 1.27 | ||||
| cxcr7b−/−; SDF1a-TP | 21 | 21 | 496 | 23.62 | 100 |
| Mean | 22.25 | ||||
| SD | 6.35 | ||||
| SEM | 1.35 |
Mispositioned neurons were defined as neurons anterior or adjacent to the MHB at the 10-somite stage. Severity is defined as the average number of mispositioned neurons per embryo. Penetrance is defined as the frequency of embryos with mispositioned neurons.
Cxcr7b is required for TgSN migration
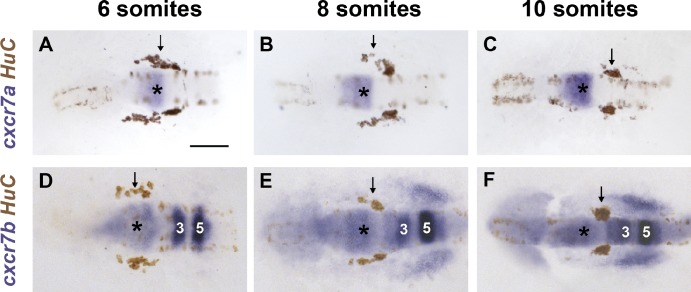
The requirement of miR-430–mediated SDF1a transcript clearance suggests that correct TgSN migration requires tight spatial and temporal control of chemokine distribution. Although miR-430 clears SDF1a transcripts from past sites of expression, it does not clear SDF1a protein that has already been translated and secreted. CXCR7, the second SDF1 receptor, has been reported to sequester SDF1 protein during germ cell and cortical interneuron migration (Boldajipour et al., 2008; Naumann et al., 2010; Sánchez-Alcañiz et al., 2011). This suggests that CXCR7 could aid miR-430 in refining SDF1a protein expression during TgSN migration. We analyzed the expression of the two zebrafish CXCR7 orthologues, cxcr7a and cxcr7b (Miyasaka et al., 2007), in relation to the TgSN marker HuC. Cxcr7a and cxcr7b are both expressed in the central nervous system during TgSN migration: cxcr7a is expressed in parts of the midbrain and the MHB (Fig. 4, A–C; and Fig. S2 A), whereas cxcr7b is expressed in the midbrain, MHB, and rhombomeres 3, 5, and 6 of the hindbrain (Fig. 4, D–F; and Fig. S2 B). Additionally, we observed low levels of ubiquitous cxcr7b expression in the tissues through which TgSNs migrate (Fig. 4, E and F). Thus, both cxcr7a and cxcr7b are expressed in the vicinity of migrating TgSNs. In contrast to cxcr4b, however, neither of the cxcr7 paralogues were detected in the TgSNs themselves (Fig. 4, arrows).
Figure 4.
cxcr7a and cxcr7b are expressed during TgSN migration. (A–C) cxcr7a (blue) and HuC (brown) mRNA distribution in 6-, 8-, and 10-somite-stage embryos. HuC stains TgSNs (arrows). (D–F) cxcr7b (blue) and HuC (brown) mRNA distribution in 6-, 8-, and 10-somite-stage embryos. Dorsal view, anterior to the left. MHB is labeled with asterisks. Hindbrain rhombomeres 3 and 5 are indicated by “3” and “5.” Bar, 100 µm. See also Fig. S2.
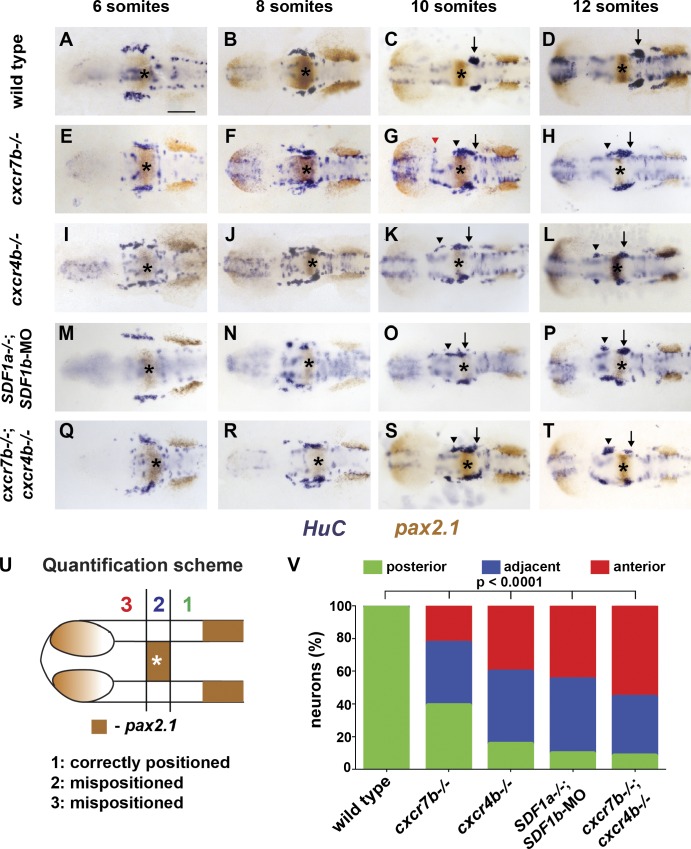
To test whether cxcr7a or cxcr7b is required for TgSN migration, we reduced Cxcr7a function using antisense morpholinos (Nasevicius and Ekker, 2000) and Cxcr7b function using a cxcr7b mutant allele, in which the seven-transmembrane receptor is truncated after its first transmembrane domain (Fig. S3, A and B; Busch-Nentwich et al., 2010). In cxcr7a morpholino-injected embryos, TgSN migration was indistinguishable from control wild-type embryos (not depicted; Fig. 5, A–D). In cxcr7b−/− embryos, however, TgSNs were born in a pattern indistinguishable from wild-type embryos (Fig. 5 E), but anterior-born neurons failed to migrate to join posterior-born neurons at the ganglion assembly site, resulting in stretched, disorganized clusters (Fig. 5, F–H). These neuronal clusters stretched from the normal ganglion assembly site (Fig. 5 G, black arrow) to positions anterior to the MHB (Fig. 5 G, black arrowhead), delineating the migratory route of the neurons. In some embryos, the stretched clusters were split, and some neurons were found adjacent to the eyes at the site where olfactory neurons assemble in response to SDF1a (Fig. 5 G, red arrowhead). Ectopic neurons persisted to later stages and extended axons (Fig. S4), which suggests that Cxcr7b activity is not required for neuron differentiation. These observations indicate that cxcr7b is expressed near TgSNs and is required for correct migration.
Figure 5.
TgSN migration is disrupted in embryos deficient in chemokine signaling. 6-, 8-, 10-, and 12-somite-stage embryos in A–T were stained with HuC (blue) and pax2.1 (brown) mRNA. MHB is marked with asterisks. Arrows and arrowheads denote correctly and incorrectly positioned TgSNs, respectively. The red arrowhead in G denotes mispositioned neurons located close to the eye. Dorsal view, anterior to the left. Bar, 100 µm. (A–D) Wild-type embryos. (E–H) cxcr7b−/− embryos. (I–L) cxcr4b−/− embryos. (M–P) SDF1a−/− embryos injected with SDF1b morpholino. (Q–T) cxcr7b−/−; cxcr4b−/− embryos. (U) Quantification of cell migration defects at the 12-somite stage. Zone 1 represents correctly positioned neurons located posterior to the MHB. Zones 2 and 3 represent mispositioned neurons located adjacent or anterior to the MHB, respectively. This approach was also used for quantification of neuron migration defects in Figs. 3, 6, 7, and 8. (V) Summary of the neuron positioning defects. The y axis corresponds to the percentage of neurons per embryo that are located in each zone. n ≥ 20 embryos for each genotype. Contingency table analysis using the χ2 test was applied to determine statistical significance. See also Table 2 and Figs. S3 and S4.
Cxcr7b and Cxcr4b mediate neuron migration in distinct ways
Depending on the context, CXCR7 has been suggested to mediate cell guidance directly (Balabanian et al., 2005; Valentin et al., 2007; Wang et al., 2011) or indirectly through clearance of SDF1 protein (Boldajipour et al., 2008; Naumann et al., 2010; Sánchez-Alcañiz et al., 2011). To distinguish between these two roles for Cxcr7b during TgSN migration, we first compared the cxcr4b and cxcr7b expression patterns. Although cxcr4b is expressed in the migrating neurons (Knaut et al., 2005), cxcr7b is expressed near the neurons and in the tissues the neurons traverse (Fig. 4, D–F). Second, we compared the distribution of TgSNs along the anterior-posterior axis in wild-type, cxcr7b−/−, cxcr4b−/−, and SDF1b morpholino–injected SDF1a−/− embryos (Fig. 5, A–P). For this comparison, the neurons were grouped into three categories based on the location of each neuron relative to the MHB: neurons located posterior to the MHB at the correct ganglion assembly site, neurons adjacent to the MHB, and neurons anterior to the MHB (Fig. 5, U and V; and Table 2; n ≥ 20 embryos). This analysis revealed that more TgSNs reached the ganglion assembly site in cxcr7b−/− embryos (Fig. 5, E–H) than in cxcr4b−/− embryos (Fig. 5, I–L) or SDF1b morpholino–injected SDF1a−/− embryos (Fig. 5, M–P), which suggests that Cxcr7b and Cxcr4b facilitate TgSN migration in distinct ways.
Table 2.
Quantification of neuron positioning defects in different chemokine signaling mutants
| Genotype | Position relative to MHB | n | |||
| Anterior | Adjacent | Posterior | Neurons | Embryos | |
| Wild type | 1 | 10 | 767 | 778 | 20 |
| mean | 0.05 | 0.5 | 38.35 | ||
| SD | 0.22 | 0.69 | 2.28 | ||
| SEM | 0.05 | 0.15 | 0.51 | ||
| cxcr7b−/− | 461 | 773 | 787 | 2,021 | 39 |
| mean | 11.82 | 19.82 | 20.18 | ||
| SD | 5.93 | 6 | 5.47 | ||
| SEM | 0.95 | 0.96 | 0.88 | ||
| cxcr4b−/− | 541 | 586 | 205 | 1,332 | 27 |
| mean | 20.04 | 21.7 | 7.59 | ||
| SD | 6.83 | 9.08 | 5.39 | ||
| SEM | 1.31 | 1.75 | 1.04 | ||
| cxcr7b−/−; cxcr4b−/− | 1,211 | 777 | 177 | 2,165 | 39 |
| mean | 31.05 | 19.92 | 4.54 | ||
| SD | 10.29 | 5.8 | 4.78 | ||
| SEM | 1.65 | 0.93 | 0.77 | ||
| SDF1a−/−; SDF1b-MO | 500 | 502 | 106 | 1,108 | 22 |
| mean | 22.73 | 22.82 | 4.82 | ||
| SD | 8.21 | 6.43 | 3.95 | ||
| SEM | 1.75 | 1.37 | 0.84 | ||
Neurons were categorized according to their position with respect to the MHB at the 12-somite stage as diagrammed in Fig. 5 U.
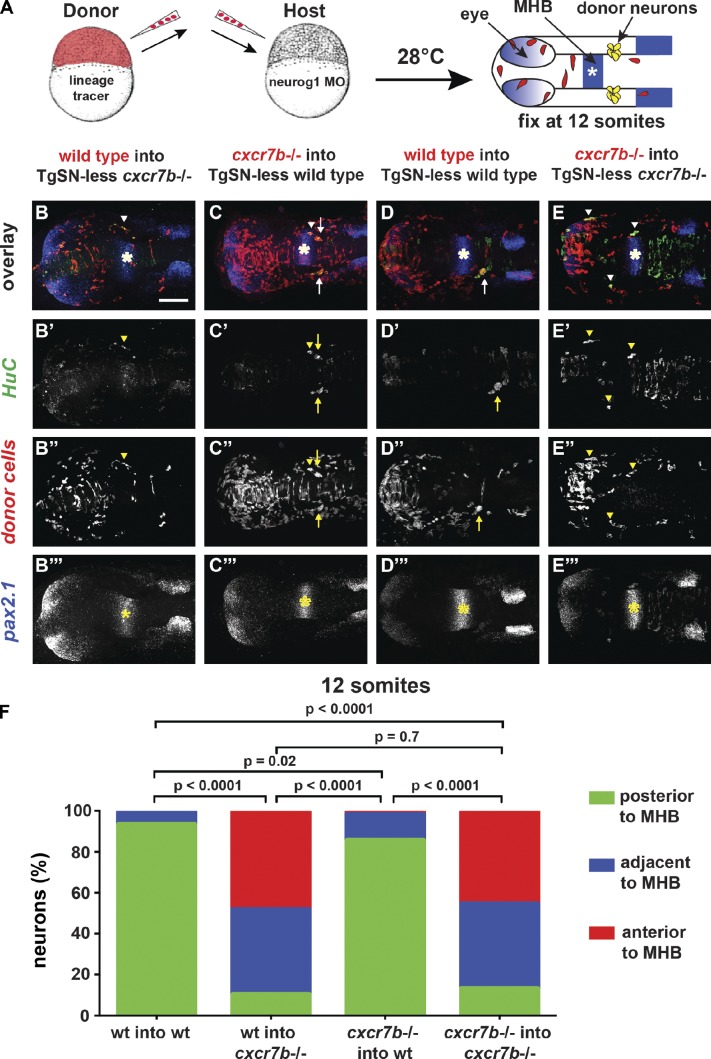
If Cxcr7b acts as an SDF1a protein clearance receptor, it should be required in the migration substrate. Conversely, if Cxcr7b acts as a cell guidance receptor, it should be required in the migrating neurons. To distinguish between these two possibilities, we generated genetic chimeras by cell transplantation and analyzed TgSN positioning at the 12-somite stage (15 hpf; Fig. 6 A). One caveat to this approach is that TgSNs interact and influence each other’s position (Knaut et al., 2005). This neuron–neuron interaction would confound cell autonomy analysis because one could not distinguish whether donor-derived neurons are located at a particular position because of their genotype or because of their interaction with host-derived neurons of a different genotype. To circumvent this problem, we blocked the development of TgSNs in host embryos by morpholino-mediated knockdown of the transcription factor neurog1, which is necessary for the specification of TgSNs in zebrafish (Andermann et al., 2002; Cornell and Eisen, 2002; Blader et al., 2003). This approach enabled us to study the behavior of donor-derived TgSNs in a host embryo devoid of endogenous TgSNs (TgSN-less host). Upon transplantation of wild-type cells into TgSN-less cxcr7b−/− hosts, most TgSNs were located at ectopic positions (90% ectopic; Fig. 6, B and F; and Table 3), similar to cxcr7b−/− neurons placed into TgSN-less cxcr7b−/− hosts (87% ectopic; Fig. 6, E and F; and Table 3). Conversely, most cxcr7b−/− TgSNs transplanted into TgSN-less wild-type hosts were located at the correct position (15% ectopic; Fig. 6, C and F; and Table 3), similar to wild-type neurons placed into TgSN-less wild-type hosts (7% ectopic; Fig. 6, D and F; and Table 3). These observations suggest that Cxcr7b primarily functions in the migration substrate.
Figure 6.
cxcr7b is required cell nonautonomously during TgSN migration. (A) Strategy for generating genetic chimeras. 12-somite-stage chimeric embryos were fluorescently stained for HuC (green, neurons), pax2.1 mRNA (blue, asterisks, MHB), and the lineage tracer (red). Arrows and arrowheads in B–E denote correctly positioned and mispositioned donor-derived TgSNs (yellow), respectively. (B–B‴) Wild-type donor cells in a cxcr7b−/− host. (C–C‴) cxcr7b−/− cells in a wild-type host. (D–D‴) Wild-type donor cells in a wild-type host. (E–E‴) cxcr7b−/− cells in a cxcr7b−/− host. Bar, 100 µm. (F) Summary of TgSN positioning. The y axis corresponds to the percentage of neurons per embryo that are located in each zone as diagrammed in Fig. 5 U. Transplanted neurons ≥121 for each scenario. A contingency table analysis using the χ2 test was applied to determine statistical significance. Dorsal view, anterior to the left in A–E‴. See also Table 3.
Table 3.
Quantification of neuron positioning defects in genetic chimeras composed of wild-type and cxcr7b−/− embryos
| Genotype | Position relative to MHB | n (neurons) | Ectopic | ||
| Anterior (ectopic) | Adjacent (ectopic) | Posterior (correct) | |||
| % | |||||
| Wild type into TgSN-less wild type | 0 | 11 | 154 | 165 | 6.70 |
| Wild-type into TgSN-less cxcr7b−/− | 94 | 81 | 20 | 195 | 89.70 |
| cxcr7b−/− into TgSN-less wild type | 7 | 48 | 328 | 383 | 14.40 |
| cxcr7b−/− into TgSN-less cxcr7b−/− | 55 | 50 | 16 | 121 | 86.80 |
Neurons were categorized according to their position with respect to the MHB at the 12-somite stage as diagrammed in Fig. 5 U.
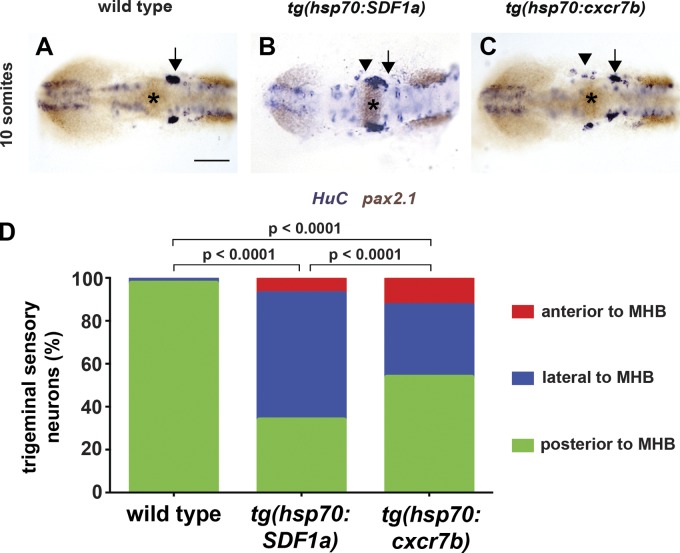
If TgSN migration is perturbed in cxcr7b−/− embryos because SDF1a protein is not inactivated or cleared, then increasing SDF1a protein levels in wild-type embryos should perturb neuron migration in a manner similar to what is observed in cxcr7b−/− embryos. Conversely, increasing Cxcr7b protein levels in wild-type embryos should decrease the levels of SDF1a protein and perturb neuron migration in a manner similar to what is observed in cxcr4b−/− embryos. To test the first prediction, we expressed SDF1a under the control of a heat shock promoter (tg(hsp70:SDF1a)) during neuron migration. In such embryos, TgSN distribution was similar to what we observed in cxcr7b−/− embryos (Fig. 7, B and D; and Fig. 5 G) but distinct from cxcr4b−/− embryos (Fig. 5 K). TgSN migration was rarely affected in heat-shocked wild-type embryos (Fig. 7, A and D; and Table 4). To test the second prediction, we expressed cxcr7b under the control of a heat shock promoter (tg(hsp70:cxcr7b)) during neuron migration. Such embryos show a TgSN migration defect (Fig. 7, C and D; and Table 4), but this defect is not as severe as the defect observed in embryos that lack cxcr4b or SDF1a and SDF1b function (Fig. 5, K and O; and Table 2).
Figure 7.
Cxcr7b activity perturbs TgSN migration. (A) Heat-shocked 10-somite-stage wild-type embryo stained for HuC (blue, neurons) and pax2.1 (brown). Asterisks, MHB. (B) Similarly treated tg(hsp70:cxcr7b) transgenic embryo. (C) Similarly treated tg(hsp70:SDF1a) transgenic embryo. Bar, 100 µm. (D) Summary of neuron positioning depicted as in Fig 6. n ≥ 20 embryos for each genotype. Contingency table analysis using the χ2 test was applied to determine statistical significance. Dorsal view, anterior to the left. Arrows and arrowheads denote correctly positioned and mispositioned TgSNs, respectively. See also Table 4.
Table 4.
Quantification of neuron positioning defects in embryos overexpressing SDF1a or Cxcr7b
| Genotype | Position relative to MHB | n | |||
| Anterior | Adjacent | Posterior | Neurons | Embryos | |
| Wild type, heat shocked | 3 | 18 | 830 | 851 | 20 |
| Mean | 0.15 | 0.9 | 41.5 | ||
| SD | 0.49 | 1.21 | 3.12 | ||
| SEM | 0.11 | 0.27 | 0.7 | ||
| hsp70:SDF1a | 102 | 813 | 467 | 1,382 | 27 |
| Mean | 3.78 | 30.11 | 17.3 | ||
| SD | 3.31 | 8.91 | 13.16 | ||
| SEM | 0.64 | 1.71 | 2.53 | ||
| hsp70:cxcr7b | 113 | 293 | 471 | 877 | 22 |
| Mean | 5.14 | 13.32 | 21.41 | ||
| SD | 3.89 | 5.34 | 8.75 | ||
| SEM | 0.18 | 0.24 | 0.4 | ||
Neurons were categorized according to their position with respect to the MHB at the 12-somite stage as diagrammed in Fig. 5 U.
Next, we compared TgSN positioning in cxcr7b−/−; cxcr4b−/− embryos (Fig. 5 Q-T) to cxcr7b−/− and cxcr4b−/− embryos. The migration defect at 12 somites in double mutant embryos (Fig. 5 T) resembled that of cxcr4b−/− embryos (Fig. 5 L) rather than cxcr7b−/− embryos (Fig. 5 H), with TgSNs frequently forming ectopic clusters anterior to the MHB, and relatively few neurons located at the ganglion assembly site (Fig. 5 T and Table 2). Although quantification suggests that the loss of Cxcr4b and Cxcr7b leads to a slightly enhanced migration defect (Fig. 5 V), this observation is consistent with the idea that both receptors function primarily in the same pathway.
Cxcr7b activity is required for TgSNs to follow a shifting SDF1 expression domain
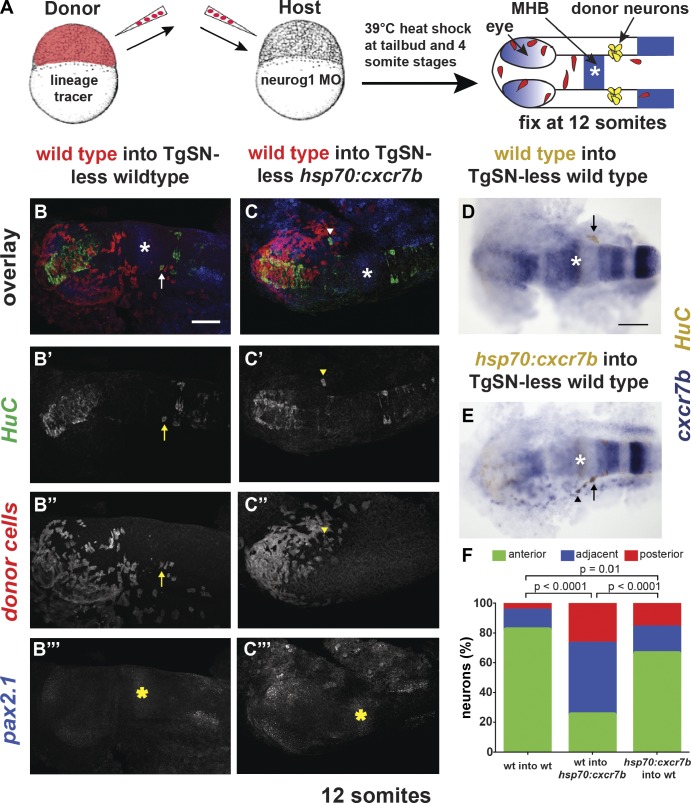
To determine if elevated Cxcr7b levels outside of the neurons perturb migration, we generated genetic chimeras composed of wild-type and tg(hsp70:cxcr7b) cells and induced cxcr7b expression during neuron migration (Fig. 8 A). Analysis with the neuronal marker HuC and the MHB marker pax2.1 revealed that the majority of wild-type TgSNs failed to reach the ganglion assembly site in host embryos that overexpress cxcr7b upon heat shock (75% ectopic; Fig. 8, C and F; and Table 5), whereas most wild-type neurons reached the assembly site in heat-shocked, nontransgenic hosts (18% ectopic; Fig. 8, B, D, and F; and Table 5). In the converse experiment, we analyzed the distribution of tg(hsp70:cxcr7b) transgenic neurons in TgSN-less wild-type embryos. In this scenario, most neurons reached the assembly site (34% ectopic; Fig. 8, E and F; and Table 5). Injection of wild-type cxcr7b mRNA restored TgSN migration in cxcr7b−/− embryos (Fig. S3, C–I; and Table 6). These observations are consistent with the idea that too much Cxcr7b outside of the neurons lowers active SDF1 protein levels and causes TgSNs to lose their association with the shifting chemokine expression domain.
Figure 8.
Misexpression of Cxcr7b in the tissue surrounding migrating neurons perturbs neuron migration. (A) Strategy for generating genetic chimeras. 12-somite-stage embryos were stained for HuC (green in B and C and brown in D and E; neurons) and pax2.1 mRNA (blue in B and C and brown in D and E). MHB is marked with asterisks. Donor cells overexpressing cxcr7b (blue) in wild-type hosts were detected with a cxcr7b antisense RNA probe. (B–B‴) Wild-type donor cells in a wild-type host. Arrows and arrowheads denote correctly positioned and mispositioned donor-derived TgSNs, respectively, in B–E. (C–C‴) Wild-type donor cells in a tg(hsp70:cxcr7b) host. (D) Wild-type donor cells in a wild-type host. (E) Tg(hsp70:cxcr7b) cells in a wild-type host. (F) Summary of TgSN positioning as depicted in Fig 6. n ≥ 40 transplanted neurons for each scenario. Contingency table analysis using the χ2 test was applied to determine statistical significance. Dorsal view, anterior to the left. Bars, 100 µm. See also Table 5.
Table 5.
Quantification of neuron positioning defects in genetic chimeras composed of wild-type and hsp70:cxcr7b cells
| Genotype | Position relative to MHB | n (neurons) | ectopic | ||
| Anterior(ectopic) | Adjacent(ectopic) | Posterior(correct) | |||
| % | |||||
| Wild type into TgSN-less wild type | 6 | 14 | 92 | 112 | 17.90 |
| Wild type into TgSN-less hsp70:cxcr7b | 11 | 19 | 10 | 40 | 75.00 |
| hsp70:cxcr7b into TgSN-less wild type | 19 | 20 | 76 | 115 | 33.90 |
Neurons were categorized according to their position with respect to the MHB at the 12-somite stage as diagrammed in Fig. 5 U.
Table 6.
Quantification of neuron positioning defects in cxcr7b−/− embryos rescued by cxcr7b mRNA injection
| Genotype | cxcr7b mRNA dose | Mispositioned neurons per embryo | n (embryos) |
| cxcr7b−/−, pair no. 1, 10 somites | Uninjected | 26.3 | 9 |
| cxcr7b−/−, pair no. 1, 10 somites | 150 pg | 4.8 | 12 |
| cxcr7b−/−, pair no. 2, 10 somites | Uninjected | 25.3 | 17 |
| cxcr7b−/−, pair no. 2, 10 somites | 150 pg | 8.2 | 23 |
| cxcr7b−/−, pair no. 3, 12 somites | Uninjected | 18.9 | 14 |
| cxcr7b−/−, pair no. 3, 12 somites | 150 pg | 4.4 | 15 |
Neurons were categorized according to their position with respect to the MHB at the 10- or 12-somite stage. Mispositioned neurons were defined as neurons anterior or adjacent to the MHB. This table is related to Fig. S3.
miR-430 and Cxcr7b act synergistically during neuron migration
If SDF1a availability is modulated on a transcript level by miR-430 and on a protein level by Cxcr7b, miR-430 and Cxcr7b should synergize in guiding TgSNs to the ganglion assembly site. We thus asked if disrupting miR-430 regulation of SDF1a transcripts in embryos with reduced (cxcr7b−/+) or absent (cxcr7b−/−) Cxcr7b activity makes TgSNs more susceptible to misdirection. First, we found that more TgSNs were mislocalized in cxcr7b−/+ embryos injected with SDF1a-TP than in wild-type embryos injected with SDF1a-TP (Fig. 3, C, E, and L; and Table 1). Neuron migration was not affected in cxcr7b−/+ embryos injected with SDF1a-ctrl-TP (Fig. 3 F). Moreover, lowering the levels of SDF1a protein in cxcr7b−/+; SDF1a-TP embryos by injecting suboptimal amounts of a translation-blocking SDF1a morpholino (SDF1a-AUG) improved neuron migration (Fig. 3, G and L; and Table 1), which is consistent with the idea that SDF1a levels are too high in cxcr7b−/+; SDF1a-TP embryos. Second, we found that neuron migration is more severely affected in cxcr7b−/−; SDF1a-TP embryos than in cxcr7b−/−; SDF1a-ctrl-TP embryos (Fig. 3, I, J, and L; and Table 1). Importantly, Cxcr7b activity is not required for the refinement of SDF1a mRNA expression (unpublished data). These genetic interactions support the idea that SDF1a transcript clearance through miR-430 and SDF1a protein inactivation through Cxcr7b synergize to ensure precise cell guidance by a dynamic chemokine expression domain.
miR-430 and Cxcr7b modulate SDF1-Cxcr4b signaling levels along the migratory route
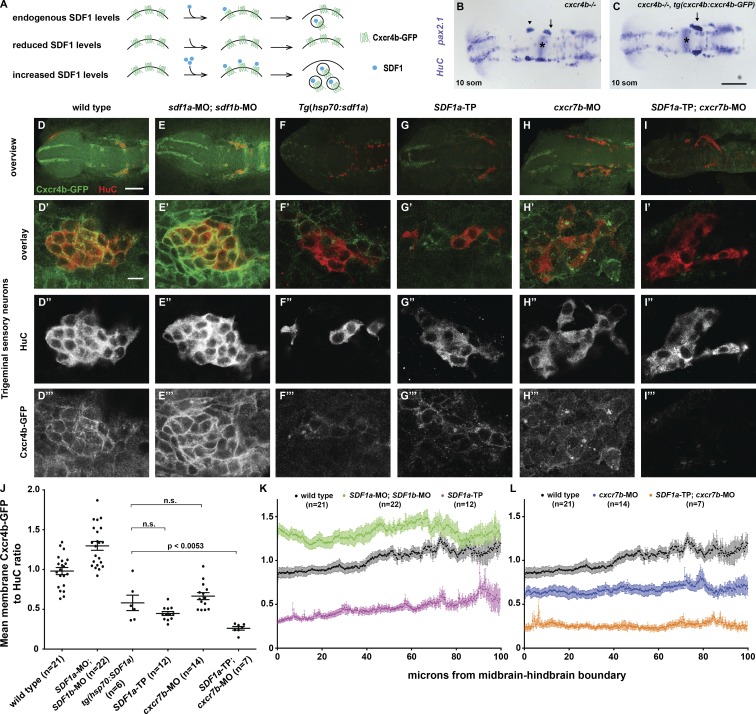
Our genetic analysis suggests that miR-430 and Cxcr7b refine SDF1a protein expression during TgSN migration. To test this idea more directly, we developed a tool to measure SDF1a-Cxcr4b signaling in the neurons. Because SDF1 triggers CXCR4 internalization and degradation (Marchese and Benovic, 2001; Marchese et al., 2003; Minina et al., 2007), measuring the degree of CXCR4 internalization should be a readout of the levels of extracellular SDF1 (Fig. 9 A). Based on this concept, we created transgenic zebrafish in which the cxcr4b promoter drives Cxcr4b-GFP expression from a 69-kb genomic DNA fragment (tg(cxcr4b:cxcr4b-GFP)). This transgene drives Cxcr4b-GFP expression in the TgSNs (Fig. 9 D) and restores neuron migration in cxcr4b mutant embryos (Fig. 9, B and C). To assess whether Cxcr4b-GFP internalization correlates with the levels of extracellular SDF1a, we measured the neuronal uptake of Cxcr4b-GFP in embryos with reduced or increased SDF1 levels by immunofluorescence against GFP and the neuronal marker HuC. Cxcr4b-GFP internalization was measured by normalizing the Cxcr4b-GFP fluorescence to the HuC fluorescence on the membranes of the neurons using ImageJ-based automated image analysis. Cxcr4b-GFP/HuC ratios were then normalized to ratios from wild-type or heat-shocked wild-type control embryos to reflect the fold change (see Materials and methods). Tg(cxcr4b:cxcr4b-GFP) embryos injected with morpholinos against SDF1a and SDF1b displayed higher Cxcr4b-GFP/HuC ratios on the membrane (Fig. 9, E and J; 1.3 ± 0.06) than wild-type neurons (Fig. 9, D and J; 1.0 ± 0.04). Conversely, in heat-shocked tg(cxcr4b:cxcr4b-GFP); tg(hsp70:SDF1a) embryos, about half as much Cxcr4b-GFP was found on the membrane (Fig. 9, F and J; 0.6 ± 0.1) compared with heat-shocked control embryos. This suggests that Cxcr4b-GFP internalization correlates with SDF1 protein activity.
Figure 9.
TgSNs are exposed to elevated SDF1 protein levels when Cxcr7b or miR-430 activity is reduced. (A) Conceptual overview of quantification of SDF1-Cxcr4b signaling. (B) 12-somite cxcr4b−/− nontransgenic sibling embryo stained in blue for HuC and pax2.1 mRNA to visualize TgSNs in relation to the MHB (asterisks). (C) 12-somite cxcr4b −/−; tg(cxcr4b:cxcr4b-GFP) sibling embryo. Arrows and arrowhead denote correctly and incorrectly positioned TgSNs, respectively. (D–I) Overview of a representative embryo of the indicated genotype stained for TgSNs with HuC (red) and for Cxcr4b-GFP (green). (D′–I‴) High magnification of embryos stained as indicated in D–I. (D′–I′) Overlay of Cxcr4b-GFP and HuC Fluorescence. (D″–I″) HuC fluorescence. (D‴–I‴) Cxcr4b-GFP fluorescence. All embryos are eight somites. (D–D‴) Wild-type embryo. (E–E‴) SDF1a and SDF1b morpholino–injected embryo. (F–F‴) Heat-shocked tg(hsp70:SDF1a) embryo. (G–G‴) SDF1a-TP embryo. (H–H‴) cxcr7b morpholino–injected embryo. (I–I‴) SDF1a-TP and cxcr7b morpholino–injected embryo. (J) Mean Cxcr4b-GFP/HuC ratios ± SEM on the outer membranes of TgSNs in indicated genetic scenarios at the eight-somite stage. All differences between the different genetic scenarios are statistically significant, with P < 0.0001, except where noted otherwise. n.s., not significant. (K–L) Mean Cxcr4b-GFP/HuC ratios ± SEM on the outer membranes of TgSNs along the migration route in the indicated genetic scenarios at the eight-somite stage. Because of the neuron dispersal in heat-shocked tg(hsp70:SDF1a) embryos, quantification could not be resolved along the migratory route. 0 µm corresponds to the MHB. Bars: (B and C) 100 µm; (D–I) 100 µm; (D′–I‴) 10 µm.
Using Cxcr4b-GFP internalization as a readout, we asked if TgSNs encounter higher SDF1 protein levels in embryos lacking miR-430–mediated SDF1a mRNA clearance or Cxcr7b activity. In tg(cxcr4b:cxcr4b-GFP) embryos injected with SDF1a-TP or a morpholino against cxcr7b, we observed very little Cxcr4b-GFP on the membrane relative to SDF1a-ctrl-TP–injected or wild-type embryos (Fig. 9, G, H, and J; 0.4 ± 0.02 and 0.7 ± 0.04, respectively). Co-injection of SDF1a-TP and cxcr7b morpholinos into tg(cxcr4b:cxcr4b-GFP) embryos enhanced the degree of Cxcr4b-GFP internalization (Fig. 9, I and J; 0.3 ± 0.02). Overexpression of Cxcr7b from a heat shock promoter had no discernable effect on the Cxcr4b-GFP/HuC ratio on the membrane (not depicted). These observations corroborate the idea that mir-430 and Cxcr7b synergize to regulate the activity of SDF1a.
To assess whether miR-430 and Cxcr7b modulate the SDF1a protein distribution, we quantified the Cxcr4b-GFP/HuC ratio on the membrane of the TgSNs at the eight-somite stage when the neurons are still dispersed along their migratory route. In wild-type embryos, the Cxcr4b-GFP/HuC ratio on the membranes of neurons furthest away from the ganglion assembly site was close to the Cxcr4b-GFP/HuC ratio on neurons in embryos injected with morpholinos against SDF1a and SDF1b, and lower in neurons located closer to the ganglion assembly site (Fig. 9 K). In SDF1a TP embryos, the Cxcr4b-GFP/HuC ratio in the neurons along the migratory route was uniformly lowered but remained higher in neurons further away from the ganglion assembly site than in neurons closer to the assembly site (Fig. 9 K). Similarly, in cxcr7b morpholino-injected embryos, the Cxcr4b-GFP/HuC ratios were decreased along the migratory route (Fig. 9 L). However, in contrast to SDF1a TP embryos, there was no apparent difference in the Cxcr4b-GFP/HuC ratios between neurons close to or further away from the ganglion assembly site. Reducing the activity of both miR-430 and Cxcr7b lowered the Cxcr4b-GFP/HuC ratios further (Fig. 9 L). Because low Cxcr4b-GFP/HuC ratios correlate with high SDF1 activity and high ratios correlate with low SDF1 activity, these observations are consistent with the ideas that TgSNs are exposed to a graded distribution of SDF1 activity along their migratory route, and that SDF1 availability is sculpted through the combined activities of miR-430 and Cxcr7b.
Discussion
We used zebrafish TgSN migration as a model system to understand how one cue can simultaneously guide multiple populations of cells to distinct locations during development. Our results suggest an “attractive path” model in which a dynamic domain of SDF1a expression that is closely associated with the TgSNs leads these cells to their target and prevents them from being attracted to inappropriate sources of SDF1a. This close association between the expression domain of the guidance cue and the neurons is ensured through two synergistic mechanisms that refine the dynamic SDF1a expression domain (Fig. 10). First, the miRNA miR-430 eliminates persisting SDF1a transcripts from tissues that no longer actively transcribe the chemokine. Concurrently, the atypical chemokine receptor Cxcr7b inactivates, possibly through chemokine clearance, SDF1a protein from tissues that no longer express the chemokine. It is conceivable that these mechanisms tailor the expression domains of other shared cues in the nervous system, such that migrating neurons follow the correct source of an attractant and are not misguided by other nearby sources of the same attractant.
Figure 10.
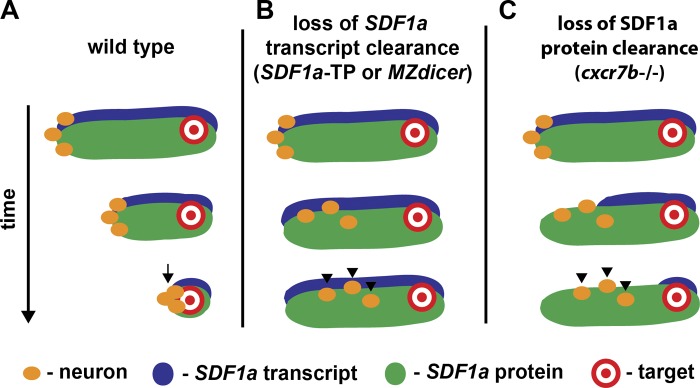
An attractive path leads neurons to their destination.
Dynamic SDF1a expression paves an attractive path for neurons that is refined through Cxcr7b and miR-430
TgSNs are born amid multiple SDF1 expression domains and need to follow the correct one to reach their destination. Our analysis shows that TgSNs closely follow a dynamic SDF1a expression domain. This domain is closely associated with TgSNs during migration, and its anterior border shifts posteriorly toward the future site of ganglion assembly as migration proceeds. When migration is complete, the SDF1a mRNA expression domain is restricted to a compact patch slightly posterior to the assembled ganglion. These observations suggest that the close association of the migrating TgSNs with a shifting source of attractant ensures that the neurons follow the correct SDF1a expression domain and ignore the others. To create an attractive path, SDF1a protein needs to be inactivated at sites of expression that the neurons have passed. Without inactivation, extracellular pools of active chemokine would persist at sites that no longer express SDF1a, resulting in misdirection of migrating neurons. Our observations suggest that the tight control of SDF1a expression is achieved via SDF1a transcript and protein inactivation by miR-430 and Cxcr7b, respectively. First, the SDF1a mRNA expression domain fails to refine toward the ganglion assembly site after either the collective loss of most miRNAs or the specific loss of miR430-mediated regulation of SDF1a transcripts, resulting in displacement of neurons along the migratory route. Second, disruption of Cxcr7b activity prevents the TgSNs from assembling at the appropriate site. Instead, they are found along the migratory route, a defect that is also found in embryos with elevated levels of SDF1a. Although reducing Cxcr7a function did not perturb TgSN migration, Cxcr7a may play a redundant role in this context. Third, removing cxcr7b function in cxcr4b-deficient embryos only mildly enhances the neuronal migration defect seen in cxcr4b mutants, which suggests that cxcr7b and cxcr4b act primarily in the same pathway. Fourth, Cxcr7b activity is required in the tissues the neurons traverse rather than in the neurons themselves. Fifth, increased Cxcr7b activity in the migration substrate impairs neuronal migration. Sixth, SDF1a-Cxcr4b signaling in the TgSNs is enhanced in the absence of miR-430–mediated SDF1a mRNA clearance and in cxcr7b mutant embryos. Together, these observations suggest that miR-430 and Cxcr7b clear SDF1a mRNA and inactivate SDF1a protein, possibly through chemokine clearance, from sites that no longer actively transcribe the SDF1a gene. Without these two mechanisms, active SDF1a inappropriately persists in tissues that have ceased transcription of SDF1a. Such persistence could cause normally separate SDF1a expression domains to overlap, putting migrating cells at risk of following the incorrect path. Indeed, in cxcr7b mutant and MZdicer mutant embryos, TgSNs are sometimes found close to the eyes (red arrowhead in Fig. 5 G and Fig. 2 E, respectively), where a different SDF1a expression domain assembles cxcr4b-expressing neurons into the olfactory placode (Miyasaka et al., 2007).
Other SDF1-guided cells also follow a dynamic chemokine expression domain similar to the one we observed for TgSNs. This suggests that the cells in these cases may also rely on an attractive path for guidance. For example, dynamic expression of SDF1 seems to underlie the clustering of olfactory neurons (Miyasaka et al., 2007), the assembly of sensory neurons into dorsal root ganglia (Belmadani et al., 2005), and the positioning of primordial germ cells (Doitsidou et al., 2002; Knaut et al., 2003). In each context, the migrating cells express the receptor CXCR4 and closely follow a refining SDF1 expression domain. However, other SDF1-guided migration events seem to use a different mechanism. For example, Cajal-Retzius cells and interneurons of the cortex migrate over a constant SDF1 expression domain that does not refine (Daniel et al., 2005; Tiveron et al., 2006; Stumm et al., 2007; Li et al., 2008; López-Bendito et al., 2008; Sánchez-Alcañiz et al., 2011; Wang et al., 2011). In these cases, it is thought that SDF1 acts as a retention signal that restricts the dispersal of the migrating neurons to the chemokine expression domain.
Intriguingly, germ cells and cortical interneurons also require CXCR7 for their migration (Boldajipour et al., 2008; Sánchez-Alcañiz et al., 2011; Wang et al., 2011). In both cases, CXCR7 activity is required outside the migrating cells, although interneurons also require CXCR7 cell autonomously (Wang et al., 2011). Although CXCR7 is necessary to clear SDF1 protein from the tissues where it is not needed during primordial germ cell migration (Boldajipour et al., 2008), its role during interneuron migration appears to involve both SDF1 protein clearance (Sánchez-Alcañiz et al., 2011) and intracellular signaling (Wang et al., 2011). During germ cell migration, CXCR7-mediated chemokine clearance is thought to sharpen the SDF1 gradient. In contrast, it seems that interneurons use CXCR7 to lower the ambient concentrations of SDF1 to levels that are suitable for CXCR4-mediated chemotaxis; in the absence of CXCR7, the ambient SDF1 concentration becomes high enough to induce complete internalization of CXCR4, impairing the interneurons’ ability to respond to SDF1 (Sánchez-Alcañiz et al., 2011).
Although the regulation of SDF1 levels through CXCR7 is similar in germ cell, TgSN, and interneuron migration, there are also interesting differences in how CXCR7 contributes to these three migration events. In the absence of CXCR7, cortical interneurons in the marginal zone or intermediate/subventricular zones prematurely exit a constant stripe of SDF1 expression, causing them to enter the cortical plate, a prominent site of CXCR7 expression, too early. Primordial germ cells and TgSNs similarly fail to stay closely associated with their SDF1 expression domains in cxcr7b mutant embryos, but the reason for this failure seems to be different. Whereas cortical interneurons in CXCR7 mutants appear to migrate actively away from an SDF1 expression domain, primordial germ cells and TgSNs in cxcr7b mutants seem to fall off of a shifting SDF1a expression domain. This distinction follows from the contrasting rationales for non-cell-autonomous CXCR7 activity in these contexts. Interneurons require CXCR7 for retention in a specific location, whereas primordial germ cells and TgSNs require Cxcr7b to follow a shifting expression domain without being misdirected by former sites of expression or domains intended for other SDF1-responsive cells.
Our observation that SDF1a transcript clearance through the ubiquitously expressed miR-430 (Giraldez et al., 2006) contributes to correct TgSN migration suggests that miRNA regulation might also ensure precision in other chemokine-guided cell migration events. In the contexts discussed in the previous paragraphs, SDF1 is expressed either dynamically or in a constant domain. In both cases, transcript availability needs to rapidly reflect changes in transcriptional activity at the SDF1 locus. For instance, removal of SDF1 transcript from past sites of expression can make a dynamic expression domain more precise. A prominent example of dynamic chemokine expression is the SDF1a domain that guides primordial germ cells in zebrafish (Doitsidou et al., 2002; Knaut et al., 2003). Similar to TgSNs, the refinement of this expression domain requires miR-430 activity (Staton et al., 2011). However, in contrast to TgSNs, primordial germ cells seem to require miR-430 activity to be retained at their target, as blocking miR-430 regulation of SDF1a transcripts causes germ cells to migrate past their destination (Staton et al., 2011).
Although it is conceptually clear how transcript clearance could help shape dynamic expression domains, it is less clear how transcript clearance could contribute to constant expression domains. In principle, cells neighboring constant SDF1 expression domains might express low levels of SDF1 because of leaky promoters or shared progenitors. In this case, clearance of SDF1 transcripts could contribute to the refinement of the chemokine expression domain. In the cortex, for example, the cortical plate does not express SDF1, but it is sandwiched between two SDF1-expressing layers. Cajal-Retzius cells and interneurons rely on the expression of SDF1 in these layers for proper migration (Stumm et al., 2003; Borrell and Marín, 2006; Paredes et al., 2006). Thus, it is intriguing to speculate that miRNA-mediated clearance of SDF1 transcripts in the cortical plate may confine SDF1 expression to the marginal zone and intermediate/subventricular zones to prevent erroneous neuronal guidance. Indeed, in silico and in vitro approaches have shown that mammalian SDF1 transcripts contain miRNA target sites and are regulated by miRNAs (Pillai et al., 2010; Staton et al., 2011).
In summary, our observations suggest that TgSNs are guided to the ganglion assembly site through their close association with the dynamic expression domain of the attractant SDF1. This process requires the clearance of SDF1a transcripts and the inactivation of SDF1a protein by miR-430 and Cxcr7b, respectively, from past sites of expression to ensure that subtle changes in transcriptional activity are precisely reflected at the protein level. This combination of dynamic gene expression, transcript clearance, and protein inactivation paves an attractive path of SDF1 protein (Fig. 10), ensuring that TgSNs follow the correct SDF1 source. Using such an attractive path mechanism allows the animal to use a shared guidance cue to guide different cells to different positions, a strategy that may be used by other shared guidance cues involved in organizing the developing embryo.
Materials and methods
Zebrafish strains
Embryos were staged as described previously (Kimmel et al., 1995). In brief, for stages in which somites were used as the unit of measurement for developmental age, embryos were examined with a dissecting microscope (Stemi SV11; Carl Zeiss), and the number of somites was counted based on either a lateral or dorsal view. For stages in which hours postfertilization was used as the unit of measurement for developmental age, one-cell-stage embryos (0 hpf) were incubated at 28.5°C until the indicated number of hours postfertilization had elapsed. Embryos containing mutant alleles of cxcr7b, cxcr4b, SDF1a, and MZdicer were generated by inbreeding homozygous adults to obtain 100% homozygous mutant embryos or by inbreeding heterozygous adults to obtain 25% wild-type, 50% heterozygous, and 25% homozygous mutant embryos for synchronized development. In the latter case, heterozygous and homozygous mutant embryos were identified by PCR-based amplification of the mutant locus followed by sequencing. cxcr4b; cxcr7b double mutant embryos were generated by inbreeding mutant embryos that were homozygous for both mutations. The cxcr7bsa16 allele contains a nonsense mutation that results in a premature stop at codon 76 (Busch-Nentwich et al., 2010). The cxcr4bt26035 allele contains a nonsense mutation that results in a premature stop at codon 239 (Knaut et al., 2003). The SDF1at30516 allele contains a nonsense mutation that results in a premature stop at codon 33 (Valentin et al., 2007). The dicerhu715 allele contains a nonsense mutation that results in a premature stop at codon 1427 (Wienholds et al., 2003). MZdicer mutants were generated through germ line replacement (Ciruna et al., 2002) and have been described previously (Giraldez et al., 2005). Transgenic zebrafish carrying the zebrafish heat-shock promoter (Halloran et al., 2000) driving SDF1a or Cxcr7b expression have been previously described (Knaut et al., 2005) or were generated in this study, respectively. The hsp70:SDF1a transgene contains a genomic fragment spanning 1.5 kb upstream of the hsp70 start codon (Halloran et al., 2000) fused to the coding sequence of SDF1a followed by the SV40pA signal.
Generation of transgenic animals
For the hsp70:cxcr7b transgene, a genomic fragment spanning 1.5 kb upstream of the hsp70 start codon (Halloran et al., 2000) was fused to the coding sequence of cxcr7b followed by the SV40pA signal and cloned into a vector that contains I-SceI sites (Thermes et al., 2002). For transgenesis, 25 ng/µl of this construct was coinjected with the I-SceI enzyme (New England Biolabs, Inc.) into one-cell-stage embryos. Transgenic fish were identified by in situ hybridization against cxcr7b after a 20 min heat shock at 37°C. For the cxcr4b:cxcr4b-GFP transgene, the bacterial artificial chromosome (BAC) clone DKEY-169F10 was modified in two ways by recombineering (Warming et al., 2005). First, the Tol2 (exon 4)-FRT-GalK-FRT-Tol2 (exon 1)-α-Crystallin-dsRed cassette was inserted into the BAC, replacing nucleotides 729–760 of its pIndigo-356 backbone using GalK as a selection marker. GalK was removed by Flippase-mediated recombination. The arms of homology were 320-bp fragments corresponding to nucleotides 409–728 and 761–1,080 of the pIndigo-356 backbone, respectively. Second, a cassette consisting of EGFP, an internal ribosome entry site (IRES) from the encephalomyocarditis virus, and kate2-CaaX followed by FRT-kanamycin-FRT flanked by 1,457 bp and 812 bp of homology upstream and downstream of the cxcr4b stop codon, respectively, was inserted between the last amino acid and the stop codon of cxcr4b using the kanamycin resistance gene as a selection marker. The kanamycin resistance gene was removed by Flippase-mediated recombination. The final BAC was characterized by restriction digest, PCR amplification, and BAC-end sequencing. It was then purified with the nucleobond BAC 100 kit (Takara Bio Inc.) and coinjected with tol2 transposase mRNA into one-cell-stage zebrafish embryos. The full name of this transgenic line is tg(cxcr4b:cxcr4b-EGFP-IRES-kate2-CaaX)p1.
Whole-mount in situ hybridization and antibody staining
Preparation of RNA probes and in situ hybridization were performed as described previously (Ober and Schulte-Merker, 1999). For nonfluorescent double in situ hybridizations, RNA probes against cxcr7a, cxcr7b, cxcr4b, SDF1a, SDF1b, HuC, and krox20 were labeled with DIG (Roche), and RNA probes against pax2.1 and HuC were labeled with DNP (Mirus). Probes were detected with anti–DIG-AP antibody (1:5,000; Roche) and NBT/BCIP (Roche) or anti-DNP-HRP antibody (1:1,000; PerkinElmer) with tyramide signal amplification (TSA) Plus DNP (HRP) amplification (PerkinElmer) and the DAB peroxidase substrate kit (Vector Laboratories), respectively. For fluorescent in situ hybridization, DIG-labeled probes were detected with anti–DIG-HRP antibody (1:1,000; Roche) and TSA FITC (PerkinElmer), and DNP labeled probes were detected with anti–DNP-HRP (1:1,000; PerkinElmer) antibody with TSA DNP amplification and TSA Cy3 (PerkinElmer). Antibody staining against HNK-1 (zn-12; Developmental Studies Hybridoma Bank; 1:1,000) was performed as described previously (Trevarrow et al., 1990). In brief, embryos were fixed overnight at 4°C in 4% PFA (Sigma-Aldrich), dehydrated in 100% methanol for at least 1 h at −20°C, rehydrated in PBS containing 0.1% Tween-20 (PBS-T; Sigma-Aldrich), permeabilized in a 1:1,000 dilution of proteinase-K (Sigma-Aldrich) in PBS-T for 5 min, blocked for 1 h in a solution of PBS-T containing 2% BSA (B-PBS-T; Sigma-Aldrich), incubated overnight at 4°C in a 1:1,000 dilution of HNK-1 antibody in B-PBS-T, washed four times for 20 min with PBS-T, incubated overnight at 4°C in a 1:2,000 dilution of biotinylated anti–mouse IgG (Vector Laboratories), washed four times for 20 min with PBS-T, incubated for 2 h at room temperature with a solution of avidin/biotinylated enzyme complex (ABC kit; Vector Laboratories) in PBS-T, washed four times for 20 min with PBS-T, and stained with the DAB peroxidase substrate kit (Vector Laboratories). Embryos were stored in 4% PFA at 4°C until the time of image acquisition. Antibody stainings against Cxcr4b-GFP (mix of rabbit anti-GFP from Invitrogen and Torrey Pines Biolabs, Inc.; 1:500 each) and HuC (mouse anti-HuC; Invitrogen; 1:500) were performed as described in this paragraph with the following changes: 90-min 4% PFA fixation at room temperature, donkey anti–rabbit-Cy3 (Jackson ImmunoResearch Laboratories, Inc.; 1:500) and goat anti–mouse-647 (Invitrogen; 1:500) secondary antibodies, and exclusion of the methanol-mediated dehydration and proteinase K–mediated permeabilization steps. To exclude differences in antibody staining, embryos of different genotypes were stained in the same tube.
Morpholino injections
Morpholinos were injected into one-cell-stage embryos. SDF1a morpholino (Doitsidou et al., 2002) was injected at a concentration of 0.02 mM and a volume of 1 nl; the cxcr7a morpholinos were injected at different concentrations of 0.2, 0.5, and 1 mM and a volume of 1 nl; the cxcr7b morpholino (Boldajipour et al., 2008) was injected at a concentration of 1.2 mM and a volume of 1 nl; the SDF1b morpholino (Knaut et al., 2003) was injected at a concentration of 2 mM and a volume of 2 nl; the neurog1 morpholino (Andermann et al., 2002; Cornell and Eisen, 2002) was injected at a concentration of 0.5 mM and a volume of 1 nL; and SDF1a-TP and SDF1a-control-TP (Staton et al., 2011) were injected at a concentration of 0.2 mM and a volume of 1 nl.
RNA injections
cxcr7b was amplified from cDNA generated from 10-somite-stage embryos (SuperScript III cDNA Synthesis kit; Invitrogen) and subcloned into the pCS2+ vector. Forward primer, 5′-GGCCAGATCTATGAGTGTGAACGTGAATGATTTC-3′; reverse primer, 5′-CCGGCTCGAGTCATAATGGTCCCTGGTTTTCCAC-3′. mRNA was synthesized using the mMESSAGE mMACHINE SP6 kit (Applied Biosystems) and injected into cxcr7b−/− embryos at concentrations of 75, 150, and 300 ng/µl and a volume of 1 nl. miR-430 duplexes were obtained from Integrated DNA Technologies and injected at a concentration of 10 µM and a volume of 1 nl into MZdicer mutant embryos (Giraldez et al., 2005). Injected and uninjected embryos were fixed at the 10- or 12-somite stages. In situ hybridization with RNA probes against pax2.1 and HuC was performed as described previously.
Mosaic analysis
For mosaics involving the cxcr7b mutant, one-cell-stage donor embryos were injected with lysine-fixable biotin-dextran to label donor cells (Invitrogen). One-cell-stage host embryos were injected with 1 nL of 0.5 mM neurog1 morpholino to block differentiation of endogenous TgSNs (Andermann et al., 2002; Cornell and Eisen, 2002). At the 1,000-cell to sphere stage, ∼50 donor cells were transplanted into recipient embryos of an equivalent stage. Embryos were incubated at 28.5°C and fixed at the 10- or 12-somite stages. TgSNs were identified with a DIG-labeled RNA probe against HuC and the MHB was identified with a DNP-labeled probe against pax2.1. The probes were detected with anti–DIG-HRP (Roche) and TSA Fluorescein (PerkinElmer) or anti–DNP-HRP (PerkinElmer), TSA Plus DNP (HRP) amplification (Perkin Elmer), and TSA Cyanine 3 (Perkin Elmer), respectively. Biotin-dextran–containing donor-derived cells were identified using HRP-coupled streptavidin (ABC kit) and TSA coumarin (PerkinElmer). For wild-type into hsp70:cxcr7b mosaics, one-cell-stage donor embryos were injected with lysine-fixable biotin-dextran to label donor cells (Invitrogen). One-cell host embryos were injected with 1 nL of 0.5 mM neurog1 morpholino to block differentiation of endogenous TgSNs. Transplantation was performed as described in the beginning of this paragraph. Embryos were incubated at 28.5°C, heat-shocked at the tail bud stage for 20 min and the four-somite stage for 20 min in a 39°C water bath, and fixed at the 10- or 12-somite stages. Transgenic hosts were identified by PCR-based amplification of part of the transgenic locus (outer PCR: forward primer, 5′-TGAGCATAATAACCATAAATACTA-3′, reverse primer, 5′-GAGGCCAATGATGAAGAGGAAGAT-3′; inner PCR: forward primer, 5′-AGCAAATGTCCTAAATGAAT-3′; reverse primer, 5′-CTCTGGCTGAAGGTGCTGTG-3′). TgSNs, the MHB, and donor-derived cells were detected as described for cxcr7b mutant mosaics. For hsp70:cxcr7b into wild-type mosaics, donor embryos were left uninjected, and one-cell-stage host embryos were injected with 1 nL of 0.5 mM neurog1 morpholino to block differentiation of endogenous TgSNs. Transplantation was performed as described in the beginning of this paragraph. Embryos were incubated at 28.5°C, heat-shocked at the tail bud stage for 20 min and the four-somite stage for 20 min in a 39°C water bath, and fixed at the 10- or 12-somite stages. Transgenic donor cells in host embryos were identified by in situ hybridization with a DIG-labeled RNA probe against cxcr7b that was detected with an anti–DIG-AP antibody (Roche) and NBT/BCIP (Roche). HuC and pax2.1 RNA probes labeled with DNP were detected with anti–DNP-HRP antibody, TSA DNP amplification (PerkinElmer), and the DAB peroxidase substrate kit (Vector Laboratories).
SDF1a and Cxcr7b misexpression
For ubiquitous misexpression of SDF1a and Cxcr7b, we used tg(hsp70:SDF1a) and tg(hsp70:cxcr7b) transgenic lines, respectively. Transgenic adults were bred with wild-type adults to yield 50% transgenic embryos and 50% wild-type embryos. Tg(hsp70:SDF1a) embryos and their wild-type siblings were raised at 28.5°C, heat-shocked at the six-somite stage for 20 min in a 39°C water bath, and fixed at the 10- or 12-somite stage. tg(hsp70:cxcr7b) embryos and their wild-type siblings were raised at 28.5°C, heat shocked at the tail bud and four-somite stages for 20 min in a 39°C water bath, and fixed at the 10- or 12-somite stage. Transgenic embryos were identified by in situ hybridization against SDF1a or cxcr7b or by PCR-based amplification of part of the transgenic locus (for primer pairs, see earlier in this section). All embryos were stained with antisense RNA probes against HuC and pax2.1 as described earlier in this section.
Quantification of TgSN position
For quantification of the position of TgSNs in loss-of-function, gain-of-function, and mosaic experiments, neurons were categorized and quantified based on position relative to the MHB (see schematic in Fig. 5 U). The counts were performed at the 12-somite stage, which is 60 min after the trigeminal sensory ganglion has assembled in wild-type embryos. This time point was chosen to ensure that slight staging errors were not mistaken for migration defects. Embryos were dissected, flat mounted, and analyzed using an epifluorescence microscope (Axioplan; Carl Zeiss) for chromogenic stainings and a confocal laser scanning microscope (SP5; Leica) for fluorescent stainings. Significance values were determined using a two-tailed, unpaired Student’s t test. All error bars represent SEM.
Image acquisition
For colorimetric in situ hybridizations and antibody stainings, embryos were dissected and flat mounted on glass microscopy slides in a 2:1 mixture of benzyl benzoate (Sigma-Aldrich) and benzyl alcohol (Sigma-Aldrich), respectively. Images were acquired on an Axioplan microscope (10× air objective lens, NA 0.50) equipped with an AxioCam camera and AxioVision 3.0 software (all from Carl Zeiss). Images were processed using Photoshop CS5 (Adobe). The only operation used was adjustment of input levels to eliminate unused intensity values. Gamma was not adjusted in any of the images. For fluorescent in situ hybridizations used for mosaic analysis, embryos were dissected and flat mounted on glass microscopy slides in 50% glycerol. Images were acquired on a confocal microscope (TCS SP5 II; 10× air objective lens, NA 0.30, and 20× air objective lens, NA 0.70) equipped with photomultiplier tubes for detection and Leica Application Suite Advanced Fluorescence software (all from Leica). Images were processed using Photoshop CS5. The only operation used was adjustment of input levels to eliminate unused intensity values. Gamma was not adjusted in any of the images. For immunofluorescence of Cxcr4b-GFP and HuC, embryos were dissected and flat mounted on glass microscopy slides in 50% glycerol. Images were acquired on a confocal microscope (TCS SP5 II; 10× air objective lens, NA 0.30, and 63× oil-immersion objective lens, NA 1.40–0.60) equipped with two hybrid detectors (HyD) for photon counting and Leica Application Suite Advanced Fluorescence software (all from Leica). 20× images were processed using the maximum intensity projection function in ImageJ (National Institutes of Health). 63× images were processed using ImageJ as described in the following section. All imaging described in this section was performed at room temperature.
Quantification of Cxcr4b-GFP internalization
Using ImageJ, a custom ImageJ macro language script was written to automate the measurement of the mean ratio of membrane Cxcr4b-GFP fluorescence to membrane HuC fluorescence along the anterior–posterior axis in HuC-positive TgSNs (supplemental text file). In brief, a mask was applied to the HuC channel to selectively mark HuC-positive TgSNs using thresholding algorithms. The membrane of the TgSNs was defined as a 1-voxel/240-nm-wide circumference of the HuC mask. This mask includes only membranes between neurons and surrounding tissues but excludes all membranes between neurons. The membrane mask was then applied to a ratio image of Cxcr4b-GFP fluorescence divided by HuC fluorescence to isolate the values that correspond to the ratio on the surface of the TgSNs. For ease of comparison, the Cxcr4b-GFP/HuC ratio of each genetic scenario was normalized to the mean Cxcr4b-GFP/HuC ratio of wild-type controls. For measuring the Cxcr4b-GFP/HuC ratio along the migratory route, a group of HuC-positive neurons at the position of the MHB was used as a landmark to define the position of the ganglion assembly site.
Online supplemental material
Fig. S1 shows three pairs of embryos that show refinement of the SDF1a mRNA expression domain that delineates the migratory route of the TgSNs. Fig. S2 shows reference markers for identifying anatomical regions where cxcr7a and cxcr7b are expressed in the central nervous system. Fig. S3 shows information about the cxcr7b mutation and the ability of cxcr7b mRNA to rescue the neuron positioning defect in cxcr7b−/− embryos. Fig. S4 shows the TgSN positioning defects through 24 hpf in different mutants affecting chemokine signaling. A text file featuring an image-processing script is also available. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207099/DC1.
Supplementary Material
Acknowledgments
We thank J. Torres-Vazquez, F. Schnorrer, J. Nance, and J. Hubbard for critical comments; F. Fuentes and A. Hannon for excellent fish care; and Yan Deng from the Microscopy Core of New York University Langone Medical Center for microscopy guidance. For providing the zebrafish knockout allele cxcr7bsa16, we thank the Sanger Institute Zebrafish Mutation Resource, sponsored by the Wellcome Trust (WT 077047/Z/05/Z). For providing the zebrafish knockout allele dicerhu715 we thank the Hubrecht laboratory and the Sanger Institute Zebrafish Mutation Resource, (ZF-MODELS Integrated Project; contract number LSHG-CT-2003-503496; funded by the European Commission) also sponsored by the Wellcome Trust (WT 077047/Z/05/Z). The HNK-1 antibody (zn-12), developed by B. Trevarrow, was obtained from the Developmental Hybridoma Studies Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biology (Iowa City, IA 52242).
This work was supported by National Institutes of Health grants NS069839 (to H. Knaut) and HD007520 (to S.W. Lewellis and D. Nagelberg).
Footnotes
Abbreviations used in this paper:
- BAC
- bacterial artificial chromosome
- hpf
- hours postfertilization
- MHB
- midbrain–hindbrain boundary
- TgSN
- trigeminal sensory neuron
- TSA
- tyramide signal amplification
References
- Andermann P., Ungos J., Raible D.W. 2002. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev. Biol. 251:45–58 10.1006/dbio.2002.0820 [DOI] [PubMed] [Google Scholar]
- Baker C.V., Bronner-Fraser M. 2001. Vertebrate cranial placodes I. Embryonic induction. Dev. Biol. 232:1–61 10.1006/dbio.2001.0156 [DOI] [PubMed] [Google Scholar]
- Balabanian K., Lagane B., Infantino S., Chow K.Y.C., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., Bachelerie F. 2005. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280:35760–35766 10.1074/jbc.M508234200 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Belmadani A., Tran P.B., Ren D., Assimacopoulos S., Grove E.A., Miller R.J. 2005. The chemokine stromal cell-derived factor-1 regulates the migration of sensory neuron progenitors. J. Neurosci. 25:3995–4003 10.1523/JNEUROSCI.4631-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 409:363–366 10.1038/35053110 [DOI] [PubMed] [Google Scholar]
- Blader P., Plessy C., Strähle U. 2003. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech. Dev. 120:211–218 10.1016/S0925-4773(02)00413-6 [DOI] [PubMed] [Google Scholar]
- Blaser H., Eisenbeiss S., Neumann M., Reichman-Fried M., Thisse B., Thisse C., Raz E. 2005. Transition from non-motile behaviour to directed migration during early PGC development in zebrafish. J. Cell Sci. 118:4027–4038 10.1242/jcs.02522 [DOI] [PubMed] [Google Scholar]
- Bleul C.C., Fuhlbrigge R.C., Casasnovas J.M., Aiuti A., Springer T.A. 1996. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J. Exp. Med. 184:1101–1109 10.1084/jem.184.3.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldajipour B., Mahabaleshwar H., Kardash E., Reichman-Fried M., Blaser H., Minina S., Wilson D., Xu Q., Raz E. 2008. Control of chemokine-guided cell migration by ligand sequestration. Cell. 132:463–473 10.1016/j.cell.2007.12.034 [DOI] [PubMed] [Google Scholar]
- Borrell V., Marín O. 2006. Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat. Neurosci. 9:1284–1293 10.1038/nn1764 [DOI] [PubMed] [Google Scholar]
- Busch-Nentwich E., Kettleborough R., Fenyes F., Herd C., Collins J., Winkler S., Brand M., de Bruijn E., van Eeden F., Cuppen E., Stemple D.L. 2010. Sanger Institute Zebrafish Mutation Resource targeted knock-out mutants phenotype and image data submission. Sanger Institute Zebrafish Mutation Resource, MPI Dresden, and Hubrecht Laboratory; ZFIN Direct Data Submission. http://zfin.org/cgi-bin/webdriver?MIval=aa-fxallfigures.apg&OID=ZDB-PUB-100504-23 (accessed January 15, 2012) [Google Scholar]
- Cheloufi S., Dos Santos C.O., Chong M.M.W., Hannon G.J. 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 465:584–589 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.-Y., Giraldez A.J., Schier A.F. 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 318:271–274 10.1126/science.1147535 [DOI] [PubMed] [Google Scholar]
- Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D., et al. 2010. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 328:1694–1698 10.1126/science.1190809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B., Weidinger G., Knaut H., Thisse B., Thisse C., Raz E., Schier A.F. 2002. Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc. Natl. Acad. Sci. USA. 99:14919–14924 10.1073/pnas.222459999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell R.A., Eisen J.S. 2002. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 129:2639–2648 [DOI] [PubMed] [Google Scholar]
- Daniel D., Rossel M., Seki T., König N. 2005. Stromal cell-derived factor-1 (SDF-1) expression in embryonic mouse cerebral cortex starts in the intermediate zone close to the pallial-subpallial boundary and extends progressively towards the cortical hem. Gene Expr. Patterns. 5:317–322 10.1016/j.modgep.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Davies A.M. 1988. The trigeminal system: an advantageous experimental model for studying neuronal development. Development. 103(Suppl):175–183 [DOI] [PubMed] [Google Scholar]
- Doitsidou M., Reichman-Fried M., Stebler J., Köprunner M., Dörries J., Meyer D., Esguerra C.V., Leung T., Raz E. 2002. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 111:647–659 10.1016/S0092-8674(02)01135-2 [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 308:833–838 10.1126/science.1109020 [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 312:75–79 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 106:23–34 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- Halloran M.C., Sato-Maeda M., Warren J.T., Su F., Lele Z., Krone P.H., Kuwada J.Y., Shoji W. 2000. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 127:1953–1960 [DOI] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan J., Pasquinelli A.E., Bálint E., Tuschl T., Zamore P.D. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 293:834–838 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- Kim C.H., Ueshima E., Muraoka O., Tanaka H., Yeo S.Y., Huh T.L., Miki N. 1996. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216:109–112 10.1016/0304-3940(96)13021-4 [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Knaut H., Werz C., Geisler R., Tübingen 2000 Screen Consortium, and Nüsslein-Volhard C. 2003. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 421:279–282 10.1038/nature01338 [DOI] [PubMed] [Google Scholar]
- Knaut H., Blader P., Strähle U., Schier A.F. 2005. Assembly of trigeminal sensory ganglia by chemokine signaling. Neuron. 47:653–666 10.1016/j.neuron.2005.07.014 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Fjose A. 1991a. Expression pattern of zebrafish pax genes suggests a role in early brain regionalization. Nature. 353:267–270 10.1038/353267a0 [DOI] [PubMed] [Google Scholar]
- Krauss S., Johansen T., Korzh V., Fjose A. 1991b. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 113:1193–1206 [DOI] [PubMed] [Google Scholar]
- Li G., Adesnik H., Li J., Long J., Nicoll R.A., Rubenstein J.L.R., Pleasure S.J. 2008. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J. Neurosci. 28:1085–1098 10.1523/JNEUROSCI.4602-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G., Sánchez-Alcañiz J.A., Pla R., Borrell V., Picó E., Valdeolmillos M., Marín O. 2008. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J. Neurosci. 28:1613–1624 10.1523/JNEUROSCI.4651-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A., Benovic J.L. 2001. Agonist-promoted ubiquitination of the G protein-coupled receptor CXCR4 mediates lysosomal sorting. J. Biol. Chem. 276:45509–45512 10.1074/jbc.C100527200 [DOI] [PubMed] [Google Scholar]
- Marchese A., Raiborg C., Santini F., Keen J.H., Stenmark H., Benovic J.L. 2003. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell. 5:709–722 10.1016/S1534-5807(03)00321-6 [DOI] [PubMed] [Google Scholar]
- Minina S., Reichman-Fried M., Raz E. 2007. Control of receptor internalization, signaling level, and precise arrival at the target in guided cell migration. Curr. Biol. 17:1164–1172 10.1016/j.cub.2007.05.073 [DOI] [PubMed] [Google Scholar]
- Miyasaka N., Knaut H., Yoshihara Y. 2007. Cxcl12/Cxcr4 chemokine signaling is required for placode assembly and sensory axon pathfinding in the zebrafish olfactory system. Development. 134:2459–2468 10.1242/dev.001958 [DOI] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S.C. 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26:216–220 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Naumann U., Cameroni E., Pruenster M., Mahabaleshwar H., Raz E., Zerwes H.-G., Rot A., Thelen M. 2010. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS ONE. 5:e9175 10.1371/journal.pone.0009175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober E.A., Schulte-Merker S. 1999. Signals from the yolk cell induce mesoderm, neuroectoderm, the trunk organizer, and the notochord in zebrafish. Dev. Biol. 215:167–181 10.1006/dbio.1999.9455 [DOI] [PubMed] [Google Scholar]
- Oberlin E., Amara A., Bachelerie F., Bessia C., Virelizier J.L., Arenzana-Seisdedos F., Schwartz O., Heard J.M., Clark-Lewis I., Legler D.F., et al. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 382:833–835 10.1038/382833a0 [DOI] [PubMed] [Google Scholar]
- Paredes M.F., Li G., Berger O., Baraban S.C., Pleasure S.J. 2006. Stromal-derived factor-1 (CXCL12) regulates laminar position of Cajal-Retzius cells in normal and dysplastic brains. J. Neurosci. 26:9404–9412 10.1523/JNEUROSCI.2575-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai M.M., Yang X., Balakrishnan I., Bemis L., Torok-Storb B. 2010. MiR-886-3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PLoS ONE. 5:e14304 10.1371/journal.pone.0014304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Alcañiz J.A., Haege S., Mueller W., Pla R., Mackay F., Schulz S., López-Bendito G., Stumm R., Marín O. 2011. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 69:77–90 10.1016/j.neuron.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Staton A.A., Knaut H., Giraldez A.J. 2011. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 43:204–211 10.1038/ng.758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R.K., Zhou C., Ara T., Lazarini F., Dubois-Dalcq M., Nagasawa T., Höllt V., Schulz S. 2003. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 23:5123–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm R., Kolodziej A., Schulz S., Kohtz J.D., Höllt V. 2007. Patterns of SDF-1alpha and SDF-1gamma mRNAs, migration pathways, and phenotypes of CXCR4-expressing neurons in the developing rat telencephalon. J. Comp. Neurol. 502:382–399 10.1002/cne.21336 [DOI] [PubMed] [Google Scholar]
- Thermes V., Grabher C., Ristoratore F., Bourrat F., Choulika A., Wittbrodt J., Joly J.-S. 2002. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 118:91–98 10.1016/S0925-4773(02)00218-6 [DOI] [PubMed] [Google Scholar]
- Tiveron M.-C., Cremer H. 2008. CXCL12/CXCR4 signalling in neuronal cell migration. Curr. Opin. Neurobiol. 18:237–244 10.1016/j.conb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Tiveron M.-C., Rossel M., Moepps B., Zhang Y.L., Seidenfaden R., Favor J., König N., Cremer H. 2006. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J. Neurosci. 26:13273–13278 10.1523/JNEUROSCI.4162-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevarrow B., Marks D.L., Kimmel C.B. 1990. Organization of hindbrain segments in the zebrafish embryo. Neuron. 4:669–679 10.1016/0896-6273(90)90194-K [DOI] [PubMed] [Google Scholar]
- Valentin G., Haas P., Gilmour D. 2007. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr. Biol. 17:1026–1031 10.1016/j.cub.2007.05.020 [DOI] [PubMed] [Google Scholar]
- Van Haastert P.J.M., Devreotes P.N. 2004. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 5:626–634 10.1038/nrm1435 [DOI] [PubMed] [Google Scholar]
- Wang Y., Li G., Stanco A., Long J.E., Crawford D., Potter G.B., Pleasure S.J., Behrens T., Rubenstein J.L.R. 2011. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 69:61–76 10.1016/j.neuron.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S., Costantino N., Court D.L., Jenkins N.A., Copeland N.G. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Koudijs M.J., van Eeden F.J.M., Cuppen E., Plasterk R.H.A. 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 35:217–218 10.1038/ng1251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.