Abstract
Autophagy is an evolutionarily conserved pathway responsible for delivery of cytoplasmic material into the lysosomal degradation pathway to enable vesicular exocytosis. Interleukin (IL)-2 is produced by T-cells and its activity is important for immunoregulation. Fibroblasts are an immune competent cell type, playing a critical role in wound healing, chronic inflammation, and tumor development. Although autophagy plays an important role in each of these processes, whether it regulates IL-2 activity in fibroblasts is unknown. Here, we show that autophagy is required for IL-2-induced cell growth in fibroblasts. IL-2 significantly induced autophagy in mouse embryonic fibroblasts (MEFs) and primary lung fibroblasts. Autophagy inhibitors (e.g., 3-methylamphetamine and bafilomycin A1) or knockdown of ATG5 and beclin 1 blocked clinical grade IL-2-induced autophagy. Moreover, IL-2 induced HMGB1 cytoplasmic translocation in MEFs and promoted interaction between HMGB1 and beclin1, which is required for autophagy induction. Pharmacological and genetic inhibition of autophagy inhibited IL-2-induced cell proliferation and enhanced IL-2-induced apoptosis. These findings suggest that autophagy is an important pro-survival regulator for IL-2-induced cell growth in fibroblasts.
Keywords: IL-2, Autophagy, Apoptosis, Immunotherapy, HMGB1
INTRODUCTION
Autophagy is a highly regulated catabolic process involving the degradation of the cell’s internal components [1] and is distinguished from phagocytosis by employing a double-membrane structure as opposed to a single-membrane vesicle. [2] It is induced in response to environmental and cellular stressors such as starvation, toxins, microbial entry, chemotherapy, and immune stimuli. [3] It initiates with sequestration of cytoplasmic constituents, including macromolecules and organelles, by an isolation membrane - namely, the phagophore. After the formation of the autophagosome and the autolysosome, the contents are then degraded to be reused as cellular bioenergetics and anabolic substrates. Increasing studies suggest that autophagic dysfunction is associated with human physiological and pathological states. [4] These include aging, chronic inflammatory conditions, innate and adaptive immunity, cancer, metabolic disorders, and neurodegenerative diseases.
IL-2, first identified as a mitogenic molecule for T cells produced by activated T cells, was initially termed T cell growth factor [5]. In addition to promoting T-cell proliferation, IL-2 increases cytokine production and enhances proliferation of B cells and natural killer (NK) cells, modifying the functional properties of fibroblasts and possibly macrophages, thereby contributing to immune responses. [6, 7] IL-2 can induce either proliferation or death, depending on the cellular context and dose. [8–12] As an immunotherapy currently applied to melanoma and renal cell carcinoma patients, IL-2 promotes recognition and destruction of cancer cells [7, 13, 14]. Fibroblasts are important sentinel cells in the immune system and play a critical role in the switch from acute inflammation to adaptive immunity and tissue repair [15, 16]. Human and mouse fibroblasts express functional IL-2 receptors, including the IL-2R alpha-, beta-, and gamma- chain subunits [17–19]. IL-2-transduced fibroblasts have antitumor therapeutic effects in neuroblastoma and colorectal carcinoma [20, 21].
Our and others’ recent studies show that IL-2 induces autophagy in mouse cancer cells and human CD4+ T lymphocytes, which can either promote survival [22] or death [23]. However, the mechanism and significance of IL-2-induced autophagy remains largely unknown. In this study, we examined the role of autophagy and its molecular actions in the regulation of IL-2 activity in fibroblasts.
RESULTS
IL-2 induces autophagy in fibroblasts
Microtubule-associated protein 1 light chain 3 (LC3), a mammalian homologue of yeast Atg8, is a widely used marker of autophagy. During autophagy, the cytoplasmic form (LC3-I) is processed and recruited to the autophagosome, where LC3-II is generated by site specific proteolysis and lipidation adjacent to the C-terminus. [24] Tracking of the conversion of LC3-I to LC3-II is indicative of autophagic activity [25]. Classical autophagic stimuli, such as starvation (such as that induced by placement in serum free medium, Hank’s Balanced Salt Solution) and rapamycin induced LC3-II expression in mouse embryonic fibroblasts (MEFs) (Fig. 1A). Similarly, treatments with IL-2 at 6000 U/ml for 48 hours induce LC3-II expression in MEFs (Fig. 1A) and primary lung fibroblasts (Fig. 1B). 3-methylamphetamine (“3MA”), a class III PI3 kinase inhibitor, prevents autophagy at an early stage; while bafilomycin A1 (“Baf”), a specific inhibitor of vacuolar H+-ATPase, prevents autophagy at later stages by inhibiting fusion between autophagosomes and lysosomes. Pretreatment with 3MA significantly inhibits IL-2-induced LC3-II expression (Fig. 1A and 1B). Moreover, LC3 accumulation following IL-2 treatment is exaggerated in MEFs following treatment with Baf (Fig. 1A). Furthermore, we observed an increase of endogenous LC3 puncta formation following IL-2 treatment in the absence or presence of 3MA and Baf by immunofluorescence analysis (Fig. 1C). In addition, LC3 puncta was induced by IL-2 in a dose-dependent fashion (Fig. 1D). These findings suggest that IL-2 is sufficient to induce autophagy in fibroblasts.
Figure 1. IL-2 induces autophagy in fibroblasts.
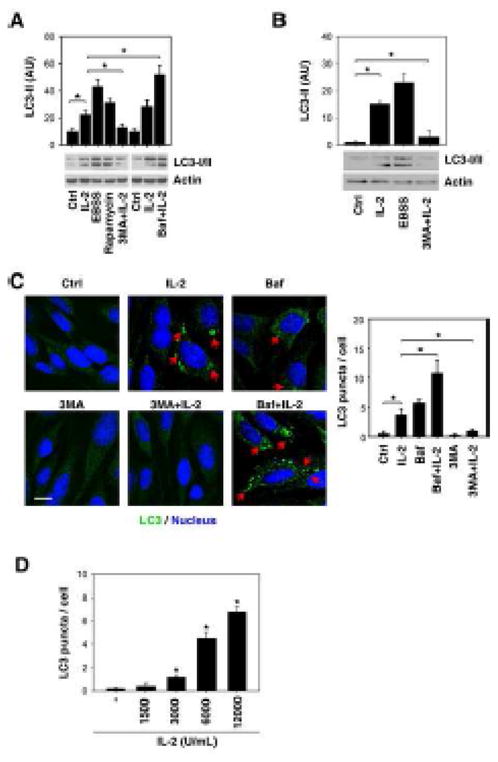
(A, B) MEFs (A) and primary lung fibroblasts (B) were treated with 6000 U/mL IL-2 for 48 hours in the presence or absence of 3-methyladenine (“3-MA”, 10 mM) or bafilomycin A1 (“Baf”, 100 nM), and the level of LC3 was assayed by Western blot. The cells were cultured in Hank’s balanced salt solution (starvation) for three hours or treated with rapamycin (1 μM) for 24 hours as a positive control. “AU”: arbitrary units (* p < 0.05). (C) In parallel, LC3 puncta per cell were assayed by confocal microscopy as described in the methods section (* p < 0.05). Representative images in MEFs are shown in the left panel. Bar=20 μm. (D) IL-2 induced a dose-dependent LC3 puncta accumulation in MEFs after treatment with 48 hours. The number of LC3 puncta per cell was calculated, and data are expressed as means ± SD of 50–100 randomly chosen cells (* p < 0.05 versus untreated group).
ATG5 and Beclin 1 are required for IL-2-induced autophagy
It is believed that ATG5 [26] and beclin1 [27] are essential genes for mammalian autophagy. ATG5 conjugates to ATG12 and associates with the isolation membrane to form a cup-shaped isolation membrane and autophagosome. Beclin-1, the mammalian orthologue of the yeast ATG6, is a critical component in the class III PI3 kinase complex (PI3KC3) that induces the formation of autophagosomes in mammalian cells [28]. To explore whether ATG5 and beclin1 are required for clinical grade IL-2-induced autophagy, we suppressed ATG5 and beclin1 expression by RNAi in MEFs (Fig. 2A). Indeed, knockdown of ATG5 and beclin 1 by specific shRNA significantly inhibited IL-2-induced LC3-II expression (Fig. 2A) as well as LC3 puncta formation (Fig. 2B). Transmission electron microscopy (TEM) is considered the gold standard for documenting autophagic vacuoles. By definition, autophagic vacuoles are limited by a double or occasionally multi-layered membrane [29] and contain cytoplasmic material or organelles. Sometimes the autophagic vacuole membrane does not have any contrast in thin sections [29]. The increase of autophagic vacuoles in fibroblasts after treatment with IL-2 was confirmed by TEM (Fig. 2C). In contrast, knockdown of ATG5 and beclin1 by shRNA inhibited IL-2 -induced autophagic vacuole formation (Fig. 2C). Collectively, these findings suggest that ATG5 and beclin 1 are required for IL-2-induced autophagy in fibroblasts.
Figure 2. ATG5 and beclin 1 are required for IL-2-induced autophagy.
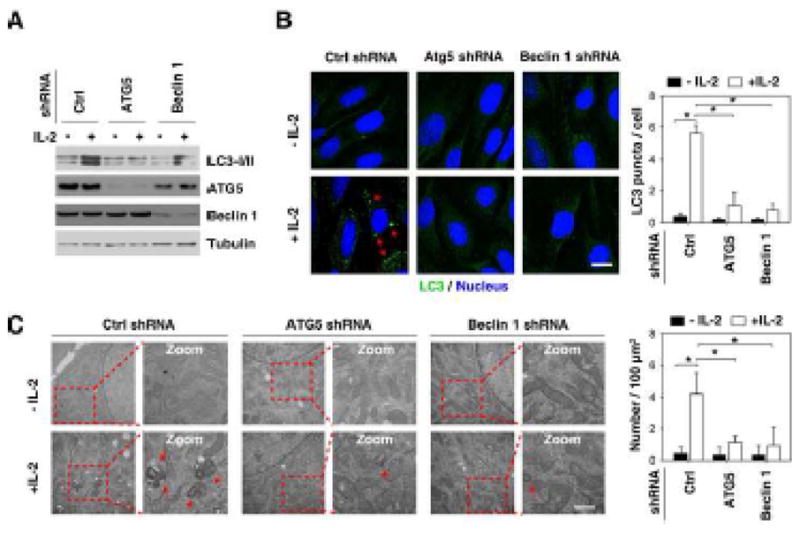
MEFs were transfected with indicated shRNA for 48 hours, and then treated with IL-2 (6000 U/ml) for 48 hours. Autophagy was assayed by Western blot analysis of LC3 expression (A) or quantitation of average number of LC3 puncta (indicated by the red arrows) per cell by confocal microscopy (B) or the number of autophagic vacuoles (indicated by the red arrows) by TEM (C) as described in the methods section (* p < 0.05).
HMGB1 is required for IL-2-induced autophagy
Our recent findings suggest that cytoplasmic high mobility group box 1 (HMGB1), a chromatin-associated nuclear protein, is a novel beclin 1-binding protein important in sustaining autophagy [30, 31]. To evaluate the possibility that HMGB1 cytoplasmic translocation ensues following IL-2 treatment, we isolated nuclear and cytoplasmic proteins. IL-2 significantly increased cytosolic HMGB1 at 24 and 48 hours as confirmed by Western blot analysis (Fig. 3A). Furthermore, we found that IL-2 increased interaction between HMGB1 and beclin 1 in MEFs as confirmed by co-immunoprecipitation analysis (Fig. 3B). In addition, we observed significant colocalization between HMGB1 and beclin 1 in the cytosol following IL-2 treatment in MEFs (data not shown). To determine whether HMGB1 is required for IL-2-induced autophagy, we assayed LC3 turnover in HMGB1+/+ and HMGB1−/− MEFs. Consistent with Western blotting for monitoring of LC3-II expression (Fig. 3C), knockout of HMGB1 in MEFs inhibited IL-2-induced LC3 puncta formation (Fig. 3D) and autophagic vacuole formation (Fig. 3E). Collectively, these data suggest that IL-2 induces HMGB1-beclin 1 complex formation and that HMGB1 is required for IL-2 induced autophagy.
Figure 3. HMGB1 is required for IL-2-induced autophagy.
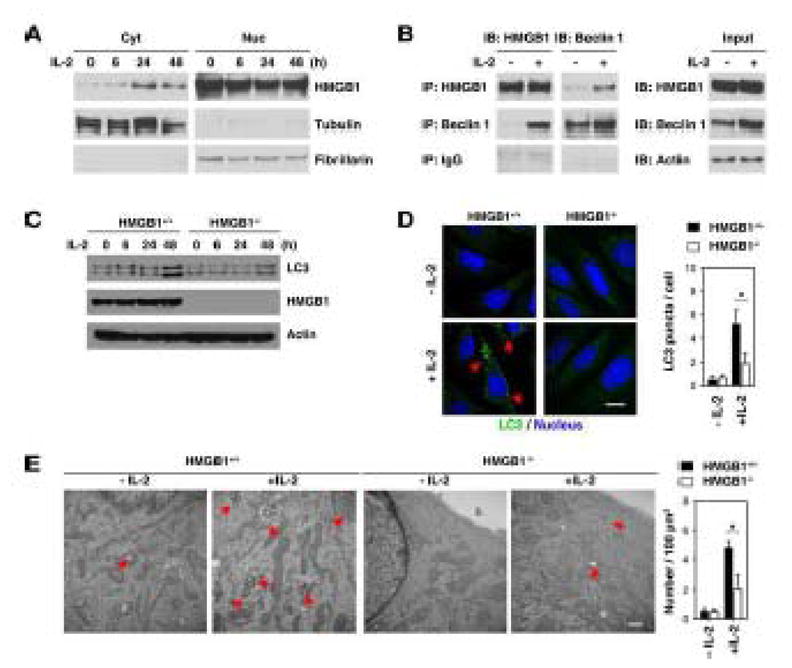
(A) MEFs were treated with IL-2 (6000 U/ml) for six to 48 hours, and HMGB1 expression in nuclear and cytosolic fraction were assayed by Western blot. Fibrillarin is a nuclear fraction control, and tubulin is a cytoplasmic fraction control. (B) In parallel, the HMGB1-beclin 1 complex at 48 hours after IL-2 (6000 U/ml) treatment was assessed as indicated by co-immunoprecipitation or Western blotting. (C-E) HMGB1+/+ and HMGB1−/− MEFs were treated with IL-2 (6000 U/ml) for 48 hours. Autophagy was assayed by Western blot analysis of LC3 expression (C) or quantitation of average number of LC3 puncta (indicated by the red arrows) per cell by confocal microscopy (D) or the number of autophagic vacuoles (indicated by the red arrows) by TEM (E) as described in the methods section (* p < 0.05).
Inhibition of autophagy switches IL-2-induced proliferation to apoptosis
IL-2 at 6000 U/mL time-dependently induces fibroblast proliferation and autophagy (Fig. 4A). To explore whether enhanced autophagy is required for IL-2-induced proliferation, we treated MEFs with IL-2 for six-48 hours with or without the autophagic inhibitor 3MA. 3MA inhibited IL-2-induced cell proliferation (Fig. 4B), suggesting that autophagy promotes cell survival in response to IL-2. We further confirmed that the pro-proliferative effect of autophagy was due to the genetic inhibition of ATG5, beclin 1, and HMGB1. Knockdown or knockout of these proteins in MEFs inhibited IL-2-induced cell proliferation (Fig. 4C). Cell proliferation and apoptosis pathways are closely linked. To explore whether inhibition of autophagy switches IL-2-induced proliferation to apoptosis, we performed staining with Annexin V for flow cytometric detection of phosphatidylserine expression on MEFs. IL-2 at 6000 U/mL cannot induce apoptosis in MEFs (Fig. 4A). However, application of the autophagy inhibitor 3MA (Fig. 5A), knockdown of ATG5/beclin 1 (Fig. 5B), and knockout of HMGB1 (Fig. 5B) restore sensitivity of fibroblasts to apoptosis following IL-2 treatment. Caspase-3 is activated in the apoptotic cell both by extrinsic (death ligand) and intrinsic (mitochondrial) pathways. As expected, inhibition of autophagy by 3MA increased IL-2-induced caspase-3 activity. Moreover, knockout of HMGB1 diminished IL-2-induced autophagy (Fig. 3C–E) and increased cleaved-caspase-3 and cleaved- poly (ADP ribose) polymerase (PARP), as confirmed by Western blot analysis (Fig. 5C). These findings suggest that autophagy plays a pro-survival role in response to IL-2 in murine fibroblasts.
Figure 4. Inhibition of autophagy decreases IL-2-induced proliferation.
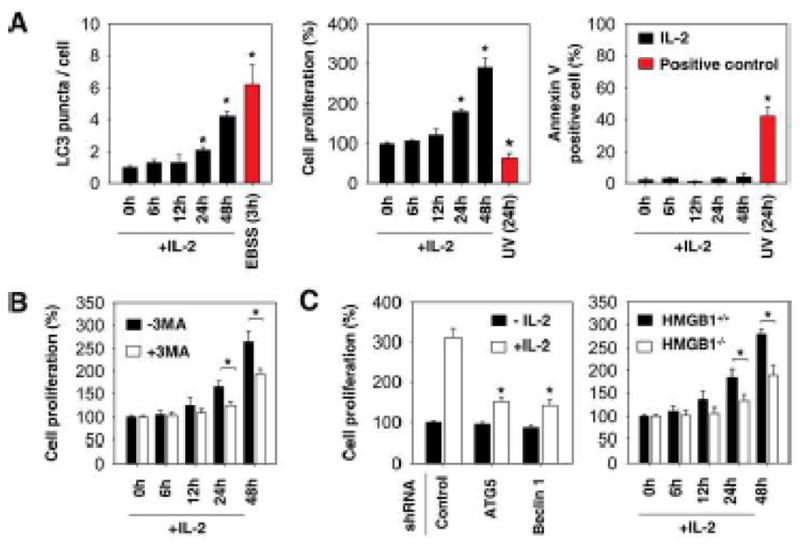
(A) MEFs were treated with IL-2 (6000 U/ml) for six-48 hours. Autophagy was assayed by quantitation of average number of LC3 puncta per cell by confocal microscopy. Cell proliferation was assayed by cell counting kit – 8 (CCK-8). Cell apoptosis was assayed by Annexin V-FITC apoptosis detection kit by flow cytometric analysis. Cells were cultured in Hank’s balanced salt solution (starvation) for three hours or treated with UV light for 24 hours as a positive control. * p < 0.05 versus untreated group. (B) MEFs (wild type or HMGB1 knockouts) were treated with 6000 U/mL IL-2 for 48 hours in the presence or absence of 3-methyladenine (“3-MA”, 10 mM), and cell proliferation was assayed by cell counting kit - 8. (C) Indicated autophagy deficient MEFs were treated with 6000 U/mL IL-2 for six-48 hours, and cell proliferation was assayed by cell counting kit - 8.
Figure 5. Inhibition of autophagy increases IL-2-induced apoptosis.
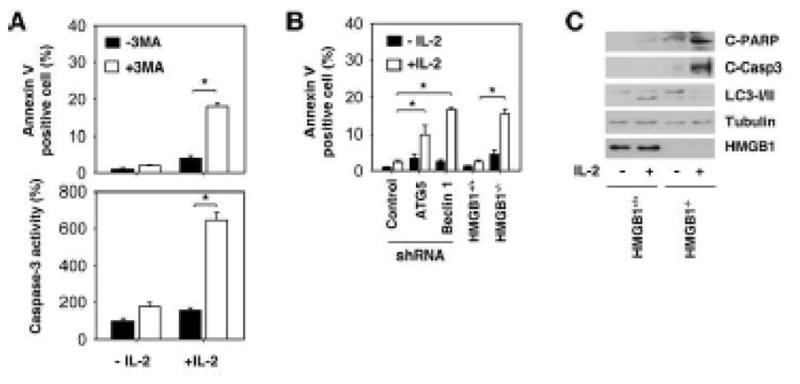
(A) MEFs were treated with 6000 U/mL IL-2 for 48 hours in the presence or absence of 3-methyladenine (“3-MA”, 10 mM), and cell apoptosis was assayed by flow cytometric analysis or by caspase-3 activity assays as described in the methods section (* p < 0.05). (B–C) Indicated autophagy deficient MEFs were treated with 6000 U/mL IL-2 for 48 hours, and cell apoptosis was assayed by Annexin V-FITC apoptosis detection kit by flow cytometric analysis (B) or by Western blot analysis of cleaved-PARP (“C-PARP”) and cleaved-caspase 3 (“C-Casp3”).
DISCUSSION
The present study provides evidence for the importance of autophagy in regulation of IL-2’s activity to promote growth of fibroblasts. We demonstrated that the ATG5-beclin 1-HMGB1-mediated autophagy pathway is required for IL-2-induced cell growth. Inhibition of autophagy switches IL-2-induced cell proliferation to cell apoptosis. These findings provide important evidence supporting the existence of a complex interplay between autophagy, cell proliferation, and cell death during fibroblast activation (Fig. 6).
Figure 6. Autophagy is involved in regulation of IL-2-induced cell growth, survival, and death in fibroblasts.
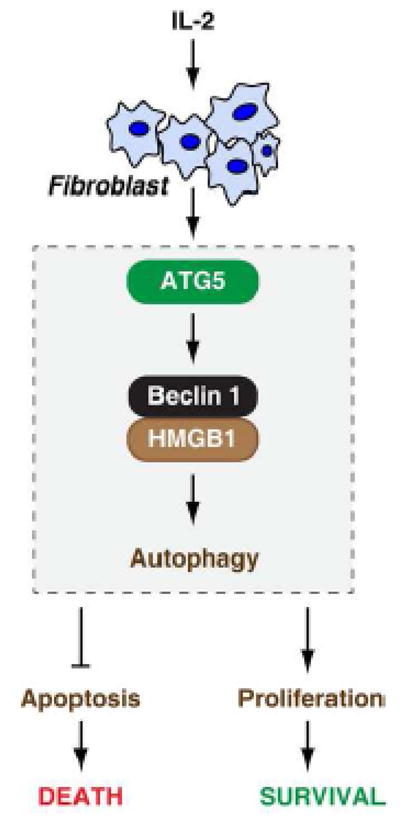
IL-2 induces autophagy in fibroblasts by an ATG5-beclin 1-HMGB1 dependent pathway. Enhanced autophagy from IL-2 treatment promotes proliferation and limits apoptosis in fibroblasts.
Fibroblasts synthesize extracellular matrix and collagen, the structural framework (stroma) for animal tissues, and play a critical role in wound healing [32], chronic inflammation [15, 16], and tumor development [33]. Fibroblast activation leads to production of cytokines, chemokines, and prostanoids, and fibroblast dysfunction causes chronic persistent inflammation [15]. Fibroblasts are major cellular components of the tumor microenvironment, influencing tumor cell behavior directly and indirectly through the secretion of growth regulators and angiogenic factors, extracellular matrix proteins, and proteases [34]. These new findings have important therapeutic implications. Manipulation of fibroblasts and their biologically active products is an emerging therapeutic target in cancer therapy. Moreover, it is likely to provide a novel method to achieve improved control of chronic inflammatory disease. In this study, we demonstrated that IL-2-mediated autophagy promotes fibroblast proliferation. Pharmacological (e.g., 3MA) and genetic inhibition of autophagy markedly enhances IL-2-induced cell death in fibroblasts. In addition, we recently demonstrated that the autophagy inhibitor chloroquine markedly enhances IL-2 immunotherapy efficacy and limits toxicity in an advanced murine metastatic liver tumor model [35]. These findings suggest that combinations with the autophagy inhibitor and IL-2 not only promote death of cancer cells, but also that of fibroblasts within the tumor microenvironment. Thus, our studies provide a novel clinical strategy to enhance the efficacy of IL-2 immunotherapy for cancer patients.
Autophagy is regulated via a group of genes called autophagy related genes (ATGs) and is executed at basal levels in virtually all cells as a homeostatic mechanism for maintaining cellular integrity. Autophagy confers stress tolerance, limits damage, and sustains viability under adverse conditions. Older studies regarded autophagy as a form of programmed cell death (type II) distinct from apoptosis (type I), depending on the context in which it was observed [36]. Current studies have shown that autophagy and apoptosis are predominantly opposed biologic programs, with cross-talk between these two processes. [37, 38] Defects in autophagy have been implicated in many diseases, including neurodegeneration, aging, liver disease, and importantly, cancer [4, 27, 39–43]. Moreover, autophagy has been implicated in conferring resistance to chemotherapy, radiation therapy, and immunotherapy in cancer cells [38, 40, 43, 44]. The precise molecular mechanism by which autophagy induces drug resistance in cancer cells has not been clearly defined. In this study, we demonstrated that ATG5 and beclin 1 are required for IL-2-induced autophagy, suggesting that the classic autophagic pathway is activated by IL-2 in fibroblasts. In addition, we found that IL-2 induced not only LC3-II expression, but also LC3-I expression, suggesting that different mechanisms at transcriptional level may be involved in IL-2 induced autophagy. Recent study indicates that the PI3K/Akt signalling pathway regulates the expression of LC3-I in dasatinib-induced autophagy in glioma [45]. IL-2 has been reported to active PI3K/Akt signalling pathway in several cells [46, 47]. Thus it is possible that PI3K/Akt signalling pathway mediates IL-2 -induced LC3 turnover as well as LC3-I expression.
HMGB1, a nuclear DNA chaperone and extracellular DAMP molecular, has been implicated in several inflammatory disorders and cancer [48–50]. Our recent studies demonstrate that HMGB1 is an important regulator of autophagy. The mechanism of HMGB1-mediated autophagy is dependent on its subcellular localization. Nuclear HMGB1 regulates HSPB1 expression in mitochondrial autophagy, which is required for mitochondrial quality control [51]. Cytosolic HMGB1 competes with Bcl-2 for interaction with beclin 1 and regulates beclin 1-PI3KC3 core complex formation in autophagy [30, 52]. Extracellular HMGB1 promotes autophagy in a RAGE-dependent manner [53]. Current studies demonstrate that IL-2 promotes expression of cytosolic HMGB1 as well as HMGB1-beclin 1 complex formation. Knockout of HMGB1 diminishes autophagy and increases apoptosis following IL-2 treatment in fibroblasts, suggesting that targeting HMGB1-mediated autophagy may improve the immunotherapeutic efficacy of IL-2.
In summary, we demonstrate here for the first time that autophagy is required for IL-2-mediated fibroblast growth. We hypothesize that this ATG5-beclin 1-HMGB1 dependent programmed cell survival pathways provides a means for fibroblasts to gain a selective growth advantage.
MATERIALS AND METHODS
Cell culture and reagent
HMGB1+/+ and HMGB1−/− immortalized MEFs were a kind gift from Dr. Marco E. Bianchi (San Raffaele Institute, Italy) [54] and cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum in a humidified incubator with 5% CO2 and 95% air. Primary culture of lung fibroblasts from mice was performed as previously described. [55] Clinical grade rIL-2 was a kind gift from Prometheus Laboratories, Inc. The antibodies to actin (#3700), beclin 1 (#3738), cleaved-PARP (#9544), and cleaved-caspase 3 (#9661) were obtained from Cell Signaling Technology. The antibodies to LC3 (NB100-2220) and ATG5 (NB110-53818) were obtained from Novus. The antibody to tubulin (T5293) was obtained from Sigma. The antibody to fibrillarin and HMGB1 were from Abcam (ab18256). Other chemical reagents were from Sigma.
RNAi by shRNA
Beclin1-shRNA (SHCLNG-NM_019584), ATG-5 shRNA (SHCLNG-NM_053069), and control shRNA (from Sigma) were transfected into cells using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s instructions. The medium over the cells was changed at the end of the 48 hour shRNA treatment and before the addition of IL-2.
Western blotting
Proteins in cell lysates were resolved on 4–12% Criterion XT Bis-Tris gels (#345-0123, Bio-Rad) and transferred to a nitrocellulose membrane. After blocking, the membrane was incubated overnight at 4°C with various primary antibodies. After incubation with peroxidase-conjugated secondary antibodies for one hour at 25°C, signals were visualized by enhanced chemiluminescence detection according to the manufacturer’s instructions. The relative band intensities were quantified using the Gel-pro Analyzer® software (Media Cybernetics, Bethesda).
Immunoprecipitation analysis
Cells were lysed at 4°C in ice-cold modified radioimmunoprecipitation (RIPA) lysis buffer (Cell Signaling Technology, #9806), and cell lysates were cleared by centrifugation (12000 g, 10 min). Prior to immunoprecipitation, samples containing equal amount of proteins were pre-cleared with Protein A or Protein G agarose/sepharose (4°C, 3 h) and subsequently incubated with various irrelevant IgG or specific antibodies (2–5 μg/ml) in the presence of Protein A or G agarose/sepharose beads for two hours or overnight at 4°C with gentle shaking. Following incubation, agarose/sepharose beads were washed extensively with phosphate buffered saline, and proteins were eluted by boiling in 2 × sodium dodecyl sulfates (SDS) sample buffer before SDS-PAGE electrophoresis.
Autophagy assays
The percentage of cells with LC3 puncta was determined by quantifying the number of positively stained cells from 50–100 randomly chosen cells by Image-Pro Plus 5.1 software (Media Cybernetics) as previously described. [30, 56] Autophagic flux assays were performed by Western blotting for LC3-I/II with or without 3-methylamphetamine or bafilomycin A1. TEM assessment of autophagic vacuoles was performed as previously described. [51]
Apoptosis assays
Cell apoptosis was assessed using the Annexin V-FITC Apoptosis Detection Kit (#556570, BD Pharmingen) by flow cytometric analysis. Caspase-3 activity assays were performed using Caspase-3 Colorimetric Assay Kit from Calbiochem (#235419). Cleaved-PARP and cleaved-caspase 3 were measured by Western blot analysis.
Cell proliferation assay
Cells were plated at a density of 2×104 cells/well on 96-well plates in 100 μl RPMI. Cell number was determined by WST-8 (2-(2-methoxy-4-nitrophenyl) - 3 - (4-nitrophenyl) - 5 - (2, 4-disulfophenyl) - 2 H - tetrazolium, monosodium salt), assay using a Cell Counting Kit - 8 (CCK-8) from Dojindo Laboratories (Tokyo, # CK04-01) according to the manufacturer’s instructions.
Statistical analysis
Data are expressed as mean ± SD. One-way ANOVA was used for comparison among the different groups. When the ANOVA was significant, post hoc testing of differences between groups was performed using a Fisher’s Least Significant Difference (LSD) test. A p-value < 0.05 was considered significant.
Acknowledgments
We thank Christine Heiner (University of Pittsburgh, Department of Surgery) for critical reading of the manuscript. This project was supported by grants from the National Institutes of Health (P01 CA 101944 to M.T.L, R01CA160417 to D.T.) and the University of Pittsburgh (D.T, and H.J.Z.). This project used University of Pittsburgh Cancer Institute (UPCI) shared resources that are supported in part by award P30CA047904. We also thank Drs. Donna Stolz and Simon Watkins for providing the confocal microscopy facilities at the Center for Biologic Imaging at the University of Pittsburgh School of Medicine.
Footnotes
CONFLICT OF INTEREST
Authors declare no conflicts of interest or financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang D, Vernon P. Eat-Me: Autophagy, Phagocytosis, and ROS Signaling. Antioxid Redox Signal. 2012 doi: 10.1089/ars.2012.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon J, MacLean LD. A lymphocyte-stimulating factor produced in vitro. Nature. 1965;208:795–796. doi: 10.1038/208795a0. [DOI] [PubMed] [Google Scholar]
- 6.Malek TR. The biology of interleukin-2. Annual review of immunology. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 7.Chavez AR, Buchser W, Basse PH, Liang X, Appleman LJ, Maranchie JK, Zeh H, de Vera ME, Lotze MT. Pharmacologic administration of interleukin-2. Ann N Y Acad Sci. 2009;1182:14–27. doi: 10.1111/j.1749-6632.2009.05160.x. [DOI] [PubMed] [Google Scholar]
- 8.Gombert W, Borthwick NJ, Wallace DL, Hyde H, Bofill M, Pilling D, Beverley PC, Janossy G, Salmon M, Akbar AN. Fibroblasts prevent apoptosis of IL-2-deprived T cells without inducing proliferation: a selective effect on Bcl-XL expression. Immunology. 1996;89:397–404. doi: 10.1046/j.1365-2567.1996.d01-759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii H. Cell type-specific roles of Jak3 in IL-2-induced proliferative signal transduction. Biochem Biophys Res Commun. 2007;354:825–829. doi: 10.1016/j.bbrc.2007.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaltenberg J, Plum LM, Ober-Blobaum JL, Honscheid A, Rink L, Haase H. Zinc signals promote IL-2-dependent proliferation of T cells. Eur J Immunol. 2010;40:1496–1503. doi: 10.1002/eji.200939574. [DOI] [PubMed] [Google Scholar]
- 11.Mahaffey CL, Mummert ME. Hyaluronan synthesis is required for IL-2-mediated T cell proliferation. J Immunol. 2007;179:8191–8199. doi: 10.4049/jimmunol.179.12.8191. [DOI] [PubMed] [Google Scholar]
- 12.Marks-Konczalik J, Dubois S, Losi JM, Sabzevari H, Yamada N, Feigenbaum L, Waldmann TA, Tagaya Y. IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proc Natl Acad Sci U S A. 2000;97:11445–11450. doi: 10.1073/pnas.200363097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antony GK, Dudek AZ. Interleukin 2 in cancer therapy. Curr Med Chem. 2010;17:3297–3302. doi: 10.2174/092986710793176410. [DOI] [PubMed] [Google Scholar]
- 14.Atkins MB, Lotze MT, Dutcher JP, Fisher RI, Weiss G, Margolin K, Abrams J, Sznol M, Parkinson D, Hawkins M, Paradise C, Kunkel L, Rosenberg SA. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 15.Buckley CD, Pilling D, Lord JM, Akbar AN, Scheel-Toellner D, Salmon M. Fibroblasts regulate the switch from acute resolving to chronic persistent inflammation. Trends Immunol. 2001;22:199–204. doi: 10.1016/s1471-4906(01)01863-4. [DOI] [PubMed] [Google Scholar]
- 16.Flavell SJ, Hou TZ, Lax S, Filer AD, Salmon M, Buckley CD. Fibroblasts as novel therapeutic targets in chronic inflammation. Br J Pharmacol. 2008;153(Suppl 1):S241–246. doi: 10.1038/sj.bjp.0707487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gruss HJ, Scott C, Rollins BJ, Brach MA, Herrmann F. Human fibroblasts express functional IL-2 receptors formed by the IL-2R alpha- and beta-chain subunits: association of IL-2 binding with secretion of the monocyte chemoattractant protein-1. J Immunol. 1996;157:851–857. [PubMed] [Google Scholar]
- 18.Minamoto S, Mori H, Hatakeyama M, Kono T, Doi T, Ide T, Uede T, Taniguchi T. Characterization of the heterodimeric complex of human IL-2 receptor alpha.beta chains reconstituted in a mouse fibroblast cell line, L929. J Immunol. 1990;145:2177–2182. [PubMed] [Google Scholar]
- 19.Ozawa A, Tada H, Tamai R, Uehara A, Watanabe K, Yamaguchi T, Shimauchi H, Takada H, Sugawara S. Expression of IL-2 receptor beta and gamma chains by human gingival fibroblasts and up-regulation of adhesion to neutrophils in response to IL-2. J Leukoc Biol. 2003;74:352–359. doi: 10.1189/jlb.0103044. [DOI] [PubMed] [Google Scholar]
- 20.Barker SE, Grosse SM, Siapati EK, Kritz A, Kinnon C, Thrasher AJ, Hart SL. Immunotherapy for neuroblastoma using syngeneic fibroblasts transfected with IL-2 and IL-12. Br J Cancer. 2007;97:210–217. doi: 10.1038/sj.bjc.6603857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fakhrai H, Shawler DL, Gjerset R, Naviaux RK, Koziol J, Royston I, Sobol RE. Cytokine gene therapy with interleukin-2-transduced fibroblasts: effects of IL-2 dose on anti-tumor immunity. Hum Gene Ther. 1995;6:591–601. doi: 10.1089/hum.1995.6.5-591. [DOI] [PubMed] [Google Scholar]
- 22.Liang X, De Vera ME, Buchser WJ, de Vivar Chavez AR, Loughran P, Stolz DB, Basse P, Wang T, Van Houten B, Zeh HJ, 3rd, Lotze MT. Inhibiting Systemic Autophagy during Interleukin 2 Immunotherapy Promotes Long-term Tumor Regression. Cancer Res. 2012;72:2791–2801. doi: 10.1158/0008-5472.CAN-12-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. Journal of Immunology. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 24.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ET, Banhegyi G, Bartholomew CR, Bassham DC, Bast RC, Jr, Batoko H, Bay BH, Beau I, Bechet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen YG, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronje MJ, Cuervo AM, Cullen JJ, Czaja MJ, D’Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farre JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fesus L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, Gonzalez-Estevez C, Gonzalez-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hebert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Hohfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Hoyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jaattela M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jimenez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhasz G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovacs AL, Kraft C, Krainc D, Kramer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Kruger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, Laszlo L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, Lopez-Otin C, Lossi L, Lotze MT, Low P, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Marino G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Melendez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Muller S, Munger K, Munz C, Murphy LO, Murphy ME, Musaro A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nurnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Poggeler S, Poirot M, Poletti A, Pous C, Pozuelo-Rubio M, Praetorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Regnier-Vigouroux A, Reichert AS, Reiners JJ, Jr, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodriguez de Cordoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, Rudich A, 3rd, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schatzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schuller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Stralfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takacs-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tonges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ, 3rd, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 27.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 28.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009;452:143–164. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 30.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881–892. doi: 10.1083/jcb.200911078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang R, Livesey KM, Zeh HJ, Loze MT, Tang D. HMGB1: a novel Beclin 1-binding protein active in autophagy. Autophagy. 2010;6:1209–1211. doi: 10.4161/auto.6.8.13651. [DOI] [PubMed] [Google Scholar]
- 32.Darby IA, Hewitson TD. Fibroblast differentiation in wound healing and fibrosis. Int Rev Cytol. 2007;257:143–179. doi: 10.1016/S0074-7696(07)57004-X. [DOI] [PubMed] [Google Scholar]
- 33.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature reviews. Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 34.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang X, de Vera ME, Buchser WJ, Romo de Vivar Chavez A, Loughran P, Beer-Stolz D, Basse P, Wang T, van Houten B, Zeh HJ, Lotze M. Inhibiting Autophagy During Interleukin 2 Immunotherapy Promotes Long Term Tumor Regression. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-12-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 38.Livesey KM, Tang D, Zeh HJ, Lotze MT. Autophagy inhibition in combination cancer treatment. Curr Opin Investig Drugs. 2009;10:1269–1279. [PubMed] [Google Scholar]
- 39.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 40.White E, DiPaola RS. The double-edged sword of autophagy modulation in cancer. Clin Cancer Res. 2009;15:5308–5316. doi: 10.1158/1078-0432.CCR-07-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hippert MM, O’Toole PS, Thorburn A. Autophagy in cancer: good, bad, or both? Cancer Res. 2006;66:9349–9351. doi: 10.1158/0008-5472.CAN-06-1597. [DOI] [PubMed] [Google Scholar]
- 43.Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 44.Weiner LM, Lotze MT. Tumor-cell death, autophagy, and immunity. N Engl J Med. 2012;366:1156–1158. doi: 10.1056/NEJMcibr1114526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milano V, Piao Y, LaFortune T, de Groot J. Dasatinib-induced autophagy is enhanced in combination with temozolomide in glioma. Mol Cancer Ther. 2009;8:394–406. doi: 10.1158/1535-7163.MCT-08-0669. [DOI] [PubMed] [Google Scholar]
- 46.Ahmed NN, Grimes HL, Bellacosa A, Chan TO, Tsichlis PN. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc Natl Acad Sci U S A. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang K, Zhong B, Ritchey C, Gilvary DL, Hong-Geller E, Wei S, Djeu JY. Regulation of Akt-dependent cell survival by Syk and Rac. Blood. 2003;101:236–244. doi: 10.1182/blood-2002-04-1251. [DOI] [PubMed] [Google Scholar]
- 48.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 49.Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 51.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R, Vernon P, Cao L, Tang D. HMGB1 promotes drug resistance in osteosarcoma. Cancer Res. 2012;72:230–238. doi: 10.1158/0008-5472.CAN-11-2001. [DOI] [PubMed] [Google Scholar]
- 53.Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, Benschop R, Sparvero LJ, Amoscato AA, Tracey KJ, Zeh HJ, Lotze MT. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010;29:5299–5310. doi: 10.1038/onc.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calogero S, Grassi F, Aguzzi A, Voigtlander T, Ferrier P, Ferrari S, Bianchi ME. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–280. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 55.Yarovinsky TO, Hunninghake GW. Lung fibroblasts inhibit activation-induced death of T cells through PGE(2)-dependent mechanisms. Am J Physiol Lung Cell Mol Physiol. 2001;281:L1248–1256. doi: 10.1152/ajplung.2001.281.5.L1248. [DOI] [PubMed] [Google Scholar]
- 56.Kang R, Tang D, Schapiro NE, Livesey KM, Farkas A, Loughran P, Bierhaus A, Lotze MT, Zeh HJ. The receptor for advanced glycation end products (RAGE) sustains autophagy and limits apoptosis, promoting pancreatic tumor cell survival. Cell Death Differ. 2010;17:666–676. doi: 10.1038/cdd.2009.149. [DOI] [PMC free article] [PubMed] [Google Scholar]


