Abstract
In natural populations of the human parasite Schistosoma mansoni, parasite distribution among snail intermediate hosts is generally overdispersed, such that a small proportion of hosts harbor the majority of parasite genotypes. Within these few infected snails, researchers have found that it can be common for hosts to harbor multiple parasite genotypes, creating circumstances in which co-infecting parasites are faced with potential competition over limited host resources. Much theoretical modeling has focused on parasite competition, especially regarding the influence of co-infection on parasite exploitation strategy evolution. However, particularly in the case of intra-molluscan intermediate stages, empirical investigations of parasite-parasite competition have often hinged on the untested assumption that co-exposure produces co-infection. That is, infected hosts exposed to multiple strains have been assumed to harbor multiple strains, regardless of the true nature of the infection outcome. Here we describe a real-time quantitative PCR method to distinguish the conditions of multiple- versus single-strain infection, as well as quantify the relative larval output of co-infecting strains. We applied the method to an empirical investigation of intraspecific parasite competition between S. mansoni strains within the intermediate snail host Biomphalaria glabrata, assessing co-exposure's effects on parasite infectivity and productivity and the concomitant effects on host fitness. Overall, there was no effect of parasite co-infection on snail life history traits relative to single-strain infection. Parasite infectivity significantly increased as a result of increasing overall miracidial dose, rather than co-exposure, though strain-specific productivity was significantly reduced in co-infections in manner consistent with resource competition. Moreover, we show that less than half of infected, co-exposed hosts had patent co-infections and demonstrate the utility of this molecular tool for the study of trematode life history variation in molluscan hosts.
Keywords: parasite competition, trematodes, snails, real-time quantitative PCR, molecular detection
1. Introduction
Schistosoma mansoni is a parasite of global public health importance and, as such, has received considerable attention in both laboratory and natural field environments. Field assessments of natural genetic variation have indicated that S. mansoni genotypes tend to be overdispersed (i.e. exhibit a negative binomial distribution) in natural host populations, such that a small proportion of the hosts harbor the majority of the parasite genotypes [1–5]. Within the infected minority, the prevalence of snails hosting multiple genetic variants of parasites has been shown to be relatively high. Minchella et al. [2] found that over half of the infected B. glabrata collected from a Brazilian field site were infected by multiple miracidia, and Eppert et al. [3] showed in two independent collections of Brazilian B. glabrata that 60% and 52.2% of the infected individuals harbored multiple parasite genotypes. Because host resources are presumably finite, it is expected that these multiple parasites compete for the limited resources of their host. Evidence of direct antagonism among co-infecting parasites within molluscan hosts has been most easily observed in situations where at least one of the infecting larval stages is actively motile and predatory on other parasites in the host tissue (e.g. the redial stage of echinostomes; [6,7]). However, competition between non-predatory larval stages has also been observed, where co-infection of B. glabrata by both S. mansoni and the strigeid Cotylurus lutzi consistently resulted in degeneration of the C. lutzi sporocyst [6].
The degree of competition in multiple infection scenarios has been the subject of much theoretical modeling, particularly concerning how infection by different genetic variants of a single parasite species influences the evolution of parasite exploitation strategies (e.g. [8–13]). Infections established by a single parasite genotype are predicted to select for strategies that maximize transmission while mitigating host morbidity and/or mortality relative to co-infections with dissimilar genotypes, where increased resource competition is expected to select for more aggressive host exploitation [10].
Although theoretical attention has focused on predicting the evolutionary trajectories of parasite exploitation strategies, empirical support is limited. This is particularly true in snail-trematode systems like that of B. glabrata and S. mansoni, where absolute enumeration of infecting parasite larvae is thus far only possible at early stages of infection via host dissection. Several studies within the snail-trematode system have attempted to demonstrate that mixed-strain infections increase parasite reproduction rates (as measured by overall propagule output) and decrease host fitness, presumably as a result of increased exploitation by the competing parasite strains [14–16]. However, definitive conclusion of competition in these studies (save for [15]) is constrained by the fact that 1) infection status has been limited to a simple infected/non-infected binary, rather than a true determination of establishment by multiple parasite strains, and 2) none have clearly demonstrated that either strain of co-infecting parasite is suffering reduced fitness. By employing molecular techniques to differentiate between propagules of multiple strains within a host, we can verify successful co-infection by multiple strains and more rigorously detect and quantify the occurrence of within-host competition among parasite strains. In particular, the evolutionarily significant assessment of how or if the exploitation strategies of parasites change in response to the presence of co-infecting strains and how those changes relate to genetic distance among the strains and host condition or genotype will not only test current models, but also allow for the creation of more realistic models.
Cheesman et al. [17] validated the powerful molecular tool of real-time quantitative PCR (qPCR) as a reliable means of distinguishing and quantifying genetically distinct clones in mixed infections of the rodent malaria parasite Plasmodium chabaudi chabaudi. Numerous studies have since applied qPCR to demonstrate differential reproduction rates among co-infecting strains of Plasmodium and other intracellular parasites and to investigate the influence of various host and parasite factors on life history outcomes [18–24]. Here we describe the adaptation of allele-specific qPCR for the purpose of assessing the influence of intra-molluscan, intraspecific parasite competition on parasite exploitation strategies in a macroparasitic system, specifically the snail-trematode system of Biomphalaria glabrata and Schistosoma mansoni. Allele-specific primers targeting strain-specific single nucleotide polymorphisms (SNPs) in two well-known laboratory parasite strains (NMRI and PR) were developed, validated, and applied in a study investigating the effects of co-exposure on parasite infectivity and co-infection on parasite and snail host life history.
2. Methods
2.1 Parasite Strains and Development of Allele-Specific Primers
The S. mansoni lab strains NMRI and PR were utilized to develop discriminatory molecular markers and for the subsequent infection experiment. Both strains have been maintained in the laboratory mouse-snail life cycle for well over twenty years as isolated inbred lines. Genomic DNA from twenty adult worms (10 each of male and female) was extracted using a modified Puregene extraction method. Briefly, individual worms were digested in 100μL of cell lysis solution (100mM Tris-HCl, 10mM EDTA, 100mM NaCl, 1% SDS, 0.06mg Proteinase K, 1.5mM dithiothreitol) for 2 hours at 55°C, followed by protein precipitation with 30% v/v of 10M potassium acetate. DNA was separated from the supernatant via standard ethanol precipitation, dried, resuspended in 20 μL sterile nuclease-free water, and quantified with a NanoDrop 1000 (ThermoFisher Scientific). DNA from each worm was sequenced (forward and reverse) with BigDye Terminator v3.1 chemistry on a 3130xl automated sequencer (Applied Biosystems) over 500–800 bp corresponding to cytochrome oxidase subunit I (COI), cathepsin B1 isotype 1, NADH dehydrogenase subunits 1 and 5 (ND1 and 5), and NADH-ubiquinone oxidoreductase chain. Trace files were examined manually in SequenceScanner 1.0 (Applied Biosystems) to trim sequence ends and mask calls with low quality and/or apparent heterozygosity. Curated forward and reverse sequences from each worm were aligned with the free alignment software MEGA v5.0 to identify strain-specific polymorphisms. COI and cathepsin B1 isotype 1 produced the most consistently strain-specific variation among repeated sequencing reactions, with over 10 strain-specific single nucleotide polymorphisms (SNPs) in each, and were used for subsequent allele-specific primer design.
Allele-specific primers targeting strain-specific COI and cathepsin B1 SNPs were designed with the aim of maximizing discrimination between strains and minimizing non-specific priming. For each SNP, two forward primers were designed such that the ultimate 3' nucleotide was complementary to the polymorphic nucleotide in a strain-specific fashion (e.g. primers specific to NMRI SNP = G and PR SNP = T would end in C and A, respectively). Additionally, the penultimate 3' nucleotides were designed to be completely non-complementary to the target sequence of either strain, using the discrimination profiles reported by Li et al. [25] (Figure 1). Thus, in the event that a primer sits down on non-target DNA, the additional penultimate mismatch augments discrimination against non-specific priming relative to discrimination achieved with only a single mismatch at the targeted SNP and without significantly hampering priming of the specific target [25]. To avoid reduced amplification efficiency during real-time PCR, “universal” reverse primers for each target SNP were designed so that total amplicon length would not exceed 150bp. Primers with low self- and hetero-dimerization tendencies and with Tm ~60°C were picked using the online primer development tool Primer3Plus [26] (www.bioinformatics.nl/cgibin/primer3plus/primer3plus.cgi).
Figure 1.
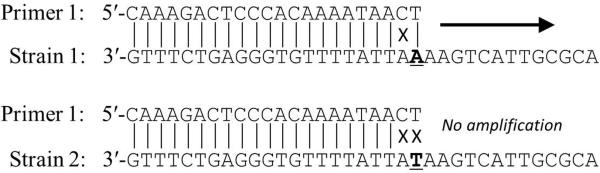
Schematic of allele-specific priming in the presence of a mismatch of the 3' penultimate nucleotide. Bold text indicates strain-specific polymorphic nucleotides.
The specificity of each primer was tested in real-time amplification reactions using 5–10ng of DNA extracted from individual worms of each strain. A primer was deemed non-specific and discarded from further consideration if more than 10% of reactions with non-target DNA resulted in cycle thresholds (CT) of 35 or lower or if post-cycling melt curve analysis indicated the presence of multiple amplification products. Real-time PCR and melt curve analyses were performed with PowerSYBR Green MasterMix chemistry on a StepOnePlus thermocycler (Applied Biosystems). Following an initial 10-minute denaturation and Taq activation at 95°C, reactions were cycled as follows, using a “stepdown” scheme to encourage reaction stringency: 2 cycles of 15s at 95°C and 60s at 60°C, 2 cycles of 15s at 95°C and 60s at 59°C, and 40 cycles of 15s at 95°C and 60s at 58°C. After the final cycle, reactions were brought down to 45°C and the temperature was increased slowly back up to 95°C for the melt curve analysis. Fluorescence levels of the SYBR Green dye relative to ROX passive reference dye were assessed in real time at each 60-second annealing step during thermal cycling and then again with every 0.3°C increase during the melt curve analysis. Primers specific to an A/T SNP on COI were chosen based on their ability to reliably discriminate the NMRI and PR parasite strains (Table 1).
Table 1.
Sequences, melting temperatures, and amplicon lengths of the COI SNP-specific primers selected for parasite strain discrimination and quantification. Bold text denotes bases conferring specific priming – the penultimate base is a mismatch to either strain and confers additional specificity in the event that the final base, which is specific to one strain or the other, is not complementary.
| Target | Sequence | Tm(°C) | Amplicon Length (bp) |
|---|---|---|---|
| NMRI-specific primer | 5' CAA AGA CTC CCA CAA AAT AAC T | 59.4 | 127 |
| PR-specific primer | 5' CCA AAG ACT CCC ATA AAA TAA GA | 59 | 128 |
| Reverse primer | 5' TGG TCT TCC TCG TCG TGT AA | 59.3 |
2.2 Strain Quantification Validation
On the one hand, the ability to simply detect the presence of multiple strains of parasite larvae within a single snail can be useful for experimental infections, particularly for confirming the assumption that exposure to multiple strains produces a multiple strain infection. Such confirmation of co-infection would not require the use of either real-time or quantitative PCR. However, the ability to reliably quantify relative proportions of the larvae from each strain can allow more nuanced investigations of coevolutionary theory. In order to verify that real-time PCR with the selected allele-specific primers would allow for realistic quantification of larval proportions, mixes of NMRI and PR cercariae were prepared in known proportions. Trials with standard “dilutions” of whole cercariae indicated that reactions experienced inhibition above 24–30 cercariae. Thus, to maintain a window for error, validation experiments were performed with a total of 12 cercariae, with each strain in proportions of 1:6, 1:3, 1:1.
Cercariae were counted under a dissecting microscope and transferred in 2–10μL of distilled water to the bottom of a well on a 96-well plate. After all cercariae were added, the plate was wrapped in a Kimwipe to prevent debris entry and left for 3–6 hours in a drying oven at 37°C to evaporate the water. DNA was extracted from dried samples with 25μL of a Chelex® 100 solution (6% Chelex®100, 0.2mg/mL proteinase K), following the method described in Valentim et al. [27]. Briefly, after addition of the Chelex® solution, samples were incubated for 2 hours at 56°C and 8 minutes at 99°C, with gentle vortexing after each incubation. Samples were cooled to room temperature, condensation was spun down via quick centrifugation, and the supernatant was transferred to a clean 96-well plate for storage at −20°C and subsequent use.
To quantify the relative proportion of each parasite strain in a sample, 10μL reactions containing 1μM of one forward primer and the reverse primer, 1× PowerSYBR Green MasterMix (Applied Biosystems), and 1μL of extracted cercarial DNA were amplified in real-time as described above. To allow for differences in amplification efficiency, the standard curve method was used to quantify samples. Cercariae from each strain were counted into individual pools of 1, 3, 6, 12, and 24 to represent a serial “dilution” for creating a strain-specific standard curve during each real-time PCR run. After samples were amplified in the presence of each allele-specific primer, the standard curve-derived quantities were summed and the proportion of each strain was calculated and compared to expected proportions.
2.3 Experimental Infections
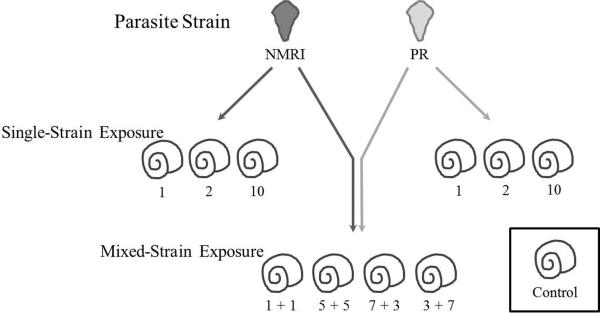
The COI allele-specific primers were used to assess the snail life history and parasite transmission outcomes of intraspecific co-infection by S. mansoni in its intermediate snail host, B. glabrata. Groups of 50 size-matched snails (5–8 mm) were assigned to one of 11 treatments – 1 control group, 6 single-strain infections, and 4 mixed-strain infections. The single-strain treatment included a group of monomiracidial exposure for each strain, and both single- and mixed-strain treatments included exposure to two or ten miracidia, with both even and uneven proportions of each strain represented in mixed-strain treatments (Figure 2). NMRI and PR strain miracidia were isolated from infected mouse livers obtained from the Biomedical Research Institute (BRI; Rockville, MD) immediately prior to snail exposure. Eggs were obtained by macerating livers in a 1.2% NaCl solution with a Waring blender, filtering the resulting slurry through a 500 micron polyvinyl mesh to remove remaining whole tissue, and then passing the filtrate through a 50 micron polyvinyl mesh to retain parasite eggs. The filter and eggs were placed in fresh water in a large petri dish under lights to induce hatching of miracidia.
Figure 2.
Experimental parasite exposure treatments. Numbers below the snails represent the number of miracidia to which snails were exposed, with all mixed-strain exposures being simultaneous. Each group consisted of 50 randomly selected, size-matched juvenile snails (5–8mm in diameter). The control group underwent identical handling as experimental groups, minus exposure to miracidia.
For exposure, snails were placed in individual wells of a 24-well cell culture plate in approximately 2 mL of water each. Given the large numbers of groups and snails and the time necessary to count out miracidia for each exposure, there is a very real possibility of diminishing miracidial quality/viability over the course of the infection procedure. To spread this risk and its effects evenly over treatment classes, exposures for each group where performed in “passes” of a single 4-well row at a time before moving on to the next treatment group. The order in which groups were exposed was random for each pass. Furthermore, the order in which strains were added to “competition” wells was random among passes to control for effects of the order of parasite penetration into the snail head/foot. Miracidia were counted and collected under a dissecting microscope and transferred to individual snail wells in 10μL of water. To ensure complete and accurate transfer, release of the miracidia into the well was observed under a second dissecting microscope. Snails remained in their individual wells with miracidia for 6 hours before being transferred to individual 9-oz jars labeled with distinct snail identification numbers, where they were kept for the next 10 weeks under a 14/10-hour light/dark cycle, fed lettuce ad libitum, and provided with Styrofoam discs to serve as egg-laying substrate. Control snails underwent the same physical treatment and isolation in wells and jars as exposed snails, minus the exposure to miracidia. During both experimental manipulation and snail maintenance, we utilized non-chlorinated well water obtained from a well at Purdue University (40°25'20”N, 86°55'3”W).
Size, reproduction, and mortality of snails were assessed weekly to investigate effects of co-infection on snail life history. Size was measured across the widest portion of the shell with Vernier calipers and reproduction was recorded as total number of eggs deposited for the week. Five weeks post-exposure and every week thereafter, snails were assessed for infection status. To induce shedding of mature cercariae, snails were placed in individual wells of 24-well cell culture plates in 2mL of non-chlorinated well water and exposed to fluorescent light for 3 hours. After shedding, snails were returned to their jars and, for infected snails, the 2mL of water containing cercariae was transferred to two 1.5mL microcentrifuge tubes labeled with the associated snail ID number and stored at −80°C for future analyses.
To assess parasite productivity (as number of cercariae released) the contents of each pair of 1.5mL microcentrifuge tubes were mixed in a small, 6mL borosilicate glass tube, where uniform suspension of cercariae was maintained with gentle agitation. Four 50μL aliquots of the suspension were transferred to individual wells of a spot plate and cercariae were stained with a 1:10 dilution of Lugol's iodine. Number of cercariae per 50μL aliquot were counted under a dissecting microscope, averaged, and then multiplied by 40 to extrapolate the total number of cercariae per 2mL of water. This was done for all cercarial samples, regardless of treatment class (i.e. single- or mixed-strain snail exposure). For mixed-strain treatments, cercariae collected at weeks 5 and 7 post-exposure were subjected to real-time qPCR assessment of strain proportion relative to the total cercarial output. (Week 5 was selected to represent early patency and week 7 provided a second time point in which snail mortality had not reduced treatment numbers excessively.) Three aliquots per snail of ten to twenty cercariae were separated out at random under a dissecting microscope and transferred to a 96-well plate in 10–20μL of distilled water. Water was evaporated, DNA extracted, and real-time qPCR was performed on each sample as described above, with each cercarial aliquot amplified in duplicate for each allele-specific primer. In all qPCR runs, negative controls composed of Chelex®-extracted cercaria water (without cercariae) and DNA from the alternative strain (to verify continued absence of cross-reactivity) were also run in duplicate to verify lack of contamination by snail factors (e.g. exudate and/or feces) and primer specificity, respectively.
2.4 Statistical Analysis
Snail infection status and reproductive status were treated as binary variables (i.e. infected vs. non-infected or reproductive vs. non-reproductive) and binary logistic regression and cross-tabulation were used to compare binary outcomes across and within treatment classes (e.g. infection prevalence or snail reproductive status across treatments). To correct for non-normal distribution, parasite productivity (number of cercariae) was square root-transformed and snail reproduction (number of eggs) was log-transformed. Effects of treatment group, parasite strain, and infection status on mean outcomes (snail growth, reproduction, parasite productivity) were assessed with two-way ANOVA. Assessments of within infection or reproductive classes (e.g. only infected or reproductive snails considered) were evaluated with one-way analysis of variance (ANOVA). Significant results were followed with post hoc pairwise comparisons subjected to the Bonferroni correction for multiple comparisons. Independent pairwise comparisons were assessed with the parametric Student's t statistic. Finally, Kaplan-Meier survivorship analysis was used to assess the influence of exposure treatment, infection status, infecting strain, and mixed- versus single-strain infection profiles on survival of snails over the 10-week period. All statistics were performed with IBM® SPSS® Statistics release 19.0.0.1.
3. Results
3.1 Allele-Specific Primer Validation
To evaluate both primer specificity and the quantitative power of the qPCR for reliable estimation of relative strain proportions, known mixes of NMRI and PR cercariae were subjected to Chelex® DNA extraction and qPCR with the chosen COI SNP-specific primers. In all tested mixes, observed proportions did not differ significantly from expectations (χ21 0.76 − 0.96, p ≥ 0.25; Table 2). Non-template and non-specific strain negative controls in all runs verified lack of snail-related contamination and confirmed primer specificity.
Table 2.
Expected and observed proportions of known cercarial mixes to validate qPCR as a means of discriminating between and quantifying parasite strains. Observed cercarial quantities were tested against expected quantities with Chi-square analysis.
| Expected Proportion | Mean Observed Proportion | ||||
|---|---|---|---|---|---|
|
|
|||||
| NMRI:PR cercariae | NMRI | PR | NMRI | PR | p-value |
| 10:2 | 0.83 | 0.17 | 0.91 | 0.09 | 0.84 |
| 8:4 | 0.67 | 0.33 | 0.55 | 0.45 | 0.80 |
| 6:6 | 0.50 | 0.50 | 0.35 | 0.65 | 0.76 |
| 4:8 | 0.33 | 0.67 | 0.36 | 0.64 | 0.96 |
| 2:10 | 0.17 | 0.83 | 0.11 | 0.89 | 0.89 |
3.2 Infection Outcomes
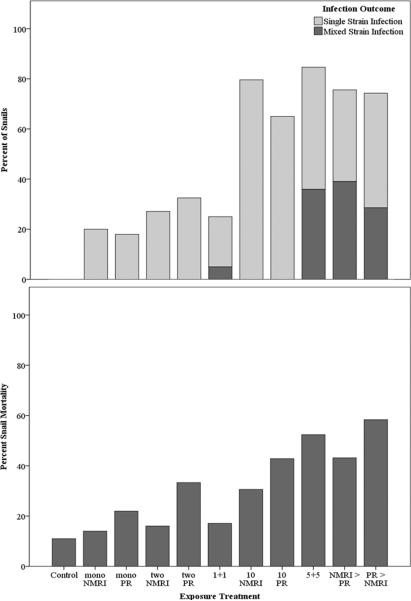
The probability of infection increased significantly with increasing miracidial dosage (Wald1 = 101.36, p < 0.001), regardless of whether exposure was to single or mixed parasite strains (in both cases Wald1 > 30.00, p < 0.001) (Figure 3). Over 70% of snails exposed to ten miracidia became infected, while less than 30% and 20% of snails exposed to two or one miracidia, respectively, developed patent infections. Average infection prevalence did not differ between groups of snails exposed to either the NMRI or PR parasite strain alone (NMRI: 42.2% and PR: 38.7%; χ21 = 0.34, p = 0.56). Likewise, when controlling for dose, there was no effect of single versus mixed strain exposure on infection status as a simple infected/non-infected binary (2 miracidia χ21 = 0.28, p = 0.68; 10 miracidia χ21 = 0.75, p = 0.39). Nor there was an effect of miracidial proportion (i.e. even versus skewed strain ratios) on infection prevalence in mixed strain exposures (even: 84.6%, NMRI>PR: 75.6%, PR>NMRI: 74.3%; χ22 = 1.42, p = 0.49).
Figure 3.
Infection prevalence and percent snail mortality for each treatment group. Each treatment group consisted of 50 size-matched B. glabrata. snails.
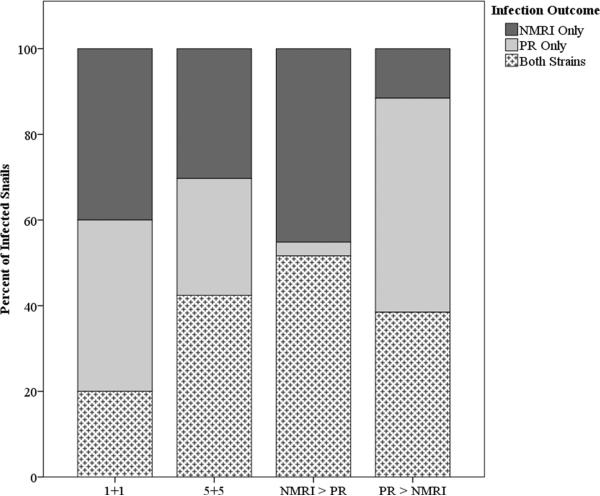
When qPCR was applied to determine the frequency with which mixed strain exposures produced mixed-strain infections, we found that overall only 42% of the infected snails harbored patent mixed strain infections. The remaining 58% of infected snails that had been exposed to both parasite strains were evenly split between the two strains (26% NMRI only and 32% PR only). However, the distribution of single- or mixed-strain infections was not independent across exposure schemes (χ26 = 19.87, p = 0.003), an outcome driven by skewed exposures. Snails exposed to even proportions of NMRI and PR miracidia shared single-strain infections evenly between the NMRI and PR strains, regardless of miracidial dose (χ22 = 1.67, p = 0.43) (Figure 4). Exposure with skewed strain proportions, on the other hand, produced significantly different outcomes (χ22 = 18.49, p < 0.001). In both skewed proportion scenarios (NMRI > PR and PR > NMRI), the outnumbered strain established sole infection less than 10% of the time (Figure 4), again pointing to the influenced of miracidial dose on infection outcomes.
Figure 4.
Percentages of co-exposed, infected snails that harbored patent single- and mixed-strain infections.
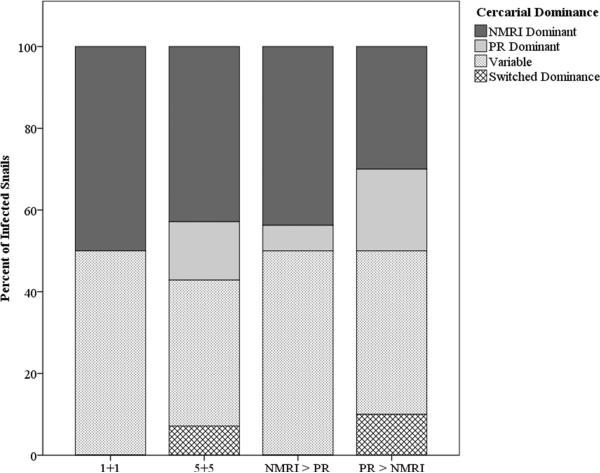
Utilizing qPCR, we were also able to observe different parasite reproduction dynamics by assessing the relative proportion of each strain in a sample of cercariae. Overall, NMRI was the dominant strain in 40.5% of confirmed mixed strain infections (i.e. NMRI parasites comprised greater than 70% of a cercarial mix), while PR was dominant in only 11.9% of mixed infections. Of the remainder, 42.9% had no clear trend of consistent dominance over the two weeks in which mixed-strain reproduction was measured and 4.8% of infections switched dominant strains from one week to the next, with no significant directional association to the dominance switching. This general trend held when co-infected snails were split according to miracidial proportion, with no significant difference in the dominance profile across exposure treatments (χ26 = 3.66, p = 0.93) (Figure 5). Within mixed-strain infections, the dominance profile did not appear to influence the total number of cercariae released from a snail (F4, 41 = 0.82, p = 0.52).
Figure 5.
Profiles of strain “dominance” in mixed strain infections. A strain was considered dominant when >70% of cercariae in a sample comprised that strain for both weeks in which cercarial proportions were assessed. “Variable” infections had no consistent patterns of dominance or non-dominance, while infections defined as “switched dominance” changed dominance from one week to the next. There was no directional trend associated with “switched dominance” infections and dominance profiles did not differ among exposure treatments (χ2 = 3.77, p = 0.88).
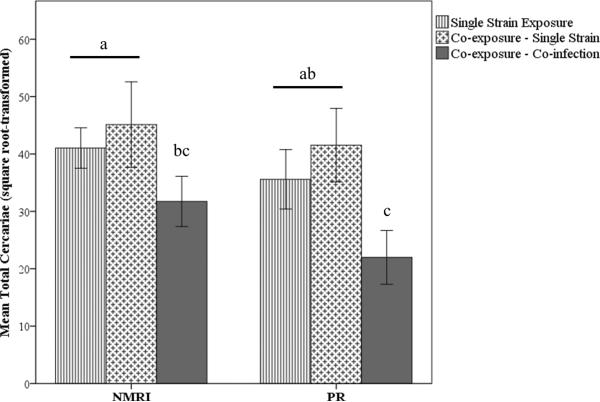
When considered across all treatments, the NMRI parasite strain had greater average total cercarial production than the PR strain (square root transformed, t238 = 3.30, p < 0.001), but the difference was not significant when the comparison was restricted to single strain infections (t156 = 1.81, p = 0.07). Overall, total cercarial production (i.e. not strain-specific) did not differ between single-strain and mixed-strain infections (t206 = −3.9, p = 0.70). Both parasite strains demonstrated a significant reduction in mean strain-specific cercarial production as a result of co-infection relative to single strain infection (F2, 240 = 20.37, p < 0.001). This was true regardless of whether the single strain infection resulted from a single- or mixed-strain exposure (Figure 6). Mean cercarial output (square root-transformed) differed significantly among miracidial doses (F2, 239 = 4.14, p = 0.02), with mean cercarial output following exposure to ten miracidia being significantly lower than exposure to two miracidia (p = 0.01), which could suggest resource competition among parasites. However, the difference among dosages was not significant when we controlled for the effect of mixed infections by removing individuals with established mixed infections (F2, 157 = 2.66, p = 0.07) and no pairwise comparisons were significantly different (all p > 0.14). Finally, there was no correlation between the mean number of cercariae produced (total across strains) and growth of the snail host, regardless of the single- or mixed-strain status of the infection (rall = 0.11, p = 0.19; rsingle = 0.08, p = 0.38; rmixed = 0.25, p = 0.17). Likewise, there was no correlation between mean total cercarial production and the reproductive output of the snail (rall = −0.20, p = 0.28; rsingle = −0.23, p = 0.25; rmixed = −0.18, p = 0.82).
Figure 6.
Mean square root-transformed cercarial productivity for each strain in established and confirmed single- versus mixed-strain infections. Different letters indicate significant differences at the α 0.05 level. Error bars = ± 2 S.E.
3.3 Snail Life History
Overall, the effects of S. mansoni infection on B. glabrata reproduction, growth, and survivorship were as expected. As with infection prevalence, there was a significant effect of miracidial dose on probability of snail mortality (Wald1 = 51.34, p < 0.001) (Figure 3). Relative to uninfected snails, infected snails were significantly less likely to reproduce (87.3% vs. 14.5%, respectively; Wald = 121.69, p < 0.001) and had significantly reduced growth (10.29mm ± 0.15 vs 8.16mm ± 0.19, respectively; t388 = 7.16, p < 0.001) and survivorship (95% vs 44%; χ21 = 189.13, p < 0.001). Pre-patent mortality was quite minimal (<4 snails) for any treatment group. Among reproductive snails, however, there was no difference in total reproduction (total egg output) between uninfected and infected snails (109.76 eggs ± 8.05 vs 77.87 eggs ± 14.14, respectively; t303 = 0.21, p = 0.84). In snails infected by only one strain of parasite, there was no difference between NMRI- or PR-infected snails in growth (9.58mm ± 0.24 vs 9.79mm ± 0.27, respectively; t221 = −0.59, p = 0.56), probability of reproduction (54.3% vs 45.2%; Wald = 2.71, p = 0.10), or total reproductive output (94.52 eggs ± 12.13 vs 112.11 eggs ± 16.26, respectively; t164 = −0.91, p = 0.36). Survivorship was significantly reduced, however, in PR-infected snails relative to NMRI-infected snails (59.9% vs 73.7%, respectively, χ21 = 8.48, p = 0.004). Finally, there was no effect of mixed- versus single-strain infection or parasite strain dominance on any of the snail life history parameters (all p ≥ 0.14).
4. Discussion
Real-time quantitative PCR with allele-specific primers was found to be a valid and reliable method for distinguishing S. mansoni larval strains and quantifying relative output of co-infecting parasites. While this method has been applied repeatedly for studies of strain competition within microparasite systems (e.g. murine malaria models [18−22]), it has not, to our knowledge, been applied to macroparasitic systems. Historically, most experiments assessing the effects of intraspecific macroparasite co-infection have had exposure as the only truly known effect for comparisons of mixed- versus single-strain treatments (though see [15]). Here we were able to verify the actual mixed- or single-strain status of established infections, as well as quantify relative parasite productivity in mixed infections and thereby take steps towards quantitatively assessing strain-specific co-infection outcomes, rather than relying solely on snail life history-based inferences.
Overall, there were several results of note. First, miracidial dose had a significant effect on infection outcomes, more so than miracidial strain diversity at the time of exposure. Second, we were able to show that mixed-strain exposure is not a reliable default predictor of mixed-strain infection. Of snails exposed simultaneously to NMRI and PR miracidia, less than half of infected snails harbored both parasite strains – a significant departure from any implicit assumption that infections produced by mixed-strain exposure are co-infections. Facilitation of infection has recently been reported in another trematode system [28], but we found no evidence here. In fact, when we control for dosage effects and calculate the probability of co-infection based on the individual strains' probabilities of infection, we find that observed co-infection rates correspond to expected co-infection rates. For example, the infection rates of single miracidia were 20% and 17.9% for NMRI and PR, respectively. The joint probability of infection by single miracidia of both strains (the product of the individual infection probabilities) is 3.6%. When snails were co-exposed to single miracidia of each strain, we observed rates of 5% coinfection. Compared to expected proportions, there was no significant difference (χ23 = 2.31, p = 0.51), indicating that the strains' infection success was not altered by the presence of another strain.
It is important to note that the term “strain” has been used to denominate two highly inbred lineages that were distinguished by 20 SNPs within a short span of the genome. As such, strain here should not be understood to imply genetic uniformity within each parasite lineage. In fact, the significant influence of miracidial dose would indicate otherwise. Regardless of the strain diversity of parasites, increasing miracidial numbers increased the probability that compatible snail-parasite combinations were formed. It has long been accepted that the host-parasite relationship encourages the genetic diversity of each, such that “…populations of hosts of unequal susceptibility are confronted by populations of parasites with unequal infectivity” [29]. There is now convincing evidence that snails are capable of mounting a specific response to particular parasite genetic variants and that this “compatibility polymorphism” is genetically based in both host and parasite [30−32]. Within S. mansoni, highly polymorphic elements associated with parasite immune response evasion have been identified [30] and have been associated with several genes located in areas of elevated recombination and with multiple known splice variants. Thus, even within presumably inbred, long-standing laboratory lines of snail and parasite, both retain the means to express immunological variability, keeping the influence of compatibility polymorphism alive and well. Moreover, this polymorphism essentially makes the outcome of each random pairing of parasite larva and snail a random event that is dependent upon whether the combination includes a suitable immunological match. The authors' personal experience with difficulties in establishing monomiracidial infections and this study's significantly reduced infection prevalence among snails exposed to one and two miracidia relative to ten miracidia support the idea that by limiting the number of miracidia in an exposure, one limits the probability of a suitable match and any subsequent infection. Moreover, this observation is supported by a much more comprehensive exploration of dose responses in lab and field parasite/snail strains, which also demonstrated that a simple host-parasite matching model created curves that closely mirrored observed dosage response curves [33]. As such, the probabilistic nature of this interaction may well explain the infection outcome trends observed here − e.g., why increased prevalence was associated with increased dosage and why single-strain infections produced by skewed mixed-strain exposures were very rarely established by the outnumbered strain. The observed power in numbers could reasonably be attributed not to either intraspecific antagonism or facilitation, but rather a simple “loading of the dice”.
By confirming co-infection and quantifying strain-specific reproductive output, this study also demonstrated a significant reduction in cercarial output from co-infections relative to single-strain infections. This is not surprising, given that there was no overall difference in total cercarial output between single- or mixed-strain infected snails. What is notable is the ability to show the strain-specific dynamics underlying the overall cercarial count. In this particular scenario we saw that both strains experienced a comparable reduction in reproductive output. On the one hand, this provides convincing support for the notion that co-infecting parasites are competing for limited resources, as (according to the classic definition of competition) both strains have suffered an apparent reproductive “hit” when sharing the same snail. Whether this outcome is due to direct inter-strain antagonism or a snail's energetic “wall” capping parasite reproductive exploitation, we cannot say. Although we were able to confirm establishment by two strains and attempted to control for parasite density by using uniform total miracidial doses across treatments, we still do not know the total number of miracidia that successfully established. Ideally, co-infections in the only instance of known miracidial establishment (those resulting from 1+1 exposure) could eliminate the concern of miracidial numbers, but the extremely low prevalence in that group precludes reliable assessment here. Thus, we are currently unable to eliminate the possibility that the apparent competition was simply a result of miracidial (and subsequently sporocyst) density, rather than a direct effect of competition or altered transmission strategies.
Finally, despite the utility of allele-specific qPCR in identifying and quantifying parasite strains and their relative rates of infection and reproduction, hosts are still part of the equation and we rely on measures of host life history in order to gain more complete understanding of whether competition for resources or adjustment to resource-use strategies is occurring. It was interesting to note that in this experiment snails seemed to suffer little, if any, negative effects that could be ascribed to the presence of co-infecting parasite strains. Infected snails in general experienced reduced growth, reproduction (as a binary event), and survivorship relative to uninfected snails (both control and exposed-uninfencted), but there was no effect of multiple infection on these traits.
Overall, we have demonstrated that there are useful empirical prospects presented by allele-specific qPCR. For example, it would be feasible to apply this method to investigations of parasite reproductive strategies at the sporocyst level and address correlations between strain-specific sporocyst quantities relative to strain-specific cercarial quantities. That said, there is still every reason to be cautious in key assumptions regarding infectivity and host-parasite life history outcomes and further steps should be taken to address these points of uncertainty. If we are to truly compare mixed- and single-strain exposure outcomes and rigorously test the theoretical predictions of virulence strategies and coevolutionary outcomes, we need to consider the verification of overall number of established miracidia, regardless of their strain. Binary markers such as the SNPs and the real-time PCR methodology used here are useful for general strain identification. However, both strain identification and assessment of more fine-scale genotypic variation (allowing for a better estimate of established miracidia quantities) could be achieved with a suite of hypervariable markers like microsatellites and/or a more complex multiplex of markers. The real-time PCR methodology used here would be an unwieldy tool for such an application, thanks to its severe multiplexing limitations. However, newer real-time platforms such as ICEPlex [34] have greatly improved multiplexing ability while simultaneously addressing the persistent real-time PCR concern of variable amplification efficiency. Application of this technology could build upon the work presented here with its ability to differentiate genetic variants as well as quantify the relative productivity of each variant. Even highly inbred and long-term laboratory strains such as PR and NMRI should provide enough genetic variation at highly variable markers to allow distinction of genetic variants [35] and schistosome microsatellites have proven amenable to multiplexing [36] and been successfully applied to pooled miracidia [37]. Though given the nature of their laboratory cycling, both the parasite and snail strains used here have regularly experienced multi-genotype infections over numerous generations. It will be interesting to assess the competitive interactions among genotypes in field-collected strains where such intensive exposures have been less likely.
Historically, one of the primary limitations to conclusions made by studies assessing intraspecific competition among the intermediate, asexually reproductive stages of trematode parasites has been the inability to confirm establishment by all strains to which snails were exposed. In these cases infection has been a binary outcome, with inferences of dosage effect and/or resource competition being made as a result of changes in host and/or parasite life history relative to controls and other treatment groups, rather than confirmation of either [e.g., 14]. With regards to confirming co-infection and subsequent strain-specific larval productivity, we have demonstrated here the utility of an allele-specific, quantitative protocol. This method was shown to perform well during validation with known mixes of parasite strains and was successfully implemented in an initial pilot study of the host and parasite life history effects of co-infection with one B. glabrata strain and two well-known S. mansoni strains. While the specific primers developed here will certainly be restricted in their broader applicability across diverse S. mansoni strains, necessitating re-design of primers appropriate to individual strains, it is not an insurmountable limitation, particularly for a well-described parasite like S. mansoni. Interrogation of only two short regions of the genome (COI and cathepsin B, ~2.3 kb) resulted in at least 20 SNPs available for potential primer development. Thus, even for application in less genetically described trematode species, we have shown here that use of general “universal primers” targeting a “barcoding” segment of the genome (i.e., COI) could easily provide the necessary variation for molecular strain discrimination.
Highlights
SNP-based RTQ-PCR to detect and distinguish competing S. mansoni strains.
Accurately identify the outcome of trematode co-exposure.
Less than half of infections following co-exposure established by both strains.
Quantify strain-specific productivity to show evidence of parasite competition.
Molecular tool for empirical assessments of coevolutionary theory.
Acknowledgments
The authors would like to thank Dr. Fred Lewis and the staff of the Biomedical Research Institute for providing the S. mansoni strains. We also thank Drs. Morris Levy, Richard Howard, J. Andrew DeWoody, and Megan D. Gall; members of the Minchella lab; and two anonymous reviewers for discussions and comments regarding the manuscript. All animal use followed protocols approved by the Purdue Animal Care and Use Committee (PACUC). This work was supported, in part, by a National Institutes of Health (USA) Research Grant (R01-A1-42768) to D.J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Woolhouse MEJ, Chandiwana SK, Bradley M. On the distribution of schistosome infections among host snails. Int J Parasitol. 1990;20:325–7. doi: 10.1016/0020-7519(90)90147-f. [DOI] [PubMed] [Google Scholar]
- [2].Minchella DJ, Sollenberger KM, Pereira de Souza C. Distribution of schistosome genetic diversity within molluscan intermediate hosts. Parasitology. 1995;111:217–20. doi: 10.1017/s0031182000064970. [DOI] [PubMed] [Google Scholar]
- [3].Eppert A, Lewis FA, Grzywacz C, Coura-Filho P, Caldas I, Minchella DJ. Distribution of schistosome infections in molluscan hosts at different levels of parasite prevalence. J Parasitol. 2002;88:232–6. doi: 10.1645/0022-3395(2002)088[0232:DOSIIM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- [4].Théron A, Sire C, Rognon A, Prugnolle F, Durand P. Molecular ecology of Schistosoma mansoni transmission inferred from the genetic composition of larval and adult infrapopulations within intermediate and definitive hosts. Parasitology. 2004;129:571–85. doi: 10.1017/s0031182004005943. [DOI] [PubMed] [Google Scholar]
- [5].Steinauer ML, Mwangi IN, Maina GM, Kinuthia JM, Mutuku MW, Agola EL, Mungai B, Mkoji GM, Loker ES. Interactions between natural populations of human and rodent schistosomes in the Lake Victoria region of Kenya: a molecular epidemiological approach. PLoS Negl Trop Dis. 2008;2:e222. doi: 10.1371/journal.pntd.0000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Basch PF. Schistosomes: Development, Reproduction, and Host Relations. Oxford University Press; New York: 1991. [Google Scholar]
- [7].Sandland GJ, Rodgers JK, Minchella DJ. Interspecific antagonism and virulence in hosts exposed to two parasite species. J Invertebr Pathol. 2007;96:43–7. doi: 10.1016/j.jip.2007.02.005. [DOI] [PubMed] [Google Scholar]
- [8].Bremermann HJ, Pickering J. A game-theoretical model of parasite virulence. J Theor Biol. 1983;100:411–26. doi: 10.1016/0022-5193(83)90438-1. [DOI] [PubMed] [Google Scholar]
- [9].Nowak MA, May RM. Superinfection and the evolution of parasite virulence. Proc Biol Sci. 1994;255:81–9. doi: 10.1098/rspb.1994.0012. [DOI] [PubMed] [Google Scholar]
- [10].van Baalen M, Sabelis MW. The dynamics of multiple infection and the evolution of virulence. Am Nat. 1995;146:881–910. [Google Scholar]
- [11].Day T. Virulence evolution and the timing of disease life-history events. Trends Ecol Evol. 2003;18:113–8. [Google Scholar]
- [12].Zhang P, Sandland GJ, Feng Z, Xu D, Minchella DJ. Evolutionary implications for interactions between multiple strains of host and parasite. J Theor Biol. 2007;248:225–40. doi: 10.1016/j.jtbi.2007.05.011. [DOI] [PubMed] [Google Scholar]
- [13].Xu D, Sandland GJ, Minchella DJ, Feng Z. Interactions among virulence, coinfection and drug resistance in a complex life-cycle parasite. J Theor Biol. 2012;304:197–210. doi: 10.1016/j.jtbi.2012.03.040. [DOI] [PubMed] [Google Scholar]
- [14].Davies CM, Fairbrother E, Webster JP. Mixed strain schistosome infections of snails and the evolution of parasite virulence. Parasitology. 2002;124:31–38. doi: 10.1017/s0031182001008873. [DOI] [PubMed] [Google Scholar]
- [15].Gower CM, Webster JP. Intraspecific competition and the evolution of virulence in a parasitic trematode. Evolution. 2005;59:544–53. [PubMed] [Google Scholar]
- [16].Blair L, Webster JP. Dose-dependent schistosome-induced mortality and morbidity risk elevates host reproductive effort. J Evol Biol. 2007;20:54–61. doi: 10.1111/j.1420-9101.2006.01230.x. [DOI] [PubMed] [Google Scholar]
- [17].Cheesman SJ, de Roode JC, Read AF, Carter R. Real-time quantitative PCR for analysis of genetically mixed infections of malaria parasites: technique validation and applications. Mol Biochem Parasitol. 2003;131:83–91. doi: 10.1016/s0166-6851(03)00195-6. [DOI] [PubMed] [Google Scholar]
- [18].de Roode JC, Culleton R, Cheesman SJ, Carter R, Read AF. Host heterogeneity is a determinant of competitive exclusion or coexistence in genetically diverse malaria infections. Proc Biol Sci. 2004;271:1073–80. doi: 10.1098/rspb.2004.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].de Roode JC, Pansini R, Cheesman SJ, Helinski MEH, Huijben S, Wargo AR, Bell AS, Chan BHK, Walliker D, Read AF, Moran NA. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci USA. 2005;102:7624–28. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].de Roode JC, Helinski MEH, Anwar MA, Read AF. Dynamics of multiple infection and within-host competition in genetically diverse malaria infections. Am Nat. 2005;166:531–42. doi: 10.1086/491659. [DOI] [PubMed] [Google Scholar]
- [21].Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution. 2006;60:1358–71. [PubMed] [Google Scholar]
- [22].Cheesman S, Raza A, Carter R. Mixed strain infections and strain-specific protective immunity in the rodent malaria parasite Plasmodium chabaudi chabaudi in mice. Infect Immun. 2006;74:2996–3001. doi: 10.1128/IAI.74.5.2996-3001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Refardt D, Ebert D. Quantitative PCR to detect, discriminate and quantify intracellular parasites in their host: an example from three microsporidians in Daphnia. Parasitology. 2006;133:11–8. doi: 10.1017/S0031182006000060. [DOI] [PubMed] [Google Scholar]
- [24].Guzmán-Franco AW, Atkins SD, Clark SJ, Alderson PG, Pell JK. Use of quantitative PCR to understand within-host competition between two entomopathogenic fungi. J Invertebr Pathol. 2011;107:155–8. doi: 10.1016/j.jip.2011.03.001. [DOI] [PubMed] [Google Scholar]
- [25].Li B, Kadura I, Fu DJ, Watson DE. Genotyping with TaqMAMA. Genomics. 2004;83:311–20. doi: 10.1016/j.ygeno.2003.08.005. [DOI] [PubMed] [Google Scholar]
- [26].Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Valentim CLL, LoVerde PT, Anderson TJC, Criscione CD. Efficient genotyping of Schistosoma mansoni miracidia following whole genome amplification. Mol Biochem Parasitol. 2009;166:81–4. doi: 10.1016/j.molbiopara.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Karvonen A, Rellstab C, Louhi KR, Jokela J. Synchronous attack is advantageous: mixed genotype infections lead to higher infection success in trematode parasites. Proc Biol Sci. 2012;279:171–6. doi: 10.1098/rspb.2011.0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wright CA. Flukes and snails. George Allen and Unwin Ltd.; London: 1971. [Google Scholar]
- [30].Roger E, Grunau C, Pierce RJ, Hirai H, Gourbal B, Galinier R, Emans R, Cesari IM, Cosseau C, Mitta G. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata) PLoS Negl Trop Dis. 2008;2:e330. doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bayne CJ. Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: a 2009 assessment. Mol Biochem Parasitol. 2009;165:8–18. doi: 10.1016/j.molbiopara.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mitta G, Adema CM, Gourbal B, Loker ES, Théron A. Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev Comp Immunol. 2012;37:1–8. doi: 10.1016/j.dci.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Théron A, Coustau C, Rognon A, Gourbière S, Blouin MS. Effects of laboratory culture on compatibility between snails and schistosomes. Parasitology. 2008;135:1179–88. doi: 10.1017/S0031182008004745. [DOI] [PubMed] [Google Scholar]
- [34].Hlousek L, Voronov S, Diankov V, Leblang AB, Wells PJ, Ford DM, Nolling J, Hart KW, Espinoza PA, Bristol MR, Tsongalis GJ, Yen-Lieberman B, Slepnev VI, Kong LI, Lee MC. Automated high multiplex qPCR platform for simultaneous detection and quantification of multiple nucleic acid targets. BioTechniques. 2012;52:316–24. doi: 10.2144/0000113852. [DOI] [PubMed] [Google Scholar]
- [35].Blank WA, Test MR, Liu SF, Lewis FA, Blanton RE. Long-term genetic stability and population dynamics of laboratory strains of Schistosoma mansoni. J Parasitol. 2010;96:900–7. doi: 10.1645/GE-2463.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Steinauer ML, Agola LE, Mwangi IN, Mkoji GM, Loker ES. Molecular epidemiology of Schistosoma mansoni: a robust, high-throughput method to assess multiple microsatellite markers from individual miracidia. Infect Genet Evol. 2008;8:68–73. doi: 10.1016/j.meegid.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hanelt B, Steinauer ML, Mwangi IN, Maina GM, Agola LE, Mkoji GM, Loker ES. A new approach to characterize populations of Schistosoma mansoni from humans: development and assessment of microsatellite analysis of pooled miracidia. Trop Med Int Health. 2009;14:322–31. doi: 10.1111/j.1365-3156.2009.02226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]