Structural analysis of a complex of tubulin and tubulin tyrosine ligase (TTL) reveals insights into TTL’s enzymatic mechanism, how it discriminates between α- and β-tubulin, and its possible evolutionary origin.
Abstract
Tubulin tyrosine ligase (TTL) catalyzes the post-translational retyrosination of detyrosinated α-tubulin. Despite the indispensable role of TTL in cell and organism development, its molecular mechanism of action is poorly understood. By solving crystal structures of TTL in complex with tubulin, we here demonstrate that TTL binds to the α and β subunits of tubulin and recognizes the curved conformation of the dimer. Biochemical and cellular assays revealed that specific tubulin dimer recognition controls the activity of the enzyme, and as a consequence, neuronal development. The TTL–tubulin structure further illustrates how the enzyme binds the functionally crucial C-terminal tail sequence of α-tubulin and how this interaction catalyzes the tyrosination reaction. It also reveals how TTL discriminates between α- and β-tubulin, and between different post-translationally modified forms of α-tubulin. Together, our data suggest that TTL has specifically evolved to recognize and modify tubulin, thus highlighting a fundamental role of the evolutionary conserved tubulin tyrosination cycle in regulating the microtubule cytoskeleton.
Introduction
Microtubules are dynamic protein filaments assembled from αβ-tubulin heterodimers that constitute key elements of the cytoskeleton. They are central to a wide variety of important cellular processes, including cell division, development, motility, and intracellular organization. The intrinsic dynamic nature of microtubules is fundamental to their function, and under tight control of a large number of microtubule-associated proteins and molecular motors that constantly act on microtubules throughout the cell cycle (Howard and Hyman, 2003).
It has been known for a long time that microtubules are subject to a number of unusual and evolutionary conserved post-translational modifications including detyrosination/tyrosination, polyglutamylation, and polyglycylation, which affect the unstructured C-terminal tail regions of αβ-tubulin located at the outer surface of microtubules (Westermann and Weber, 2003). However, it has only recently become established that these modifications have a profound impact on microtubule functions, most likely by regulating microtubule interactions with microtubule-associated proteins (Hammond et al., 2008; Janke and Bulinski, 2011). An emerging concept is that the modifications can generate distinct subpopulations of microtubules in the same cell to locally regulate microtubule-based activities (Verhey and Gaertig, 2007; Janke and Bulinski, 2011). The mechanisms that give rise to specific patterns of microtubule modifications, however, are only poorly understood.
Most of the enzymes involved in the generation and removal of post-translational modifications at the C-terminal tails of tubulin have recently been identified (Ersfeld et al., 1993; Janke et al., 2005; Ikegami et al., 2006; van Dijk et al., 2007; Rogowski et al., 2009, 2010; Wloga et al., 2009; Kimura et al., 2010). One of the best-characterized representatives is tubulin tyrosine ligase (TTL), whose activity was discovered several decades ago (Arce et al., 1975). TTL catalyzes the readdition of a tyrosine residue to the C terminus of detyrosinated α-tubulin as part of the evolutionary conserved tubulin tyrosination cycle (Murofushi, 1980; Ersfeld et al., 1993; Westermann and Weber, 2003). Remarkably, the enzyme is indispensable for cell and organism development; TTL-null mice die right after birth due to disorganized neuronal networks (Erck et al., 2005). Furthermore, TTL suppression is linked to cell transformation and correlates with poor prognosis in patients suffering from diverse forms of cancers (Lafanechère et al., 1998; Mialhe et al., 2001; Kato et al., 2004; Whipple et al., 2010). Together, these results underpin the vital role of TTL in physiological conditions and its implication in human pathologies.
On a molecular level, tubulin tyrosination controls several key proteins. For example, the microtubule plus end–tracking proteins (+TIPs) cytoplasmic linker protein-170 (CLIP-170) and large dynactin subunit p150glued use their cytoskeleton-associated protein glycine-rich (CAP-Gly) domains to bind the C-terminal tyrosine of α-tubulin (Steinmetz and Akhmanova, 2008). The sensing of the tyrosine residue is essential for CLIP-170 and p150glued for localization to growing microtubule tips, where they are involved in the regulation of microtubule dynamics and interactions of microtubules with subcellular structures (Badin-Larçon et al., 2004; Peris et al., 2006; Bieling et al., 2008). Similarly, the +TIP mitotic centromere-associated kinesin (MCAK) preferentially binds tyrosinated microtubules to trigger their disassembly (Peris et al., 2009). These molecular roles correlate with phenotypic data demonstrating that tyrosinated microtubules are less stable than detyrosinated ones (Infante et al., 2000). Moreover, differential tubulin tyrosination can also affect the behavior of motor proteins and thus intracellular trafficking. In neurons, for example, the kinesin-1 motor KIF5 senses the absence of the C-terminal tyrosine of tubulin. The higher affinity of KIF5 for detyrosinated microtubules facilitates its navigation from the cell body into axons, which are rich in detyrosinated microtubules. In contrast, the motor is less prominently localized in dendrites, which contain more tyrosinated microtubules as compared with axons (Kreitzer et al., 1999; Dunn et al., 2008; Konishi and Setou, 2009).
Despite the importance of TTL in diverse microtubule-based cellular activities, we still lack a basis for understanding the mechanism of action of this enzyme at the molecular level. Recently, the crystal structure of frog TTL has been determined (Szyk et al., 2011). The structure revealed that TTL is elongated and composed of an N-terminal domain, a central domain, and a C-terminal domain. The active site of the enzyme is formed by its three domains and comprises an adenosine nucleotide molecule. The structure of TTL has defined a conserved fold for the family of the TTL-like (TTLL) tubulin-modifying enzymes, which in addition encompasses polyglutamylases and polyglycylases (Janke et al., 2005; van Dijk et al., 2007; Rogowski et al., 2009; Wloga et al., 2009). Based on sequence conservation, critical residues for tubulin binding and enzymatic activity of TTL have been identified by mutagenesis (Szyk et al., 2011). Furthermore, using analytical ultracentrifugation and small angle x-ray scattering the presence of a moderately stable tubulin–TTL complex with a dissociation constant of 1 µM has been demonstrated (Szyk et al., 2011). However, several key questions have remained open: How does TTL, for example, discriminate between α- and β-tubulin to only tyrosinate the C terminus of α-tubulin? How does the enzyme act on different tubulin isotypes and post-translationally modified variants thereof? How is the tubulin–TTL interaction regulated? To address these fundamental and longstanding questions, we have determined several structures of TTL in complex with tubulin by x-ray crystallography. We further used biochemical and cell biological assays to demonstrate the functional role of key residues in the TTL molecule. Our results establish the first firm basis to understand the structure–function relationship of this essential tubulin-modifying enzyme.
Results
Tubulin recognition
We found that TTL binds a complex composed of two tubulin subunits and the stathmin-like domain of RB3 ([T2R]; Ravelli et al., 2004; Nawrotek et al., 2011), and tyrosinates tubulin in this complex to a similar extent as free tubulin (Fig. 1 A; Fig. S1, A and B). We obtained well-diffracting crystals of T2R in the presence of TTL and different ligands, and solved several structures of the T2R–TTL complex to high resolution (between 2.6 and 1.8 Å; Table S1). Similar to the arrangement in T2R, the two tubulin subunits in the T2R–TTL complex are aligned in a curved, head-to-tail fashion (Fig. 1 B). One TTL molecule binds to one tubulin dimer in T2R; the second apparently equivalent binding site remains free due to crystal packing restrictions (not depicted). The characteristic curved structure of tubulin, which was not significantly affected upon TTL binding (root mean square deviation [rmsd] of 0.12 Å over 650 Cα-atoms), corresponds to the conformation of unassembled, free tubulin (Buey et al., 2006; Rice et al., 2008; Barbier et al., 2010; Nawrotek et al., 2011; Pecqueur et al., 2012).
Figure 1.
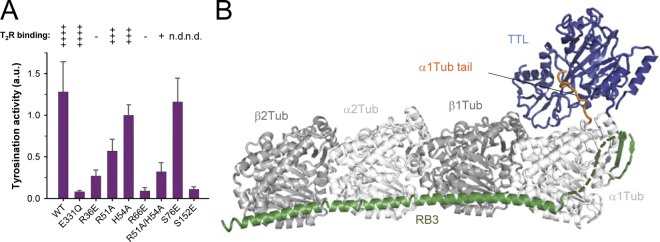
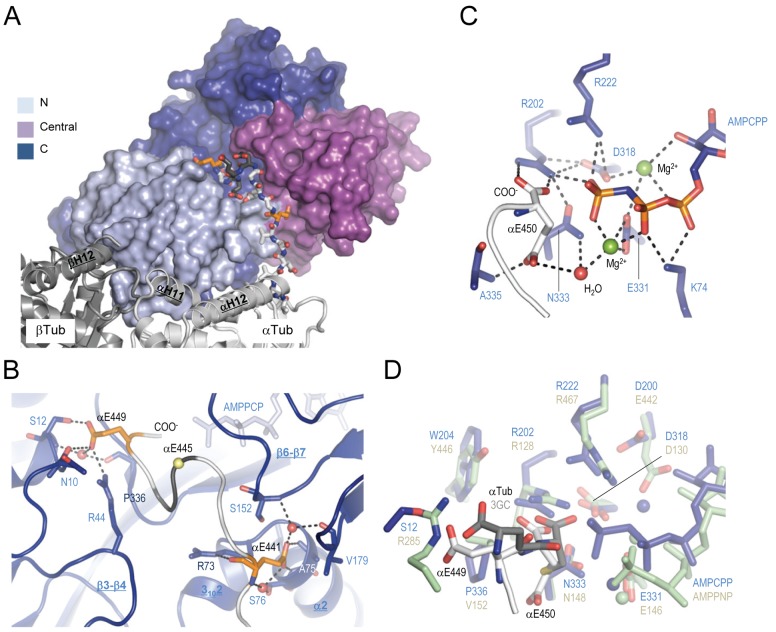
Tubulin binding, enzymatic activity, and overall T2R–TTL structure. (A) Enzyme activity tests (bars) and T2R-binding properties (symbols on top of the bars) of wild-type (WT) and different mutants of TTL. Error bars indicate SEM. The T2R-binding properties were assessed by size exclusion chromatography (see Fig. S1 A for representative data). The binding strength of TTL variants are classified from weak binding (one cross) to strong binding (four crosses). Minus sign, no binding; n.d., not determined. (B) Overall view of the T2R–TTL complex structure in ribbon representation. α-Tubulin, β-tubulin, TTL, and the stathmin-like domain of RB3 are shown in light gray, dark gray, blue, and green, respectively. The C-terminal tail region of α1-tubulin that is bound to TTL is highlighted in orange.
TTL binds tubulin adjacent to the α-β heterodimer interface, with the major part contributed by α-tubulin (∼80% of the 1912 Å2 interaction surface). This specific interface is established by three loops and one helix of TTL (β2-β3, β3-β4, β4-α2, and α6), helix H10 of β-tubulin, and helices H11 and H12 and loops H9-S8 and T5 of α-tubulin (Fig. 2 A). Comparison of the TTL structure in the free and tubulin-bound states revealed only very small conformational changes of these tubulin-interacting elements of TTL upon complex formation (rmsd of 0.5 Å over 19 Cα-atoms; Fig. S1 C). Side-chain contacts between key residues at the interface involve three pairs of histidine–arginine stacking interactions between Arg51, His54, and Arg66 of TTL and αHis393, αArg390, and αHis309 of α-tubulin, respectively, and hydrophobic interactions between Pro56 of TTL, and αPro175 and βLeu333 of αβ-tubulin (Fig. 2 B). The interaction network is complemented by hydrogen bonds formed between Arg36, Arg46, His54, Arg66, and Pro300/Ser303 of TTL, and βGln336/βAsn337, αGlu433, αGlu207, αGlu386, and αArg308 of αβ-tubulin, respectively.
Figure 2.
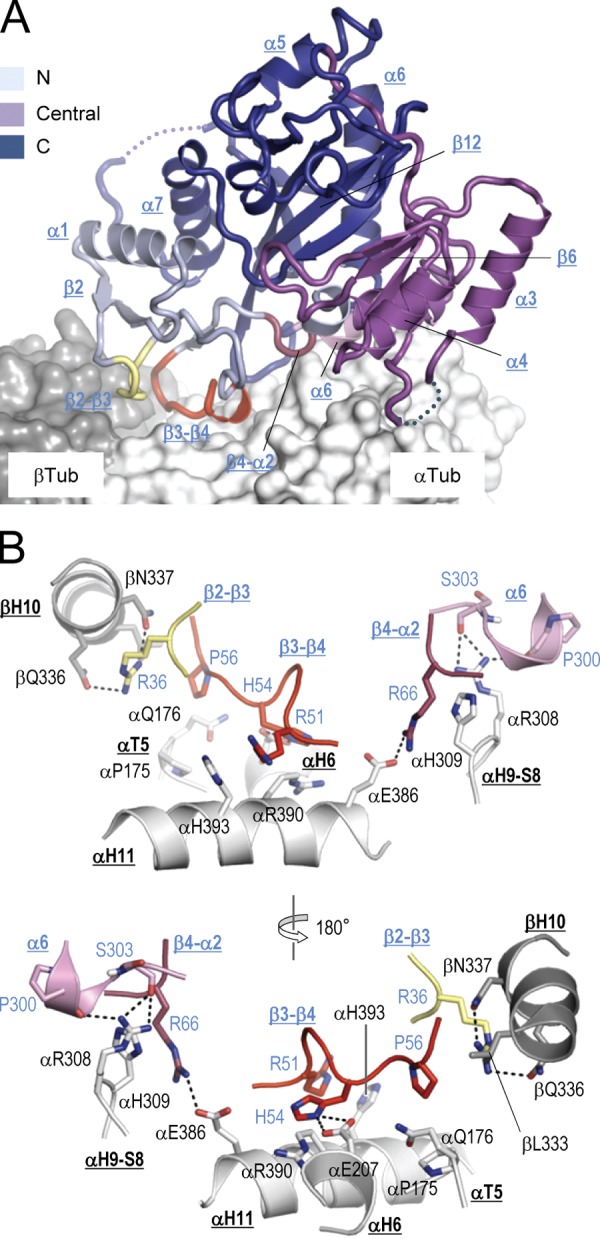
Tubulin recognition by TTL. (A) Global view of the tubulin–TTL interaction. The N-terminal (residues 1–71), central (residues 72–188), and C-terminal (residues 189–378) domains of TTL (ribbon representation) are colored in light blue, magenta, and deep blue, respectively. The interacting TTL loops β2- β3, β3- β4, and β4-α2, and the N-terminal part of helix α6 are highlighted in pale yellow, red, raspberry, and pink, respectively. The αβ-tubulin heterodimer is shown in surface representation and selected TTL elements are labeled. For simplicity, the C-terminal tail of α-tubulin is not shown. (B) Two close-up views 180° apart of the tubulin–TTL interface shown in A in ribbon representation. Interacting residues are shown in stick representation and are labeled in black (tubulin) and blue (TTL). Selected secondary structure elements of tubulin and TTL are labeled in bold black and blue underlined letters, respectively. The loops of TTL that bind tubulin are colored as in A. Oxygen and nitrogen atoms are colored in red and blue, respectively, carbon atoms in the color of the secondary structure elements depicted. Hydrogen bonds are depicted as black dashed lines. For simplicity, the Arg46-αGlu433 TTL–α-tubulin salt bridge and the interfacial water–mediated hydrogen bonding network is not depicted.
It is well established that TTL predominantly binds and modifies unassembled tubulin (Wehland and Weber, 1987; Szyk et al., 2011). To understand this preference, we superimposed the structure of α-tubulin in the tubulin–TTL complex, which is in the curved state known from unassembled tubulin, onto the straight α-tubulin conformation found in microtubules (Nogales et al., 1998, 1999). The comparison revealed that in the straight tubulin structure, the β3-β4 loop of TTL clashes into loop T5 and helix H10 of α- and β-tubulin, respectively (Fig. 3). In addition, it showed that TTL binding interferes with the formation of lateral tubulin contacts in microtubules (Fig. S1 D). These observations explain why TTL selectively modifies unassembled tubulin but not microtubules (Wehland and Weber, 1987; Szyk et al., 2011).
Figure 3.
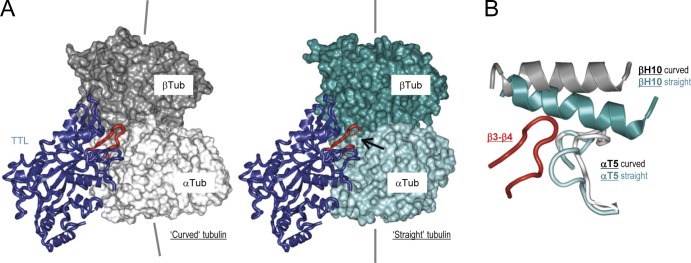
TTL recognizes the curved conformation of αβ-tubulin. (A) TTL (blue) bound to the curved (left) and straight (right) conformation of αβ-tubulin. α- and β-tubulin are shown in light and dark gray, and cyan and deep teal for the curved and straight conformations, respectively. The arrow in the right panel points to the site in which the β3-β4 loop of TTL (red) clashes into the straight conformation of tubulin. The straight tubulin structure corresponds to Protein Data Bank accession no. 1JFF (Löwe et al., 2001). (B) Superimposition of the TTL-bound curved tubulin conformation (T2R–TTL) onto the straight tubulin structure. The N-terminal nucleotide-binding and the C-terminal domains of α-tubulin were used for the superimposition (Ravelli et al., 2004). For simplicity only the T5 loop of α-tubulin (light gray and cyan for the curved and straight tubulin conformations, respectively), the H10 helix of β-tubulin (dark gray and deep teal for the curved and straight tubulin conformations, respectively), and the β3-β4 loop of TTL (red) are shown.
Collectively, these results demonstrate that a distinct set of surface-exposed residues of TTL specifically recognize the curved conformation of unassembled tubulin. Notably, the tubulin-contacting residues are conserved among TTL orthologues (Fig. S2), suggesting functional relevance. In contrast, the same residues are not conserved in different TTLLs (Fig. S2), which suggests that polyglutamylases and polyglycylases recognize tubulin by mechanisms that are distinct from TTL. Consistent with this conclusion, these two types of tubulin-modifying enzymes preferentially modify tubulin in the assembled microtubule form (van Dijk et al., 2007; Rogowski et al., 2009).
Functional importance of tubulin recognition
To investigate the importance of TTL residues engaged at the TTL–tubulin interface we performed a structure-guided mutagenesis study. Introducing single or double mutations decreased either moderately (R51A, H54A) or strongly (R36E, R66E, R51A/H54A) the binding of TTL to T2R (Fig. 1 A), consistent with previous results (Szyk et al., 2011). Concomitantly with a decrease in tubulin-binding capacity, we observed a 20–95% reduction of enzymatic activity for these mutants (Fig. 1 A). This result is consistent with activity data demonstrating that TTL can tyrosinate peptides corresponding to the detyrosinated C-terminal tail of α-tubulin; however, with a 50-fold lower efficiency compared with αβ-tubulin (Paturle et al., 1989; Rüdiger et al., 1994). It also demonstrates that the binding of TTL to the globular core of the tubulin dimer is a requirement for the overall affinity of TTL for tubulin.
Because neurons from TTL-null mice show strong developmental defects, including increased neurite extensions and premature differentiation (Erck et al., 2005), we tested whether the function of TTL in neuronal development depends on its ability to recognize and bind to the globular core of tubulin. As shown in Fig. 4, A–C, and Fig. S3, expression of wild-type TTL in cultured rat hippocampal neurons at 5 d in culture (DIV5) increased the levels of tyrosinated tubulin by a factor of about three, and led to a strong decrease in neurite outgrowth by ∼35% compared with untransfected control neurons in the same experiment. This result is in agreement with the observation that in developing neurons, microtubules are highly dynamic and readily exchange their subunits with the soluble tubulin pool (Hoogenraad and Bradke, 2009). These free tubulin subunits are apparently used as substrates in our TTL overexpression experiments. The effects obtained with wild-type TTL were completely abolished upon disruption of TTL–tubulin complex formation, as the expression of the tubulin binding–deficient mutant R66E did neither significantly alter the tubulin tyrosination status nor affect neuronal morphology. Instead, the obtained phenotype was highly similar to the expression of the catalytically dead version E331Q of TTL (van Dijk et al., 2007). Knocking down the expression of endogenous TTL by small hairpin RNA (shRNA) in turn substantially reduced the amount of tyrosinated tubulin in neurites and in the cell body by a factor of about two, which resulted in a significant increase in the length of neurites by ∼40%, consistent with the phenotype of TTL-null mice (Erck et al., 2005).
Figure 4.
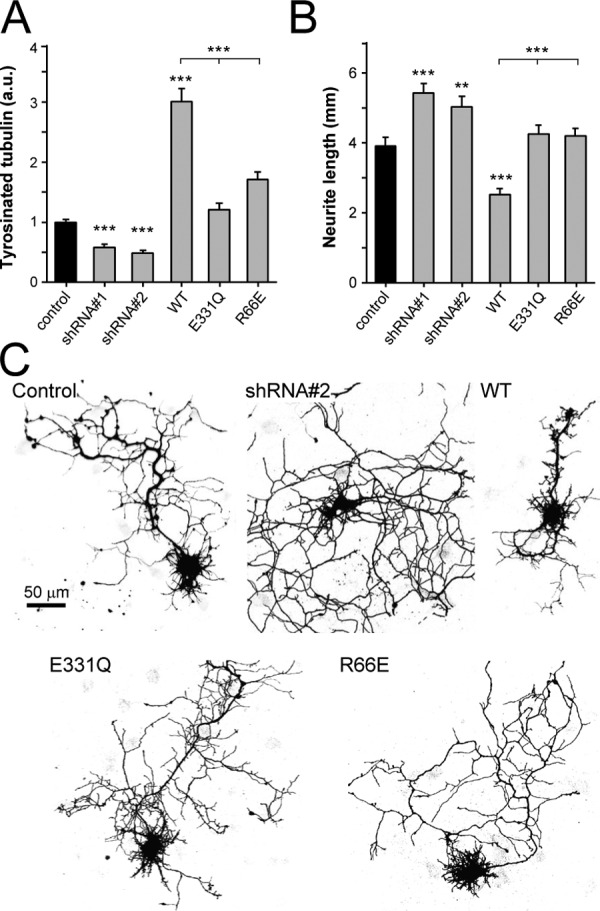
Functional analysis of TTL in hippocampal neurons. (A) Quantification of tyrosinated tubulin staining intensities in neurons transfected with GFP (control), TTL shRNA, wild-type, or mutant forms of TTL. The average intensity of the staining in the cell body was measured and the ratio of the signal in transfected versus neighboring control neurons was calculated for each image. 15–25 cells were analyzed for each condition. Error bars indicate SEM; ***, P < 0.0005. (B) Quantification of total neurite length of neurons transfected with control, TTL shRNA, wild-type, or mutant forms of TTL. 15–30 cells were analyzed for each condition. Error bars indicate SEM; *, P < 0.05; ***, P < 0.0005. (C) Representative images of hippocampal neurons (DIV5) cotransfected with β-galactosidase (to visualize morphology) and control, TTL shRNA, wild-type, or mutant forms of TTL. Bar, 50 µm.
Together, these data show that the specific recognition of the αβ-tubulin dimer is important for the enzymatic activity of TTL. They further revealed that regulating the levels of α-tubulin tyrosination is crucial for controlling neuronal development. Because it is generally accepted that detyrosinated microtubules are more stable than tyrosinated ones (Infante et al., 2000), the observed modulation of neurite length in response to differential tubulin tyrosination levels may correlate to changes in microtubule stability.
Mechanism of α-tubulin tyrosination
Besides providing detailed mechanistic insights into how TTL specifically recognized the αβ-tubulin heterodimer (see previous sections), our structure of the tubulin–TTL complex also explains how the enzyme binds and modifies the functionally important C-terminal tail of α-tubulin. As shown in Fig. 5 A, the structure revealed an extended cleft formed by the three domains of TTL that was ideally positioned to guide the C terminus of α-tubulin into the active site of the enzyme. The α-tubulin residues αGlu441 and αGlu449 anchor the tail by forming hydrogen bonds with residues Arg73, Ala75, Ser76, Ser152, and Val179, and Asn10, Ser12, Arg44, and Pro336 of TTL, respectively (Fig. 5 B). Notably, residues Ser76 and Ser152 of TTL were predicted protein kinase C phosphorylation sites, suggesting a role in regulation (Ersfeld et al., 1993; Idriss, 2000). To test the potential impact of phosphorylation on TTL activity we generated the two phospho-mimicking Ser-to-Glu mutants TTL-S76E and TTL-S152E. As shown in Fig. 1 A, whereas TTL-S76E was comparable to wild type, TTL-S152E displayed a >90% reduction in enzymatic activity. This result indicates that phosphorylation of Ser152 could play an important role in the regulation of TTL activity.
Figure 5.
Recognition of the C-terminal tail of α-tubulin by TTL. (A) Overview of the interaction between the α-tubulin tail (sticks) and TTL (surface). The two α-tubulin residues αGlu441 and αGlu449, which anchor the tail sequence of α-tubulin on TTL, are highlighted in orange. (B) Close-up view of the interaction between TTL (blue ribbon) and the C-terminal tail of α-tubulin (gray ribbon). Interacting residues are shown in stick representation. αGlu441 and αGlu449 are highlighted in orange. The position of αGlu445, the major modification site of α-tubulin, is highlighted by a yellow sphere. Three key water molecules are shown as red spheres. Note that αGlu445, αGlu446, and αGlu447 are not well defined in the electron density (see Fig. S4 A) and are thus modeled and shown in dark gray in A and B. (C) Close-up view of the active site of TTL in the T2R–TTL complex in stick representation. A water molecule and two magnesium ions are depicted as red and green spheres, respectively. (D) Superimposition of the active site of TTL (blue) bound to the nonhydrolyzable ATP analogue AMPPCP and the tail of α-tubulin (light gray) onto the one of glutathione synthase (green; Protein Data Bank accession no. 1M0W) bound to the nonhydrolyzable ATP analogue AMPPNP and gamma-glutamylcysteine (3GC; dark gray). Key residues are shown in stick representation. For simplicity only residue side chains are shown. Spheres depict magnesium ions. See Fig. 2 for additional information on labels, symbols, and color code.
When bound to TTL, the polypeptide chain of the α-tubulin tail adopted a loop-like conformation between the tail-anchoring residues αGlu441 and αGlu449. This loop allowed the side chain of αGlu445, the major polyglutamylation site of α-tubulin (Eddé et al., 1990), to be oriented away from the TTL surface (Fig. 5 B). Notably, the tail-contacting residues are well conserved among TTL orthologues but not in TTLLs (Fig. S2). This observation suggests that polyglutamylases and polyglycylases recognize the C-terminal tail sequences of αβ-tubulin by mechanisms that are distinct from TTL.
Inspection of the active site of TTL in the tubulin–TTL complex revealed that the adenosine nucleotide molecule was wedged against loops β6-β7 and β11-α5 and β-strands β6 and β13. Two magnesium ions are coordinated by the β- and γ-phosphate groups of the nucleotide, and by residues Asp318 and Glu331 of TTL (Fig. 5 C). The C-terminal residue of the detyrosinated α-tubulin tail, αGlu450, was deeply inserted into a distinct cavity located at the distal end of the TTL cleft. Both the side chain and main chain carboxylate group of αGlu450 formed hydrogen bonds with Ala335 and Arg202 of TTL, respectively. We further observed a pronounced density close to the main-chain carboxylate group of αGlu450 whose shape could fit either the αTyr451 residue of α-tubulin or a tyrosinol molecule present in the crystallization buffer (Fig. S4, A–C). Notably, the residues forming the active site are highly conserved among TTL orthologues and TTLLs (Fig. S2), suggesting a common enzymatic mechanism for the enzymes of this family.
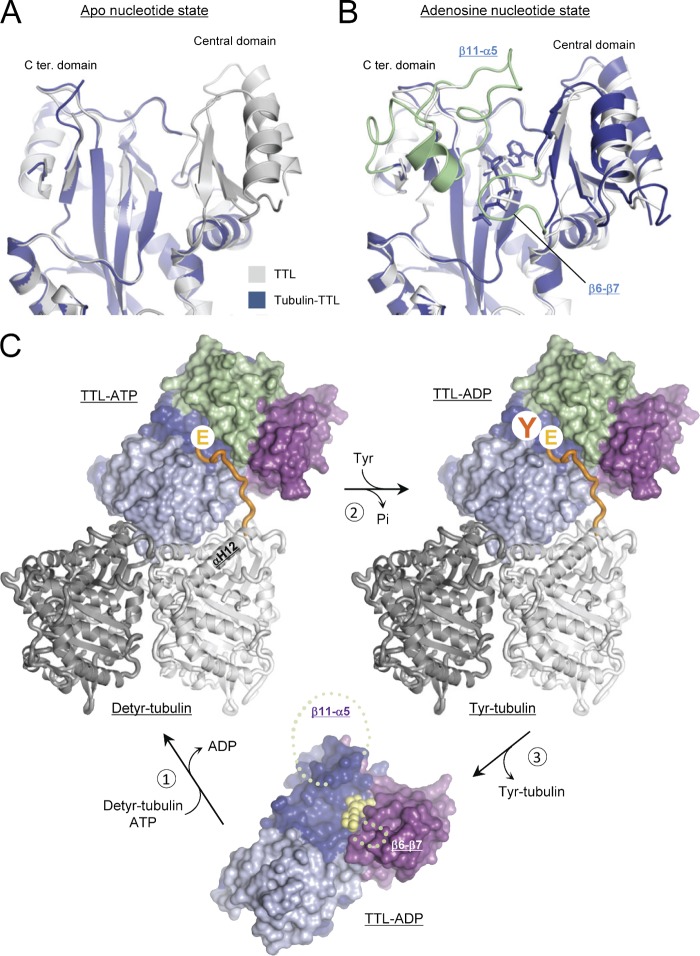
The architecture of the active site of TTL is very similar to the ones of the related ATP-grasp family members glutathione synthase (Hara et al., 1996) and d-alanine:d-alanine ligase (Fan et al., 1994). Indeed, all critical elements, including the two magnesium ions required for catalysis, superimposed well (rmsd ranging from 0.63 to 0.82 Å over >15 Cα-atoms; Fig. 5 D and Fig. S4 D). In the case of glutathione synthase it has been shown that binding of nucleotide and glutathione induces the structuring of two loops in the active site of the enzyme (Hara et al., 1996). To assess structural changes accompanying the enzymatic cycle of TTL, we solved the T2R–TTL structure in the absence of nucleotide (apo). Superimposition of the apo and nucleotide-occupied structures of TTL in their tubulin-bound (T2R–TTL) and -unbound (Szyk et al., 2011) states revealed that the β11-α5 loop and a large portion of the central domain of TTL become structured upon binding of both nucleotide and tubulin (Fig. 6, A and B; Fig. S5); as for glutathione synthase (Hara et al., 1996), these structural elements are critically involved in shaping the active site of the enzyme. Binding, tyrosination, and release of the C-terminal tail of α-tubulin thus appear coupled to nucleotide exchange, and involve a concerted structuring mechanism of TTL.
Figure 6.
Molecular mechanism of tubulin tyrosination by TTL. (A and B) Superimposition of TTL in the tubulin-bound (blue) and -unbound (gray; Protein Data Bank accession nos. 3TIG and 3TII, respectively) states in cartoon representations, and in the absence (A) or presence (B) of nonhydrolyzable ATP analogues. In B, the β11-α5 loop and a large portion of the central domain of TTL, structural elements that get structured upon binding of both tubulin and adenosine nucleotide, are highlighted in pale green. (C) Enzymatic cycle of TTL. (1) Binding of TTL-ADP (ground state) to detyrosinated tubulin (Detyr-tubulin) and exchanging ADP (in yellow sphere representation) by ATP induces the structuring of the β6-β7 and β11-α5 loops of TTL (indicated by pale green dashed lines). As a result, the ATP molecule gets buried and an extended cavity is formed, which specifically recognizes the incoming C-terminal tail of α-tubulin (highlighted in orange). (2) In the presence of tyrosine and upon ATP hydrolysis, tyrosinated tubulin (Tyr-tubulin), ADP, and inorganic phosphate (Pi) are produced. (3) Release of Tyr-tubulin from TTL restores the ground state of TTL-ADP. This could be achieved by phosphorylation or by the intrinsic thermodynamic properties of the moderately stable tubulin–TTL complex (Szyk et al., 2011), which suggests a fast dissociation rate. The αβ-tubulin heterodimer (α- and β-tubulin in light and dark gray, respectively) is shown in cartoon representation; TTL is shown in surface representation. TTL domains are colored as in Fig. 2 A.
Together, these observations underpin a conserved enzymatic mechanism shared by the ATP-grasp family of proteins (Galperin and Koonin, 1997). Accordingly, the ligation reaction of TTL is thought to proceed by the transfer of the γ-phosphate group of ATP onto the C terminus of αGlu450 to form an acyl-phosphate intermediate, followed by nucleophilic attack by the amine group of the incoming tyrosine to produce ADP, inorganic phosphate, and tyrosinated α-tubulin (Fig. 6 C).
Discussion
The data presented in this study advance our understanding of regulatory events that modify the microtubule cytoskeleton by unraveling the molecular mechanism of substrate recognition, catalysis, and modes of regulation of the essential tubulin-modifying enzyme TTL (summarized in Fig. 7). It is well known that TTL tyrosinates only the α subunit of the αβ-tubulin heterodimer (Ersfeld et al., 1993); however, the mechanism of this selectivity is not understood. Our data provide insights into how TTL discriminates between α- and β-tubulin. First, binding of TTL adjacent to the α–β tubulin interface ideally positions the active site of the enzyme to only receive the C-terminal tail of α-tubulin; modeling suggests that the tail of β-tubulin is too short to reach the same site (not depicted). Second, in contrast to β-tubulin, only α-tubulin contains two consecutive C-terminal acidic residues required for binding to the active site of TTL (Fig. 7). The characteristic bipartite αβ-tubulin–TTL and α-tubulin tail–TTL binding mode thus explains the high specificity of TTL for α-tubulin.
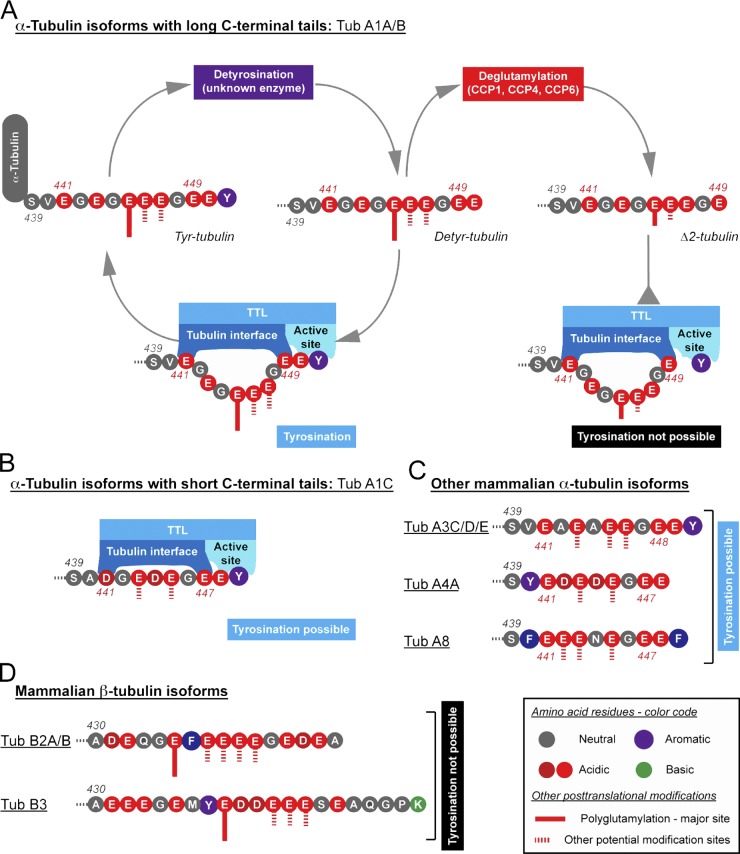
Figure 7.
Schematic representation of the interplay between different post-translational modifications and isoforms of tubulin, and the involvement of TTL. (A) α-Tubulin is detyrosinated by a yet unknown enzyme and gives rise to detyrosinated tubulin (Detyr-tubulin). The C-terminal tail of detyrosinated tubulin originating from the most prominent α-tubulin isoforms Tub A1A or Tub A1B is anchored by TTL via the acidic residues αGlu441 and αGlu449. The main site of polyglutamylation, αGlu445, is localized between these two residues and therefore does not interfere with the formation of the tubulin–TTL complex. Further transformation of detyrosinated tubulin into Δ2-tubulin by deglutamylases of the carboxy peptidase (CCP) family does not interfere with the binding of tubulin to TTL, but the tyrosination reaction is not anymore possible as the C terminus of Δ2-tubulin (αGlu449 in Tub A1A and Tub A1B) is too distant from the binding site of tyrosine. (B) A shorter isoform of α-tubulin, Tub A1C, can still bind TTL as the two binding sites on the enzyme are close enough. As in Tub A1A and A1B (A), the potential sites of polyglutamylation (which are most likely also the sites of polyglycylation) are localized in regions that are not involved in the formation of the tubulin–TTL complex. (C) Depiction of C-terminal tails of remaining mammalian α-tubulin isoforms. All these isoforms possess the two acidic “anchoring” residues necessary for TTL binding, and can therefore potentially be tyrosinated. Tub A4A is gene encoded in the detyrosinated form. Tub A8 carries a C-terminal phenylalanine instead of a tyrosine. To act as a substrate for TTL, it is necessary that phenylalanine can be enzymatically removed, which is currently not known. (D) Depiction of C-terminal tails of two major brain β-tubulin isoforms. Note that none of the β-tubulin isoforms contain the C-terminal Glu-Glu dipeptide sequence necessary for tyrosination by TTL.
The characteristic TTL binding mode of the tubulin tail further suggests that α-tubulin isotypes, despite the difference in length of their C-terminal tails (Fig. 7), can be tyrosinated by TTL. Based on our structural information we conclude that the length, amino acid composition, and additional modifications of the C-terminal tails of α-tubulin have most likely no influence on the tyrosination of the protein. In contrast, our tubulin–TTL structure also reveals why Δ2-tubulin, a post-translationally modified form of detyrosinated α-tubulin lacking αGlu450, cannot be tyrosinated (Paturle et al., 1989; Rüdiger et al., 1994); it shows that only tubulin tails with two consecutive glutamate residues on their C termini can be tyrosinated by TTL (Fig. 7). The escape of Δ2-tubulin from the tyrosination cycle is considered causative for the permanent stabilization and irreversible functional specialization of microtubule subpopulations in neurons, axonemes, and centrioles (Konno et al., 2012). Finally, the finding that the side chain of αGlu445 is oriented away from the TTL surface provides a plausible explanation why even extensive post-translational polyglutamylation of this residue does not hinder tyrosination of the α-tubulin tail (Eddé et al., 1992).
The complex bipartite interaction mode observed between tubulin and TTL reveals how the enzyme has specifically evolved to recognize and modify tubulin; they virtually preclude that the enzyme modifies additional substrates. This conclusion offers a molecular basis for understanding the lethal phenotype of TTL knockout mice (Erck et al., 2005). Our structural and cellular data implicate that the severe neuronal defects observed in these mice are primarily caused by changes in the tyrosination status of α-tubulin and not due to the modification of additional proteins by TTL. The resulting impairment of microtubule functions in TTL knockout neurons is most likely elicited either by the malfunction of microtubule-associated proteins like, for example, +TIPs, and/or molecular motors that sense the tyrosination state of α-tubulin (Janke and Bulinski, 2011). The insights gained from our study thus provide evidence for a fundamental role of the evolutionary conserved tubulin tyrosination cycle in cell and organism development.
Our finding that reduced binding of TTL to the globular core of tubulin decreases the tyrosination efficiency of the enzyme has further implications for the understanding of cytoskeleton-related human diseases. A number of neurodevelopmental disorders are linked to mutations in tubulin genes. The mechanisms by which these mutations lead to the pathology are not understood in most cases; however, it is generally assumed that they alter the function of the tubulin molecule itself (Tischfield et al., 2011). Our findings suggest that some of these mutations could have an impact on tubulin tyrosination. Indeed, two recent, independent studies describe that mutation of αArg390, an α-tubulin residue that we found to be engaged at the tubulin–TTL interface (Fig. 2 B), is linked to cases of lissencephaly (Kumar et al., 2010) and polymicrogyria (Poirier et al., 2012). Knowledge of the molecular mechanisms of tubulin-modifying enzymes may thus open new perspectives for the understanding of human pathologies linked to hereditary tubulin mutations.
Materials and methods
Cloning and protein preparation
Cloning and protein preparation of TTL samples has been described by Prota et al. (2013). In brief, chicken TTL containing a C-terminal hexahistidine tag was overexpressed in Escherichia coli BL21 (DE3) cells and purified on a HisTrap affinity column (GE Healthcare). The fractions containing TTL protein were pooled, concentrated to 5 ml using a Centriprep device (Mw cutoff 30,000; Amicon) and loaded onto a Superdex 200 16/60 column for the final purification step in 20 mM Bis-Tris propane, pH 6.5, 200 mM NaCl, 2.5 mM MgCl2, 5 mM β-mercaptoethanol, and 1% glycerol. The protein-containing fractions were collected, concentrated to ∼20 mg/ml, and frozen in aliquots in liquid nitrogen for storage.
The stathmin-like domain clone of RB3 was a gift from A. Sobel (Institut du Fer-à-Moulin, Paris, France). The protein was prepared according to Ravelli et al. (2004). Bovine and pork brain tubulin was prepared according to well established protocols (Andreu, 2007). The bovine brain tubulin was purchased from the Centro de Investigaciones Biológicas (Microtubule Stabilizing Agents Group), CSIC, Madrid, Spain. Before the reconstitution of the T2R complex, tubulin was subjected to one cycle of polymerization/depolymerization (Dorleans et al., 2007). The composition of isoforms and post-translationally modified versions of brain αβ-tubulin is as follows: β-tubulin consists of 58% Tub B2, 25% Tub B3, 13% Tub B4 (Banerjee et al., 1988), out of which Tub B2 is polyglutamylated at Glu435 (Rüdiger et al., 1992), Tub B3 at Glu438 (Alexander et al., 1991), and Tub B4 at Glu434 (Mary et al., 1994). The composition of the α-tubulin pool is less well known; however, the highly homologous members of the Tub A1 family, which are the main components of the brain tubulin pool, are modified at Glu445 (Eddé et al., 1990). The C terminus of α-tubulin in brain is present as a mixture of tyrosinated tubulin, detyrosinated tubulin, Δ2-tubulin, and perhaps other modification variants that have only recently been discovered (e.g., Δ3-tubulin; Berezniuk et al., 2012). In adult brain, ∼35–50% of the total tubulin pool cannot be retyrosinated (Paturle et al., 1989), indicating that this pool is in the Δ2 form or further modified. Moreover, 15–20% of the pool is tyrosinated tubulin, and 35–40% is detyrosinated tubulin (Barra et al., 1988). Because the aim of our biochemical and biophysical experiments was to qualitatively/semi-quantitatively compare different TTL variants, and not to provide absolute quantitative numbers, the heterogeneity of our tubulin samples is not expected to affect the interpretation of the data.
Size exclusion chromatography
Size exclusion chromatography experiments were performed in running buffer (80 mM Pipes-KOH, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) on a Superdex 75 10/60 column. 25 µl T2R (12 mg/ml) were mixed with 25 µl TTL (3 mg/ml) supplemented with 1 mM ADP and incubated on ice for 20 min. After 5 min centrifugation at 14,000 rpm on a tabletop centrifuge, 25 µl samples were loaded onto the column. All the experiments were performed at 4°C. For the control runs 25 µl of the individual components were mixed with 25 µl running buffer. Binding was assessed by monitoring the disappearance of the peak corresponding to uncomplexed TTL and by SDS-PAGE analysis of the collected peak fractions.
Enzymatic tyrosination assay
Enzymatic activities of recombinant TTL variants were determined using the incorporation of radioactively labeled tyrosine, similar to the assay commonly used for measuring glutamylase or glycylase activities (Regnard et al., 1998; Janke et al., 2005; Rogowski et al., 2009). The final reaction mixture of a total volume of 20 µl was composed of 60 mM Tris/HCl, pH 9.0, 400 µM ATP, 2.4 mM MgCl2, 625 µM DTT, 5 µM l-[3H]-tyrosine (42.6 Ci/mmol, NET12700; PerkinElmer), and 0.2 mg/ml (1.8 µM) pork brain tubulin, supplemented with various concentrations of TTL. For the experiments shown in Fig. S1 B, tubulin was preincubated with RB3 (final concentration 2.15 µM) for 15 min at 30°C in order to generate the T2R complex. All samples of one experimental series were resolved together on a single SDS-PAGE gel, specifically adapted to separate α- and β-tubulin (for a detailed description of gel system, see Lacroix and Janke, 2011). After transfer to nitrocellulose membranes and staining with Ponceau S, α- and β-tubulin bands were cut out and incorporated l-[3H]-tyrosine was quantified using a Tri-Carb 2910 TR scintillation counter and QuantaSmart software (PerkinElmer).
To set up optimal reaction conditions for this activity assay, we determined the linear range of catalysis for variable molar ratio TTL/tubulin in 45-min time course measurements. Initial speeds were derived for each series, and plotting these speeds against TTL/tubulin ratios demonstrated that for TTL/tubulin ratios between 1:1,000 and 1:200 the reactions were under nonsaturating conditions. Thus, we decided to use a TTL/tubulin ratio of 1:500 (final TTL concentration in the assay: 900 µM) for all further assays.
To compare the activities of TTL with tubulin or the T2R complex, we performed the incorporation assay using either free tubulin or T2R (molar ratio of tubulin/RB3 of 1:1.2) complex in a time-course experiment. In this context, we noted a slight increase by ∼1.2 in tyrosination efficiency of TTL for T2R compared with tubulin alone. We speculate that the dimeric nature of the T2R tubulin assembly and/or the fact that the tubulin dimers in T2R are fixed in the curved conformation are possible explanations for this slight increase in enzymatic activity. Considering that incorporation was linear throughout the experiment, we measured the enzymatic activity of all other TTL mutants by determining only the end points at 30 min (Fig. 1 A).
For statistical analysis of the incorporation experiments (at least three experiments were done for each experimental series), it was necessary to adjust total measurements because total counts tend to differ between single experiments. To adjust different experimental series for averaging (Fig. S1 B), we determined the linear slopes for each single experiment (one experiment consists of a complete time course for TTL with tubulin or with the T2R complex), determined the average between the TTL–tubulin and TTL–T2R slopes, and determined a correction factor for all experiments that we then applied to each single measurement value of the experiments (the experiment with the highest slope obtained factor 1). With these corrected values, we calculated the average values for each time point, which were then fitted linearly (Fig. S1 B). For the experiments represented in Fig. 1 A, we adjusted all measured values of one series of experiments to an average value of the four most active TTL versions (wild type, R51A, H54A, and S76E). Using those averaged values then allowed us to determine mean values from up to seven independent measurements for each TTL version, and resulted in the representation of the final results in arbitrary units.
Crystallization, data collection, and structure solution
The T2R–TTL complex was formed by mixing the individual components at a ratio of 2 tubulin, 1.3 RB3, and between 1.2 and 2.2 TTL in the absence or presence of 1 mM adenosine nucleotide, and was concentrated to 20 mg/ml before crystallization; the best quality crystals were obtained at a protein ratio of 2:1.3:1.2 (tubulin/RB3/TTL). The T2R–TTL complex crystallized by the sitting-drop vapor-diffusion method at 20°C. Crystals grew overnight in precipitant solution consisting of 4–10% PEG 4K, 4–6% glycerol, 30 mM MgCl2, 30 mM CaCl2, and 100 mM MES/imidazole, pH 6.7 and reached their maximum dimensions within one week. They belonged to the space group P212121, with one T2R–TTL complex in the asymmetric unit. Data were collected at 100 K at beamlines X06SA and X06DA at the Swiss Light Source (SLS, Paul Scherrer Institute, Villigen, Switzerland) and were processed and merged with the XDS program (Kabsch, 2010).
The T2R–TTL–apo and –ADP structures were determined by the difference Fourier method using the phases of the T2R–TTL–AMPPCP complex in the absence of ligands and solvent molecules (Protein Data Bank accession no. 4I4T; Prota et al., 2013) as a starting point for the refinement. The models were first fitted by several cycles of rigid body refinement followed by simulated annealing and restrained refinement in Phenix (Adams et al., 2010). The resulting models were further improved through iterative model rebuilding in Coot (Emsley and Cowtan, 2004) and refinement in Phenix. NCS restraints were applied in initial refinement stages and then omitted in the final cycles of refinement to account for structural variations between the ncs-related copies of α- and β-tubulin. The quality of the structures was assessed with MolProbity (Davis et al., 2004), and figures were prepared using PyMOL (The PyMOL Molecular Graphics System, version 1.4.1; Schrödinger, LLC). Data collection and refinement statistics are given in Table S1.
It should be noted that both the T2R–TTL–ADP and the T2R–TTL–apo structures displayed no well-defined electron density for the α-tubulin segment αSer439-αGlu447, most likely because of the flexibility of elements shaping the active site of TTL (Szyk et al., 2011), and maybe also because of the particularly high heterogeneity of tubulin tails due to the expression of different tubulin isotypes (Ludueña, 1993) and by extensive post-translational modifications in this sequence region (Janke and Bulinski, 2011). However, in the presence of the nonhydrolyzable ATP analogue adenylylmethylenediphosphonate or β,γ-methyleneadenosine 5′-triphosphate (AMPPCP), the tyrosine analogue tyrosinol and zampanolide (Field et al., 2009), we obtained a well-defined and continuous electron density for α-tubulin in T2R–TTL up to residue αGly444, followed by a partially fragmented density until αGly448 (Fig. S4, A–C). The overall quality of the density was sufficient to model the complete tail sequence of α-tubulin in this particular T2R–TTL complex structure. The zampanolide molecule was found to bind to the taxane site of β-tubulin, which is situated remote from the tubulin–TTL interface. Comparison of the tubulin–TTL structure in the presence and absence of the compound revealed no significant differences, demonstrating that its binding has no influence on the conformation of the complex. The zampanolide–tubulin binding mode is described in Prota et al. (2013).
Expression constructs and TTL shRNA
pβactin-HA-β-galactosidase was generated by ligating β-galactosidase in the AscI and SalI sites in a modified pβ-actin-16pl vector (Kaech et al., 1996). Wild-type and mutant chicken TTL constructs were generated by PCR and cloned in the HindIII and SalI sites in a pβactin-GFP vector (Kapitein et al., 2010). pSuper-based shRNA vectors (Brummelkamp et al., 2002) were directed against the following target sequences: rat TTL shRNA#1 (5′-CTCTGAACATTACCCTAGA-3′) and rat TTL shRNA#2 (5′-CCCCGAGTCCTATGTGATT-3′) and designed using an siRNA selection program (Yuan et al., 2004).
Hippocampal neuron experiments
Primary hippocampal cultures were prepared from embryonic day 18 (E18) rat brains (Banker and Goslin, 1988; Kapitein et al., 2010). For the different stainings we used rabbit antibody against TTL (1:200; Proteintech), rat antibody against tyrosinated α-tubulin (YL1/2; Abcam, 1:1,000), and mouse antibody against β-galactosidase (1:2,000; Promega). For secondary antibodies we used Alexa 488– and Alexa 598–conjugated goat antibodies against rabbit, rat, and mouse IgG (1:400; Molecular Probes).
Hippocampal neurons were plated on laminin (2 µg/ml) and poly-l-lysine (30 µg/ml) coated coverslips at a density of 100,000/well. Cultures were grown in neurobasal medium (NB) supplemented with B27, 0.5 mM glutamine, 12.5 µM glutamate ,and penicillin/streptomycin mix. At DIV1 or DIV2 hippocampal neurons were transfected using Lipofectamine 2000 (Invitrogen). Per well of a 12-well culture plate, 1.8 µg DNA was mixed with 200 µl of NB and 3.3 µl of Lipofectamine 2000, incubated for 30 min, and then added to the neurons in NB at 37°C in 5% CO2. After 45 min incubation, neurons were washed with NB and transferred to their original medium at 37°C in 5% CO2. After transfection (3 d for overexpression constructs and 4 d for shRNAs), neurons were fixed at DIV5 with 4% PFA/4% sucrose in PBS for 10 min at room temperature to visualize neuronal morphology and TTL staining, and with a combination of cold methanol and paraformaldehyde to stain for tyrosinated tubulin. Neurons were washed in PBS and incubated with the indicated primary antibodies in GDB buffer (0.2% BSA, 0.8 M NaCl, 0.5% Triton X-100, and 30 mM phosphate buffer, pH 7.4) overnight at 4°C. Next, cells were washed three times with PBS for 5 min and incubated with secondary antibodies in GDB for 1 h at room temperature and washed three times in PBS. Finally, coverslips were mounted using Vectashield (Vector Laboratories).
Confocal images of fixed neurons were acquired using a confocal laser microscope (model A1R; Nikon) equipped with 40× oil Plan-Fluor NA 1.3 and 60× oil Plan-Apo NA 1.4 objectives and Nikon’s NIS-Elements imaging software. Using maximum projection, z-series of images were converted into a single image. Analysis of neuron morphology was performed using SynD (Schmitz et al., 2011). Analyses of staining intensities (TTL and tyrosinated α-tubulin) were performed using MetaMorph image analysis software (Universal Imaging Corp.). Using the projection images, somas were traced of the transfected neuron (one per image) and a nontransfected neuron in the same image. Of each soma the average intensity was measured and values were imported into Excel to use for data analysis. The average background intensity (based on neighboring regions that contained no labeling) was subtracted and the ratio of transfected vs. nontransfected intensity was calculated for each image. Data were averaged over multiple cells (15–30 per condition) and a statistical analysis was performed with Student’s t test assuming a two-tailed and equal variation. Images were prepared for publication using MetaMorph and Adobe Photoshop.
Accession numbers
Coordinates have been deposited in the Protein Data Bank with accession numbers 4IIJ (T2R–TTL–apo), 4IHJ (T2R–TTL–ADP), and 4I4T (T2R–TTL–AMPPCP).
Online supplemental material
Fig. S1 shows the data for T2R binding and the tyrosination assay, and additional structural considerations of the TTL–tubulin interaction. Fig. S2 shows a multiple sequence alignment of TTL orthologues together with selected TTLLs. Fig. S3 shows the hippocampal neuron data that were used to quantify tubulin tyrosination levels. Fig. S4 shows the electron density of the active site of TTL in the T2R–TTL complex and illustrates the comparison with ATP-grasp family enzymes. Fig. S5 shows the electron density of TTL in the apo and tubulin/AMPPCP bound states. Table S1 reports the x-ray data collection and refinement statistics. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201211017/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201211017.dv.
Supplementary Material
Acknowledgments
We are indebted to F. Winkler for critical reading of the manuscript and to N. Gold for technical assistance. We thank V. Olieric and M. Wang for excellent technical assistance with the collection of x-ray data at beamline X06SA and X06DA of the Swiss Light Source.
This work was supported by grants from the Institut Curie, the CNRS, the INSERM, the French National Research Agency (08-JCJC-0007), the HFSP program (RGP 23/2008), and the EMBO Young Investigator Program (to C. Janke and C.C. Hoogenraad); by grant NWO-ALW-VICI of the Netherlands Organization for Scientific Research (to C.C. Hoogenraad); and by grant 310030B_138659 from the Swiss National Science Foundation (to M.O. Steinmetz).
Footnotes
Abbreviations used in this paper:
- +TIPs
- plus end–tracking proteins
- AMPPCP
- adenylylmethylenediphosphonate or β,γ-methyleneadenosine 5′-triphosphate
- rmsd
- root mean square deviation
- shRNA
- small hairpin RNA
- T2R
- complex of two αβ-tubulin dimers and RB3
- TTL
- tubulin tyrosine ligase
- TTLL
- TTL-like
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J.E., Hunt D.F., Lee M.K., Shabanowitz J., Michel H., Berlin S.C., MacDonald T.L., Sundberg R.J., Rebhun L.I., Frankfurter A. 1991. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc. Natl. Acad. Sci. USA. 88:4685–4689 10.1073/pnas.88.11.4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu J.M. 2007. Large scale purification of brain tubulin with the modified Weisenberg procedure. Methods Mol. Med. 137:17–28 10.1007/978-1-59745-442-1_2 [DOI] [PubMed] [Google Scholar]
- Arce C.A., Rodriguez J.A., Barra H.S., Caputo R. 1975. Incorporation of L-tyrosine, L-phenylalanine and L-3,4-dihydroxyphenylalanine as single units into rat brain tubulin. Eur. J. Biochem. 59:145–149 10.1111/j.1432-1033.1975.tb02435.x [DOI] [PubMed] [Google Scholar]
- Badin-Larçon A.C., Boscheron C., Soleilhac J.M., Piel M., Mann C., Denarier E., Fourest-Lieuvin A., Lafanechère L., Bornens M., Job D. 2004. Suppression of nuclear oscillations in Saccharomyces cerevisiae expressing Glu tubulin. Proc. Natl. Acad. Sci. USA. 101:5577–5582 10.1073/pnas.0307917101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Roach M.C., Wall K.A., Lopata M.A., Cleveland D.W., Ludueña R.F. 1988. A monoclonal antibody against the type II isotype of beta-tubulin. Preparation of isotypically altered tubulin. J. Biol. Chem. 263:3029–3034 [PubMed] [Google Scholar]
- Banker G., Goslin K. 1988. Developments in neuronal cell culture. Nature. 336:185–186 10.1038/336185a0 [DOI] [PubMed] [Google Scholar]
- Barbier P., Dorléans A., Devred F., Sanz L., Allegro D., Alfonso C., Knossow M., Peyrot V., Andreu J.M. 2010. Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers. J. Biol. Chem. 285:31672–31681 10.1074/jbc.M110.141929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra H.S., Arce C.A., Argaraña C.E. 1988. Posttranslational tyrosination/detyrosination of tubulin. Mol. Neurobiol. 2:133–153 10.1007/BF02935343 [DOI] [PubMed] [Google Scholar]
- Berezniuk I., Vu H.T., Lyons P.J., Sironi J.J., Xiao H., Burd B., Setou M., Angeletti R.H., Ikegami K., Fricker L.D. 2012. Cytosolic carboxypeptidase 1 is involved in processing α- and β-tubulin. J. Biol. Chem. 287:6503–6517 10.1074/jbc.M111.309138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I.A., van Dijk J., Janke C., Surrey T. 2008. CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183:1223–1233 10.1083/jcb.200809190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- Buey R.M., Díaz J.F., Andreu J.M. 2006. The nucleotide switch of tubulin and microtubule assembly: a polymerization-driven structural change. Biochemistry. 45:5933–5938 10.1021/bi060334m [DOI] [PubMed] [Google Scholar]
- Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32(Web Server issue):W615– W619 10.1093/nar/gkh398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorleans A., Knossow M., Gigant B. 2007. Studying drug-tubulin interactions by X-ray crystallography. Methods Mol. Med. 137:235–243 10.1007/978-1-59745-442-1_16 [DOI] [PubMed] [Google Scholar]
- Dunn S., Morrison E.E., Liverpool T.B., Molina-París C., Cross R.A., Alonso M.C., Peckham M. 2008. Differential trafficking of Kif5c on tyrosinated and detyrosinated microtubules in live cells. J. Cell Sci. 121:1085–1095 10.1242/jcs.026492 [DOI] [PubMed] [Google Scholar]
- Eddé B., Rossier J., Le Caer J.P., Desbruyères E., Gros F., Denoulet P. 1990. Posttranslational glutamylation of alpha-tubulin. Science. 247:83–85 10.1126/science.1967194 [DOI] [PubMed] [Google Scholar]
- Eddé B., Rossier J., Le Caer J.P., Promé J.C., Desbruyères E., Gros F., Denoulet P. 1992. Polyglutamylated alpha-tubulin can enter the tyrosination/detyrosination cycle. Biochemistry. 31:403–410 10.1021/bi00117a014 [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Erck C., Peris L., Andrieux A., Meissirel C., Gruber A.D., Vernet M., Schweitzer A., Saoudi Y., Pointu H., Bosc C., et al. 2005. A vital role of tubulin-tyrosine-ligase for neuronal organization. Proc. Natl. Acad. Sci. USA. 102:7853–7858 10.1073/pnas.0409626102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersfeld K., Wehland J., Plessmann U., Dodemont H., Gerke V., Weber K. 1993. Characterization of the tubulin-tyrosine ligase. J. Cell Biol. 120:725–732 10.1083/jcb.120.3.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Moews P.C., Walsh C.T., Knox J.R. 1994. Vancomycin resistance: structure of D-alanine:D-alanine ligase at 2.3 A resolution. Science. 266:439–443 10.1126/science.7939684 [DOI] [PubMed] [Google Scholar]
- Field J.J., Singh A.J., Kanakkanthara A., Halafihi T., Northcote P.T., Miller J.H. 2009. Microtubule-stabilizing activity of zampanolide, a potent macrolide isolated from the Tongan marine sponge Cacospongia mycofijiensis. J. Med. Chem. 52:7328–7332 10.1021/jm901249g [DOI] [PubMed] [Google Scholar]
- Galperin M.Y., Koonin E.V. 1997. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 6:2639–2643 10.1002/pro.5560061218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Verhey K.J. 2008. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20:71–76 10.1016/j.ceb.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Kato H., Katsube Y., Oda J. 1996. A pseudo-Michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution. Biochemistry. 35:11967–11974 10.1021/bi9605245 [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Bradke F. 2009. Control of neuronal polarity and plasticity—a renaissance for microtubules? Trends Cell Biol. 19:669–676 10.1016/j.tcb.2009.08.006 [DOI] [PubMed] [Google Scholar]
- Howard J., Hyman A.A. 2003. Dynamics and mechanics of the microtubule plus end. Nature. 422:753–758 10.1038/nature01600 [DOI] [PubMed] [Google Scholar]
- Idriss H.T. 2000. Phosphorylation of tubulin tyrosine ligase: a potential mechanism for regulation of alpha-tubulin tyrosination. Cell Motil. Cytoskeleton. 46:1–5 [DOI] [PubMed] [Google Scholar]
- Ikegami K., Mukai M., Tsuchida J., Heier R.L., Macgregor G.R., Setou M. 2006. TTLL7 is a mammalian beta-tubulin polyglutamylase required for growth of MAP2-positive neurites. J. Biol. Chem. 281:30707–30716 10.1074/jbc.M603984200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante A.S., Stein M.S., Zhai Y., Borisy G.G., Gundersen G.G. 2000. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J. Cell Sci. 113:3907–3919 [DOI] [PubMed] [Google Scholar]
- Janke C., Bulinski J.C. 2011. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 12:773–786 10.1038/nrm3227 [DOI] [PubMed] [Google Scholar]
- Janke C., Rogowski K., Wloga D., Regnard C., Kajava A.V., Strub J.M., Temurak N., van Dijk J., Boucher D., van Dorsselaer A., et al. 2005. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 308:1758–1762 10.1126/science.1113010 [DOI] [PubMed] [Google Scholar]
- Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S., Ludin B., Matus A. 1996. Cytoskeletal plasticity in cells expressing neuronal microtubule-associated proteins. Neuron. 17:1189–1199 10.1016/S0896-6273(00)80249-4 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Yau K.W., Hoogenraad C.C. 2010. Microtubule dynamics in dendritic spines. Methods Cell Biol. 97:111–132 10.1016/S0091-679X(10)97007-6 [DOI] [PubMed] [Google Scholar]
- Kato C., Miyazaki K., Nakagawa A., Ohira M., Nakamura Y., Ozaki T., Imai T., Nakagawara A. 2004. Low expression of human tubulin tyrosine ligase and suppressed tubulin tyrosination/detyrosination cycle are associated with impaired neuronal differentiation in neuroblastomas with poor prognosis. Int. J. Cancer. 112:365–375 10.1002/ijc.20431 [DOI] [PubMed] [Google Scholar]
- Kimura Y., Kurabe N., Ikegami K., Tsutsumi K., Konishi Y., Kaplan O.I., Kunitomo H., Iino Y., Blacque O.E., Setou M. 2010. Identification of tubulin deglutamylase among Caenorhabditis elegans and mammalian cytosolic carboxypeptidases (CCPs). J. Biol. Chem. 285:22936–22941 10.1074/jbc.C110.128280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y., Setou M. 2009. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12:559–567 10.1038/nn.2314 [DOI] [PubMed] [Google Scholar]
- Konno A., Setou M., Ikegami K. 2012. Ciliary and flagellar structure and function—their regulations by posttranslational modifications of axonemal tubulin. Int Rev Cell Mol Biol. 294:133–170 10.1016/B978-0-12-394305-7.00003-3 [DOI] [PubMed] [Google Scholar]
- Kreitzer G., Liao G., Gundersen G.G. 1999. Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Mol. Biol. Cell. 10:1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.A., Pilz D.T., Babatz T.D., Cushion T.D., Harvey K., Topf M., Yates L., Robb S., Uyanik G., Mancini G.M., et al. 2010. TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Hum. Mol. Genet. 19:2817–2827 10.1093/hmg/ddq182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., Janke C. 2011. Generation of differentially polyglutamylated microtubules. Methods Mol. Biol. 777:57–69 10.1007/978-1-61779-252-6_4 [DOI] [PubMed] [Google Scholar]
- Lafanechère L., Courtay-Cahen C., Kawakami T., Jacrot M., Rüdiger M., Wehland J., Job D., Margolis R.L. 1998. Suppression of tubulin tyrosine ligase during tumor growth. J. Cell Sci. 111:171–181 [DOI] [PubMed] [Google Scholar]
- Löwe J., Li H., Downing K.H., Nogales E. 2001. Refined structure of alpha beta-tubulin at 3.5 A resolution. J. Mol. Biol. 313:1045–1057 10.1006/jmbi.2001.5077 [DOI] [PubMed] [Google Scholar]
- Ludueña R.F. 1993. Are tubulin isotypes functionally significant. Mol. Biol. Cell. 4:445–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mary J., Redeker V., Le Caer J.P., Promé J.C., Rossier J. 1994. Class I and IVa beta-tubulin isotypes expressed in adult mouse brain are glutamylated. FEBS Lett. 353:89–94 10.1016/0014-5793(94)01018-8 [DOI] [PubMed] [Google Scholar]
- Mialhe A., Lafanechère L., Treilleux I., Peloux N., Dumontet C., Brémond A., Panh M.H., Payan R., Wehland J., Margolis R.L., Job D. 2001. Tubulin detyrosination is a frequent occurrence in breast cancers of poor prognosis. Cancer Res. 61:5024–5027 [PubMed] [Google Scholar]
- Murofushi H. 1980. Purification and characterization of tubulin-tyrosine ligase from porcine brain. J. Biochem. 87:979–984 [DOI] [PubMed] [Google Scholar]
- Nawrotek A., Knossow M., Gigant B. 2011. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J. Mol. Biol. 412:35–42 10.1016/j.jmb.2011.07.029 [DOI] [PubMed] [Google Scholar]
- Nogales E., Wolf S.G., Downing K.H. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 391:199–203 10.1038/34465 [DOI] [PubMed] [Google Scholar]
- Nogales E., Whittaker M., Milligan R.A., Downing K.H. 1999. High-resolution model of the microtubule. Cell. 96:79–88 10.1016/S0092-8674(00)80961-7 [DOI] [PubMed] [Google Scholar]
- Paturle L., Wehland J., Margolis R.L., Job D. 1989. Complete separation of tyrosinated, detyrosinated, and nontyrosinatable brain tubulin subpopulations using affinity chromatography. Biochemistry. 28:2698–2704 10.1021/bi00432a050 [DOI] [PubMed] [Google Scholar]
- Pecqueur L., Duellberg C., Dreier B., Jiang Q., Wang C., Plückthun A., Surrey T., Gigant B., Knossow M. 2012. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc. Natl. Acad. Sci. USA. 109:12011–12016 10.1073/pnas.1204129109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L., Thery M., Fauré J., Saoudi Y., Lafanechère L., Chilton J.K., Gordon-Weeks P., Galjart N., Bornens M., Wordeman L., et al. 2006. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J. Cell Biol. 174:839–849 10.1083/jcb.200512058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L., Wagenbach M., Lafanechère L., Brocard J., Moore A.T., Kozielski F., Job D., Wordeman L., Andrieux A. 2009. Motor-dependent microtubule disassembly driven by tubulin tyrosination. J. Cell Biol. 185:1159–1166 10.1083/jcb.200902142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K., Saillour Y., Fourniol F., Francis F., Souville I., Valence S., Desguerre I., Marie Lepage J., Boddaert N., Line Jacquemont M., et al. 2012. Expanding the spectrum of TUBA1A-related cortical dysgenesis to Polymicrogyria. Eur. J. Hum. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota A.E., Bargsten K., Zurwerra D., Field J.J., Díaz J.F., Altmann K.-H., Steinmetz M.O. 2013. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science. 10.1126/science.1230582 [DOI] [PubMed] [Google Scholar]
- Ravelli R.B., Gigant B., Curmi P.A., Jourdain I., Lachkar S., Sobel A., Knossow M. 2004. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 428:198–202 10.1038/nature02393 [DOI] [PubMed] [Google Scholar]
- Regnard C., Audebert S., Desbruyères P., Denoulet P., Eddé B. 1998. Tubulin polyglutamylase: partial purification and enzymatic properties. Biochemistry. 37:8395–8404 10.1021/bi9804131 [DOI] [PubMed] [Google Scholar]
- Rice L.M., Montabana E.A., Agard D.A. 2008. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. USA. 105:5378–5383 10.1073/pnas.0801155105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski K., Juge F., van Dijk J., Wloga D., Strub J.M., Levilliers N., Thomas D., Bré M.H., Van Dorsselaer A., Gaertig J., Janke C. 2009. Evolutionary divergence of enzymatic mechanisms for posttranslational polyglycylation. Cell. 137:1076–1087 10.1016/j.cell.2009.05.020 [DOI] [PubMed] [Google Scholar]
- Rogowski K., van Dijk J., Magiera M.M., Bosc C., Deloulme J.C., Bosson A., Peris L., Gold N.D., Lacroix B., Grau M.B., et al. 2010. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell. 143:564–578 10.1016/j.cell.2010.10.014 [DOI] [PubMed] [Google Scholar]
- Rüdiger M., Plessman U., Klöppel K.D., Wehland J., Weber K. 1992. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 308:101–105 10.1016/0014-5793(92)81061-P [DOI] [PubMed] [Google Scholar]
- Rüdiger M., Wehland J., Weber K. 1994. The carboxy-terminal peptide of detyrosinated alpha tubulin provides a minimal system to study the substrate specificity of tubulin-tyrosine ligase. Eur. J. Biochem. 220:309–320 10.1111/j.1432-1033.1994.tb18627.x [DOI] [PubMed] [Google Scholar]
- Schmitz S.K., Hjorth J.J., Joemai R.M., Wijntjes R., Eijgenraam S., de Bruijn P., Georgiou C., de Jong A.P., van Ooyen A., Verhage M., et al. 2011. Automated analysis of neuronal morphology, synapse number and synaptic recruitment. J. Neurosci. Methods. 195:185–193 10.1016/j.jneumeth.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Steinmetz M.O., Akhmanova A. 2008. Capturing protein tails by CAP-Gly domains. Trends Biochem. Sci. 33:535–545 10.1016/j.tibs.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Szyk A., Deaconescu A.M., Piszczek G., Roll-Mecak A. 2011. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat. Struct. Mol. Biol. 18:1250–1258 10.1038/nsmb.2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield M.A., Cederquist G.Y., Gupta M.L., Jr, Engle E.C. 2011. Phenotypic spectrum of the tubulin-related disorders and functional implications of disease-causing mutations. Curr. Opin. Genet. Dev. 21:286–294 10.1016/j.gde.2011.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk J., Rogowski K., Miro J., Lacroix B., Eddé B., Janke C. 2007. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol. Cell. 26:437–448 10.1016/j.molcel.2007.04.012 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Gaertig J. 2007. The tubulin code. Cell Cycle. 6:2152–2160 10.4161/cc.6.17.4633 [DOI] [PubMed] [Google Scholar]
- Wehland J., Weber K. 1987. Tubulin-tyrosine ligase has a binding site on beta-tubulin: a two-domain structure of the enzyme. J. Cell Biol. 104:1059–1067 10.1083/jcb.104.4.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Weber K. 2003. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 4:938–947 10.1038/nrm1260 [DOI] [PubMed] [Google Scholar]
- Whipple R.A., Matrone M.A., Cho E.H., Balzer E.M., Vitolo M.I., Yoon J.R., Ioffe O.B., Tuttle K.C., Yang J., Martin S.S. 2010. Epithelial-to-mesenchymal transition promotes tubulin detyrosination and microtentacles that enhance endothelial engagement. Cancer Res. 70:8127–8137 10.1158/0008-5472.CAN-09-4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D., Webster D.M., Rogowski K., Bré M.H., Levilliers N., Jerka-Dziadosz M., Janke C., Dougan S.T., Gaertig J. 2009. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev. Cell. 16:867–876 10.1016/j.devcel.2009.04.008 [DOI] [PubMed] [Google Scholar]
- Yuan B., Latek R., Hossbach M., Tuschl T., Lewitter F. 2004. siRNA Selection Server: an automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 32(Web Server issue):W130– W134 10.1093/nar/gkh366 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.