Abstract
Strain SJConT, a 2-chloro-4-nitrophenol (2C4NP) degrading bacterium, was isolated from soil collected from a pesticide-contaminated site in Punjab, India. The strain, which stained Gram positive, displayed a rod-coccus life cycle, and possessed a type A3α peptidoglycan (l-Lys–l-Ala3), MK-9(H2) as the major menaquinone, anteiso-C15 and iso-C15:0 as the major cellular fatty acids, and diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and a glycolipid as the major polar lipids, showed morphological and chemotaxonomic properties consistent with those reported for members of the genus Arthrobacter. Phylogenetic analysis of the 16S rRNA gene sequence of strain SJConT confirmed that it was a member of this genus with Arthrobacter globiformis DSM 20124T being the closest relative (sequence similarity of 97 %). The DNA G + C content of strain SJConT was 69 ± 1 mol% and DNA homology with A. globiformis DSM 20124T was 45 %, suggesting that strain SJConT represented a novel species of the genus Arthrobacter, which we have named Arthrobacter nitrophenolicus sp. nov The type strain is SJConT (=MTCC 10104T =DSM 23165T).
Keywords: Biodegradation, 2-Chloro-4-nitrophenol, 4-Nitrophenol, 3-Methyl-4-nitrophenol
Genus Arthrobacter was first proposed by Conn and Dimmick (1947) with the description of the type species Arthrobacter globiformis. Subsequently, Koch et al. (1995) emended the description with the reclassification of Micrococcusagilis as Arthrobacteragilis. The currently validated 78 members of genus Arthrobacter are members of phylum Actinobacteria, order Actinomycetales, family Micrococcaceae, and are characterized by the presence of a rod–coccus growth cycle and genomes with high G + C content (59–66 mol%) (Keddie et al. 1986). Members of genus Arthrobacter stain Gram positive are catalase positive and are sub-divided on the basis of the lysine-containing peptidoglycan into two groups, A3α and A4α (Schleifer and Kandler 1972; Stackebrandt et al. 1983; Keddie et al. 1986; Koch et al. 1995).
The primary habitat of Arthrobacter is soil and interestingly Arthrobacter strains with the ability to degrade nitrophenols and/or chlorophenols which include Arthrobacter chlorophonolicus A6 (Westerberg et al. 2000), Arthrobacter ureafaciens, strain CPR706 (Bae et al. 1996), Arthrobacter citrus (Karigar et al. 2006), Arthrobacter protophormae strain RKJ100 (Chauhan et al. 2000), Arthrobacter sp. strain JS443 (Jain et al. 1994) and Arthrobacter aurescens TW17 (Hanne et al. 1993) have all been isolated from pesticide-contaminated soil. We had previously reported for the first time on the isolation of Arthrobacter strain SJCon that degraded 2-chloro-4-nitrophenol (2C4NP) (Arora and Jain 2011). Chlorohydroquinone was identified as a major intermediate product which was further degraded via formation of maleylacetate (Arora and Jain 2011). As strain SJConT is a potential degrader of various nitrophenolic compounds including 2-chloro-4-nitrophenol, 4-nitrophenol and 3-methyl-4-nitrophenol, and it has the potential for use in the bioremediation of nitrophenolic contaminated sites. In this communication, we report on the chemotaxonomic and genotypic properties of the strain and designate it as a new species of genus, A. nitrophenolicus sp nov. (The type strain is SJConT =MTCC 10104T =DSM 23165T).
The method for isolating strain SJConT from a pesticide-contaminated soil by an enrichment method using 2-chloro-4-nitrophenol as sole carbon source (Sigma-Aldrich, GmbH, Steinheim, Germany) has been reported previously (Arora and Jain 2011).
Colony morphology was examined on Nutrient agar plates after incubation at 30 °C for 24 h. Cell morphology was examined by light microscopy (Zeiss) at ×1000 and motility was checked using the method described by Skerman (1967). Gram staining was performed using HiMedia Gram Staining kit (HiMedia, India) according to the manufacturer’s instructions. Strain SJConT had morphological characteristics consistent with members of the genus Arthrobacter. Cells exhibited a rod–coccus growth cycle, were non-motile, did not form spores and stained Gram positive.
Growth at different temperatures (between 4 and 50 °C) was determined using Nutrient agar plates. Strain SJConT was streaked on nutrient agar plates, incubated at different temperatures and the growth on the plate scored. The growth in nutrient broth containing different NaCl concentrations (0.5–7 %) was monitored by measuring optical density at 600 nm in a Lambda 35 spectrophotometer (Perkin Elmer). Growth at different pH (pH 4.5–12) was tested in nutrient broth after the pH had been adjusted using appropriate buffers as described previously (Arora et al. 2011). Growth occurred between 10 and 40 °C with the optimum temperature for growth at 30 °C. The optimum pH for growth was 7 and no growth occurred below pH 6 or above pH 10. NaCl was not required for growth, but was tolerated up to 4 %.
Hydrolysis of gelatin, casein, starch, Voges-Proskauer, methyl red, oxidation-fermentation tests, catalase and oxidase activities, growth on Simmon’s citrate and MacConkey agar, production of H2S and indole, reduction of nitrate and acid production from carbohydrates were determined as described previously (Arora et al. 2011).
A summary of the results from the phenotypic tests is presented in Table 1 and listed in the species description.
Table 1.
Differentiating characteristics of strain Arthrobacter nitrophenolicus strain SJConT and Arthrobacter globiformis DSM 20124T
| Characteristics features | Arthrobacter nitrophenolicus | Arthrobacter globiformis |
|---|---|---|
| Type strains | SJConT =MTCC 10104T =DSM 23165T | DSM 20124T |
| Sample source | Pesticide-contaminated soil | Soil |
| 16S rRNA gene accession number | GQ927310 | M23411 |
| Mol% G + C | 69 ± 1 mol% | 62.0 % |
| DNA homology (%) of strain SJConT with: | 100 | 45 |
| NaCl range | 0–4 % | 0–5 % |
| Starch hydrolysis | – | + |
| Casein hydrolysis | + | – |
| Oxidation of substrates | ||
| d-Mannitol | + | – |
| Utilization of various nitro aromatics as sole carbon and energy source | ||
| 2-Chloro-4-nitrophenol | + | – |
| 4-Nitrophenol | + | – |
| 3-Methyl-4-nitrophenol | + | – |
Both species produce white colonies, have a rod-coccus life cycle, grow between pH 6 and 10, do not reduce nitrate, hydrolyse gelatin and utilise l-arabinose and glucose
The utilization of carbon and energy sources was tested using Biolog GP2 Microplates (Hayward, CA). For this, the inoculum was prepared by re-suspending Nutrient agar grown colonies to a turbidity equivalent to 0·5 McFarland units and the GP2 Microplates inoculated following the manufacturer’s instructions. The plates were incubated at 30 °C for 24 h and the results read using a MicroPlate Reader equipped with Microlog 4.2 software. The results are listed in the species description.
Menaquinones, fatty acids, polar lipids and peptidoglycan were analyzed by standard methods (Arora et al. 2011). The major menaquinone was identified as MK-9(H2), a characteristic chemotaxonomic marker of the genus Arthrobacter. The fatty acid profile comprises C16:0 (6.48 %), C18:0 (1.61 %), iso-C14:0 (0.93 %), iso-C15:0 (16.33 %), anteiso-C15:0 (44.18 %), iso-C16:0 (9.25 %), iso-C17:0 (5.61 %) and anteiso-C17:0 (15.19 %). The major polar lipids were diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylinositol (PI) and a glycolipid. The peptidoglycan was identified as type A3α (l-Lys–l-Ala3).
DNA extraction and purification have been described previously (Arora and Jain 2011). The 16S rRNA gene was amplified using universal bacterial primers 8F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′) (Arora et al. 2011) and sequenced using n ABI automated sequencer (Applied Biosystems, USA). The sequence (1,446 bp with GenBank accession number GQ927310) was compared against sequences available in the GenBank database (version 188.0) using EzTaxon server 2.1, the sequences of the nearest phylogentic members downloaded, aligned with those of related Arthrobacter species and a phylogenetic tree constructed using the neighbour-joining method as implemented in MEGA (Tamura et al. 2007) and Bioedit (Hall 1999). During phylogenetic reconstruction, all ambiguous nucleotides were excluded from the analysis and a total of 1,270 nucleotides were used in the final analysis. Phylogenetic analysis showed that strain SJConT was a member of the genus Arthrobacter and showed the highest similarity to A. globiformis DSM 20124T and related members of the ‘globiformis’ group (average sequence similarity of 97%) (Fig. 1).
Fig. 1.
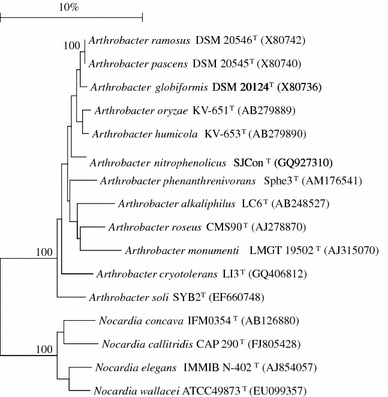
The phylogenetic position of Arthrobacter nitrophenolicus strain SJConT within the radiation of members of genus Arthrobacter based on 16S rRNA gene. The type strains are indicated with the letterT and the accession numbers are shown in brackets. Bootstrap values >95 % (expressed as percentages of 1000 replications) are indicated at the branch points
The G + C mol% content of the genomic DNA was determined in a Lambda 35 spectrophotometer (Perkin Elmer, Waltham, MA, USA) using the thermal denaturation (Tm) method and determined to be 69 ± 1 mol%.
DNA hybridization was performed using Biotin DecaLabelTM Kit and Biotin Chromogenic Detection Kit (Fermentas Life Sciences) following the manufacturer’s instruction. The DNA–DNA relatedness value of strain SJConT with A. globiformis DSM 20124T was found to be 45 %.
Strain SJConT degrades 2-chloro-4-nitrophenol, 4-nitrophenol and 3-methyl-4-nitrophenol. It possesses morphological characteristic of rod-coccus life cycle, which is consistent with that reported for members of the genus Arthrobacter. It’s affiliation to this genus is supported by the chemotaxonomic traits and 16S rRNA phylogeny. In addition, the presence of cell-wall peptidoglycan type A3α and menaquinone MK-9(H2) supports the placement of strain SJConT within the “globiformis” group. However, the large phylogenetic distance, the low DNA homology and the higher mol % G + C content and a number of phenotypic traits (Table 1) differentiate strain SJConT from A. globiformis DSM 20124T and members of the “globiformis” group. The fatty acid composition of strain SJconT was differed from the fatty acid composition of A. globiformis DSM 20124T (Table 2). The fatty acid profile of strain SJConT was comprises C16:0 (6.48 %), C18:0 (1.61 %), iso-C14:0 (0.93 %), iso-C15:0 (16.33 %), anteiso-C15:0 (44.18 %), iso-C16:0 (9.25 %), iso-C17:0 (5.61 %) and anteiso-C17:0 (15.19 %). On the basis of these polyphasic differences, strain SJConT represents a novel species of the genus Arthrobacter, for which the name Arthrobacter nitrophenolicus sp. nov. is proposed. The type strain is SJConT (=MTCC 10104T =DSM 23165T).
Table 2.
Fatty acid profile of Arthrobacter nitrophenolicus strain SJConT and Arthrobacter globiformis DSM 20124T (Data from present study)
| Fatty acid | Arthrobacter nitrophenolicus | A. globiformis |
|---|---|---|
| 14:0 iso | 0.93 | ND |
| 15:0 iso | 16.33 | 13.3 |
| 15:0 anteiso | 44.18 | 55.2 |
| 16:0 iso | 9.25 | 21.0 |
| 16:0 | 6.48 | ND |
| 17:0 iso | 5.61 | ND |
| 17:0 anteiso | 15.19 | 10.50 |
| 18:0 | 1.61 | ND |
ND Not detected
Description of Arthrobacter nitrophenolicus sp. nov.
(Nitr.o.phen.o.li.cus.M.L. adj. Nitro containing nitrogen, N.L. n. phenol phenol; M.L. adj. nitrirophenolicus relating to nitrophenol).
Cells are Gram-positive, catalase-positive, oxidase-negative, non-spore-forming, non-motile and exhibit a rod–coccus growth cycle. The optimal temperature for growth is 30 °C (temperature growth range of 10–40 °C) and pH 7.0 (pH growth range of pH 6–10). The cells grow on nutrient broth medium with 4 % NaCl, and degrade 2-chloro-4-nitrophenol, 4-nitrophenol and 3-methyl-4-nitrophenol. Chlorohydroquinone, a major intermediate product of degradation of 2-chloro-4-nitrophenol, which is further degraded via formation of maleylacetate. It hydrolyses gelatin and casein but not starch. No growth occurs on Simmon Citrate and Mac Conkey Agar. Chlorohydroquinone is negative for indole production, H2S production, nitrate reduction, Voges-Proskaur test, methyl red, and oxidation-fermentation tests; It produce acids from l-arabinose, d-glucose, and d-manitol, and oxidizes the following substrates in Biolog GN2 Microplates: arbutin, d-cellobiose, d-fructose, d-galactose, d-gluconic acid, α-d-glucose, m-inositol, α-d-lactose, maltotriose, d-mannitol, d-mannose, d-melizitose, d-ribose, salicin, d-sorbitol, sucrose, turanose, d-xylose, acetic acid, β-hydroxybutyric acid, γ-hydroxybutyric acid, α-ketoglutaric acid, l-lactic acid, l-malic acid, pyruvatic acid methyl ester, pyruvic acid, l-alaninamide, d-alanine, l-alanine, L-alanyl-Glycine, l-asparagine, l-glutamic acid, glycyl-l-glutamic acid, L-pyroglutamic acid, l-serine, putrescine, glycerol, adenosine, inosine. C16:0 (6.48), C18:0 (1.61 %), iso-C14:0 (0.93 %), iso-C15:0 (16.33 %), anteiso-C15:0 (44.18 %), iso-C16:0 (9.25 %), iso-C17:0 (5.61 %) and anteiso-C17:0 (15.19 %) are the major fatty acids. The major polar lipids are diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and a glycolipid. The major menaquinone detected is MK-9(H2) and the peptidoglycan-type is A3α. The DNA G + C content of strain SJConT is 69 ± 1 mol%.
The type strain SJConT =MTCC 10104T =DSM 23165T was isolated from a pesticide-contaminated soil sample collected in Punjab State, India.
Footnotes
This article is dedicated in the memory of Dr Rakesh Jain for his excellent contributions to the field of microbial sciences and biodegradation.
References
- Arora PK, Jain RK. Pathway for degradation of 4-chloro-2-nitrophenol by Arthrobacter sp SJCon. Curr Microbiol. 2011;63:568–573. doi: 10.1007/s00284-011-0022-2. [DOI] [PubMed] [Google Scholar]
- Arora PK, Chauhan A, Pant B, Korpole S, Mayilraj S, Jain RK. Description of Chryseomicrobium imtechense gen. nov., sp. nov., a new member of the family Planococcaceae. Int J Sys Evol Microbiol. 2011;61:1859–1864. doi: 10.1099/ijs.0.023184-0. [DOI] [PubMed] [Google Scholar]
- Bae HS, Lee JM, Lee ST. Biodegradation of 4-chlorophenol via a hydroquinone pathway by Arthrobacter ureafaciens CPR706. FEMS Microbiol Lett. 1996;45:125–129. doi: 10.1111/j.1574-6968.1996.tb08566.x. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Chakraborti AK, Jain RK. Plasmid-encoded degradation of p-nitrophenol and 4-nitrocatechol by Arthrobacter protophormiae. Biochem Biophys Res Commun. 2000;270:733–740. doi: 10.1006/bbrc.2000.2500. [DOI] [PubMed] [Google Scholar]
- Conn HJ, Dimmick I. Soil bacteria similar in morphology to Mycobacterium and Corynebacterium. J Bacteriol. 1947;54:291–303. doi: 10.1128/jb.54.3.291-303.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hanne LF, Kirk LL, Appel SM, Narayan AD, Bains KK. Degradation and induction specificity in Actinomycetes that degrade p-nitrophenol. Appl Environ Microbiol. 1993;59:3505–3508. doi: 10.1128/aem.59.10.3505-3508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Dreisbach JH, Spain JC. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl Environ Microbiol. 1994;60:3030–3032. doi: 10.1128/aem.60.8.3030-3032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karigar C, Mahesh A, Nagenahalli M, Yun DJ. Phenol degradation by immobilized cells of Arthrobacter citreus. Biodegradation. 2006;17:47–55. doi: 10.1007/s10532-005-3048-y. [DOI] [PubMed] [Google Scholar]
- Keddie RM, Collins MD, Jones D (1986) Genus Arthrobacter Conn and Dimmick 1947, 300AL. In: Sneath PHA, Mair NS, Sharpe ME, Holt JG (eds) Bergey’s Manual of systematic bacteriology, vol 2. Williams & Wilkins, Baltimore, pp 1288–1301
- Koch C, Schumann P, Stackebrandt E. Reclassification of Micrococcus agilis (Ali-Cohen 1889) to the genus Arthrobacter as Arthrobacter agilis comb. nov. and emendation of the genus Arthrobacter. Int J Syst Bacteriol. 1995;45:837–839. doi: 10.1099/00207713-45-4-837. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerman VBD. A Guide to the identification of the genera of bacteria. 2. Baltimore: Williams & Wilkins; 1967. [Google Scholar]
- Stackebrandt E, Fowler VJ, Fiedler F, Seiler H. Taxonomic studies on Arthrobacter nicotianae and related taxa: description of Arthrobacter uratoxydans sp. nov. and Arthrobacter sulfureus sp. nov. and reclassification of Brevibacterium protophormiae as Arthrobacter protophormiae comb. nov. Int J Syst Appl Microbiol. 1983;4:470–486. doi: 10.1016/S0723-2020(83)80005-8. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Westerberg K, Elväng AM, Stackebrandt E, Jansson JK. Arthrobacter chlorophenolicus sp. nov., a new species capable of degrading high concentrations of 4-chlorophenol. Int J Syst Evol Microbiol. 2000;6:2083–2092. doi: 10.1099/00207713-50-6-2083. [DOI] [PubMed] [Google Scholar]


