Abstract
Terminologies and ontologies are increasingly prevalent in health-care and biomedicine. However they suffer from inconsistent renderings, distribution formats, and syntax that make applications through common terminologies services challenging. To address the problem, one could posit a shared representation syntax, associated schema, and tags. We identified a set of commonly-used elements in biomedical ontologies and terminologies based on our experience with the Common Terminology Services 2 (CTS2) Specification as well as the Lexical Grid (LexGrid) project. We propose guidelines for precisely such a shared terminology model, and recommend tags assembled from SKOS, OWL, Dublin Core, RDF Schema, and DCMI meta-terms. We divide these guidelines into lexical information (e.g. synonyms, and definitions) and semantic information (e.g. hierarchies.) The latter we distinguish for use by informal terminologies vs. formal ontologies. We then evaluate the guidelines with a spectrum of widely used terminologies and ontologies to examine how the lexical guidelines are implemented, and whether our proposed guidelines would enhance interoperability.
Keywords: Biomedical Ontology, Terminology, W3C, OWL, RDF, Ontology Representation Guidelines
1. Introduction
Healthcare and biomedicine, more than most other sciences, has become dependent upon controlled terminologies and ontologies for interoperability, inference, and knowledge integration [6, 5, 8]. However, many terminologies and ontologies have independently emerged, some with overlapping or conflicting content. Additionally, virtually all of these terminologies and ontologies have their own representation schema, semantic tags, or term-element relationships which for example might indicate that a given text string is a synonym for a particular concept. This paper does not address the content overlap and conflict problem between and among ontologies, terminologies, and vocabularies, which is well characterized and largely understood if not solved [13]. This paper examines terminology syntax, representation, and tagging, which has not received anywhere near the same attention yet is probably equally important to content in practical usage. While many writers might assume the syntax problem largely solved with the introduction of OWL (Web Ontology Language) [1] or certainly now with OWL2 [25], we maintain that while elegant for description logic assertions, flavors of OWL are incomplete for practical distinction of a definition from a usage note. In addition, OWL does not provide modeling constructs to harmonize between ontologies: a well-defined abstraction layer is required for specifying how it is to be used [18]. We propose some guidelines and tags for the representation of terminologies, and separate these into structural or lexical information (e.g. preferred terms, synonyms, definition, and provenance) vs. semantic information (e.g. hierarchies). The latter renderings are divided into elements for informal terminologies, and elements for formal ontologies.
2. Challenge Illustration
By examining ontologies hosted in the NCBO BioPortal [7], we found that representation inconsistencies continue to flourish in biomedical ontologies. As an example, one community may publish the definition of a concept as an rdfs:comment, while a second may use the tag DEF, and yet another may use definition. Add to this the fact that the citation for the definition is sometimes found embedded in XML fragments inside a resource, as a secondary data property for a reified resource, or in many other creative and incompatible solutions. Only recently has the Semantic Web community begun to converge on what might be considered a more standard set of tags2. At the moment, however, these tags are still scattered across a variety of specifications such as Resource Description Framework (RDF) Schema (RDFS), Simple Knowledge Organization System (SKOS), SKOS eXtension for Labels (SKOS-XL), Web Ontology Language (OWL), Ontology Metadata Vocabulary (OMV), and many others [28, 29, 31, 32, 1, 24]. To compound this problem, many of the tags are close to identical or having overlapping semantics. Let us use “description” as an example. RDFS provides rdfs:comment –“an instance of rdf:Property that may be used to provide a human-readable description of a resource.” SKOS specifies skos:note – “Notes are used to provide information relating to SKOS concepts … it could be plain text, hypertext, or an image; it could be a definition, information about the scope of a concept, editorial information, or any other type of information.” The OMV provides omv:description –“Free text description of an ontology”, (a tag that must be present in any OMV compliant description.) BioPortal’s prototype RDF model uses “bio-portal:description” (http://rest.bioontology.org/bioportal/virtual/rdf/1321/NEMO_spatial:NEMO_0000024), while the Neural ElectroMagnetic Ontologies (NEMO) define their own properties, including “nemo:comment” (http://nemo.nic.uoregon.edu/ontologies/NEMO_annotation_properties.owl). All of these examples are perfectly well-formed RDF and yet, without additional information and transformations, none of the resulting content would be recognized as similar to a software program that was not specifically configured to recognize these as such.
In addition, we also want to ensure that the OWL semantic assertion and definition capacities are used in a semantically correct way. Many thesauri or classification schemes in the biomedical domain were designed to describe information through natural languages and define information in informal means. They mainly define a set of concepts, as well as associations and hierarchies among these concepts. In this case, it is impossible to represent the relations between the concepts using OWL Description Logic (DL) [15]3 without making further assumptions.
There are many biomedical terminologies and ontologies that are not originally represented in semantic web notations. Just to name a few, the Open Biological and Biomedical Ontologies (OBO) foundry hosts more than 100 ontologies in the OBO format [23]. Classification Mark-up Language (ClaML) [34], as another example, is an European norm (CEN/TS 14463) adopted by the WHO to share its classifications such as the International Classification of Diseases (ICD). When transforming ontologies or terminologies from other formats to OWL, we need to understand the potential issues on adding or changing the semantics from the original intents. Many biomedical terminologies or classification schemes do not provide a mechanism for a formal logic-based representation. In order to convert them to a formal logic-based representation, the convertor has to re-engineer the information and interpret it to generate formal axioms or facts. Since there are no formal semantics defined in the original representation, it is improbable that all translation would converge upon a single reliable interpretation [30]. Kashyap et al [17], for example, discussed multiple interpretations of a single relation. A relation Bacteria cause Infection can have 5 possible interpretations:
All Bacteria cause {each/only/some) Infection
Some Bacteria cause {all/some} Infection
Since the original source only specifies a relation Bacteria cause Infection without any further assertions, it is impossible to know which interpretation faithfully represents the original meaning. Many existing approaches chose to use owl:someValuesFrom as the value constraint to link the restriction to the class description [3, 21, 22, 19]. With this assumption, the converted ontologies would choose one single interpretation: all [ClassA] [relation] some [ClassB]. The Bacteria cause Infection example can be therefore interpreted as: each bacterium must cause some infection or it is not an instance of bacteria, which is not necessarily true. Therefore, we can argue that there cannot be a general solution for choosing one single interpretation when converting relations to OWL. How to correctly interpret the relations and/or the restriction semantics depends on each individual case. SKOS, on the another hand, offers a pragmatic solution for representing associations between two concepts in thesauri or classification schemes [26]. SKOS concepts are first order resources (OWL individuals). We can define predicates between two SKOS concepts directly (e.g., “Bacteria cause Infection”). We can further add qualifiers such as “might” to this relation using annotation properties (see Section 5.5 for details) to specify that Bacteria might cause Infection. In addition, if the object property “cause” is defined as symmetric, then we can further infer that “Disease might be caused by bacteria”. This cannot be achieved by using OWL existential restrictions, which are not symmetric.
It is important for ontology designers to understand when to use OWL or SKOS based on their own applications. SKOS is designed primarily for human users to define or navigate lexical features of terminology resources. SKOS defines concepts as OWL individuals and therefore is able to relate two concepts by using object properties directly. Using SKOS, the problem of ambiguity can be left to human readers who can tolerate ambiguities well. OWL, on the other hand, is designed primarily for automatic machine processing and reasoning. OWL defines concepts on the class level and these classes are usually related by restrictions using object properties. OWL relies on well-defined formal semantics which allows no ambiguities. In summary, it will be helpful to have guidelines according to which users can decide to use OWL or SKOS in their own applications.
These problems and challenges motivated us to propose a set of guidelines that ontology engineers can reference when creating OWL ontologies or converting ontologies from other formats to OWL.
3. Terminology Representation Guidelines
3.1. Rationale
The guidelines introduced in this paper fall into two broad categories: guidelines for the representation of human readable information such as comments, designations, definitions; and guidelines for the representation of “semantic” information that can be used by automated tooling to classify and reason across ontological contents. We will refer the first category as ”Lexical information” and the second as “Semantic information”. Our overall philosophy for designing the guidelines is: (1) using unified and standard tags to represent common lexical information; and (2) using OWL’s semantic assertions only when needed. Ontology designers need to avoid assuming semantic assertions that are not provided by the original sources when transferring terminologies or thesauri from other formats to OWL/RDF.
The first step in the proposal of terminology representation guidelines was to identify a set of commonly-used elements in biomedical ontologies and terminologies. These elements were identified based on our experience with the Common Terminology Services 2 (CTS2) Specification [10] as well as the Lexical Grid (LexGrid) [4] project. CTS2 is an Object Management Group (OMG) Standard for defining the functional requirements of service interfaces that allow the representation, access, and maintenance of terminology contents either locally, or across a federation of terminology service nodes. CTS2 contains a computational model where the common services are specified, as well as an information model where common elements in biomedical terminologies or ontologies are defined. The LexGrid project built upon a set of common tools, data formats, and read/update mechanisms for storing, representing and querying biomedical ontologies and vocabularies. The primary goal of LexGrid was to accommodate multiple vocabulary and ontology distribution formats and support of multiple data stores for a federated vocabulary and ontology access. Other than CTS2 and LexGrid, we also took the OMV (Ontology Metadata Vocabulary) into consideration when identifying the common elements. The set of elements for which we propose guidelines essentially cover two parts (1) metadata about the ontologies and terminologies, their versions and provenance; and (2) the content of the ontologies and terminologies, which includes concepts, common annotations of the concepts, and common relationships between concepts.
After identifying the common elements, we developed canonical mappings from these elements to a collection of “RDF-centric” (RDF, RDFS, SKOS, OWL, etc.) tags for representing terminological information using the appropriate World Wide Web Consortium (W3C) standards. The standard W3C notations we evaluated and include in our proposed guidelines are: the Resource Description Framework (RDF) [28], RDF Schema [29], the Web Ontology Language (OWL) [1], OWL 2 [25], and Simple Knowledge Organization System (SKOS) [31]; all are W3C recommendations. Additionally, SKOS eXtension for Labels (SKOS-XL) [32] is a W3C candidate recommendation, where labels are defined as resources to allow descriptions and associations to be added to these labels. In addition to the above W3C recommendations, we also include the Dublin Core Metadata Element Set (dc) [11], which is widely used to describe digital materials. The Dublin Core Metadata Element Set includes fifteen properties for use in resource description maintained by the the Dublin Core Metadata Initiative (DCMI). Table 1 lists the prefixes of these resources and their URIs. Finally, we used the ISO TC37 Data Category Registry (http://www.isocat.org/) for representing lexical tags such as synonyms and acronyms.
Table 1.
Prefix Index of the Standard Resources Used in the Proposed Guidelines
3.2. Lexical-information Guidelines
Lexical information plays an important role in biomedical ontologies, serving to identify the intent, purpose and meaning of the elements that constitute an ontology. Table 2 lists a set of guidelines for representing lexical information. Each guideline carries an identifier for easy reference in the subsequent discussion (column 1 in Table 2). Column 2 in Table 2 introduces their intended purposes and the tag or tags that we propose to represent them. And in column 3 we give examples of how to represent information according to the guidelines. In Table 3, we list sample lexical representations from a few different ontologies (detailed information about the ontologies are listed in Table 4) and if the guidelines are illustrated in these representations. In the rest of the section, we use these examples to explain how we propose to represent relevant information according to the guidelines.
Table 2.
Guidelines for Lexical Information: Column 1 shows the guideline numbers with a short description of the purpose for each guideline; Column 2 describes the guidelines; and Column 3 gives some examples of how to use the tags to present the corresponding information
| No./Purpose | Guideline | Example or Possible Tags |
|---|---|---|
| L1 | ||
| Version | Use owl:versionInfo to capture the version information, typically of an ontology. It can also be applied to properties and classes. | <owl:versionInfo>Revision 1.2</owl:versionInfo> |
|
| ||
| L2 | ||
| Author | Use dc:creator to represent a person, an organization, or a service that is primarily responsible for making the resource [11] | <dc:creator>Cui Tao</dc:creator> |
|
| ||
| L3 | ||
| Contributor | Use dc:contributor to represent a person, an organization, or a service that is responsible for making contributions to the resource [11] | <dc:contributor>BSI, Mayo Clinic </dc:contributor> |
|
| ||
| L4 | ||
| Copyright | Use dc:rights for copy right information | <dc:rights>(c) Mayo Clinic, 2010 </dc:rights> |
|
| ||
| L5 | ||
| Source | Use dc:source to describe the resource from which the described resource is derived [11] | <dc:source> |
|
| ||
| L6 | ||
| Preferred Label | Use skos:prefLabel for the preferred label for a resource | <skos:prefLabel> |
|
| ||
| L7 | ||
| Other Label | Use skos:altLabel for alternative label for a resource. Additionally, use ISOcat:acronymFor, ISOcat:abbreviationFor, and ISOcat:synonym 4 when representing acronyms, synonyms, and abbreviations. | use <skos:altLabel> and appropriate ISO tags to annotate information such as shortName (AA), singleLetterName(AA), abbrev (BIRNLex), synonym (BRINLex), and abbreviation (SAO) |
|
| ||
| L8 | ||
| Language | Use dc:language to identify the language used for the ontology itself; Use language tag [27] to identify languages in other lexical annotations | <dc:language>en</dc:language> when using Dublin Core to identify the human language used in the ontology; or @en when using language tag to describe the human language used for a particular resource: <rdfs:label>wine@en</rdfs:label> |
|
| ||
| L9 | ||
| Definition | Use skos:definition to provide a plain text definition on any type of resource | <skos:definition> |
|
| ||
| L10 | ||
| Note | Use skos:note and its sub-properties (except skos:definition) to define different kind of comments | <skos:note>, <skos:changeNote>, <skos:editorialNote>, <skos:example>, <skos:historyNote>, <skos:scopeNote> |
Table 3.
Exemplary Illustrations of Lexical Representations: Column 1 lists sample lexical representations in their original source formats; Column 2 shows their corresponding sources; Column 3 indicates the relation between the sample representation and its corresponding guidelines where “illustrated” means that the guide is illustrated in the example and “recommend” means that the guideline is not illustrated in the example, but we recommend using the listed guidelines.
| Example in Original Format | Source | Relation to Guidelines | |
|---|---|---|---|
| 1 | <owl:versionInfo> Version 1.2, copyright The University of Manchester, Nick Drummond, Georgina Moulton, Robert Stevens, Phil Lord </owl:versionInfo> | AA | recommend L1–L3 |
| 2 | <dc:rights>free, no license required </dc:rights> | OBI | L4 illustrated |
| 3 | <dc:source>Barry Smith: ”Against Fantology”</dc:source> | BFO | L5 illustrated |
| 4 | <preferred_label>Carollia</preferred_label> | BIRNLex | recommend L6 |
| 5 | xmlns:core=”http://www.w3.org/2004/02/skos/core#”<core:prefLabel>Drug Delivery Device</core:prefLabel> | BFO | L6 illustrated |
| 6 | <npo:preferred_Name>aluminium atom</npo:preferred_Name> | NPO | recommend L6 |
| 7 | <abbrev>cislt</abbrev> | BIRNLex | L7 not fulfilled |
| 8 | <synonyms>Cisterna lamina terminalis|Lamina Terminalis Cistern</synonyms> | BIRNLex | recommend L7 |
| 9 | <definition>The series of events in which a sensory light stimulus is received and converted into a molecular signal. [GO:ai] (GO)</definition> | BIRNLex | recommend L9 |
| 10 | xmlns:desc=”http://bioontology.org/ontologies/biositemap.owl#”<desc:definition>As defined by the USA, http://en.wikipedia.org/wiki/Medical_device</desc:definition> | BRO | recommend L9 |
| 11 | <rdfs:comment>Definition: The biological source of an entity (e.g. protein, RNA or DNA). Some entities are considered source-neutral (e.g. small molecules), and the biological source of others can be deduced from their constituents (e.g. complex, pathway). Examples: HeLa cells, human, and mouse liver tissue.</rdfs:comment> | BioPax | recommend L9–L10 |
Table 4.
Detailed Information for Ontologies in Table 3: Column 1 shows the acronym of each ontology; Column 2 shows the full names; Column 3 shows their URIs and Column 4 shows the versions of the ontologies we used to evaluate
| Acronym | Name | Source URI | Version |
|---|---|---|---|
| AA | Amino Acid Ontology | http://www.coode.org/ontologies/amino-acid/2006/05/18/amino-acid.owl | 2.0 |
| BFO | Basic Formal Ontology | http://www.ifomis.org/bfo/1.1 | 1.1 |
| BIRNLex | BIRNLex (Biomedical Informatics Research Network controlled terminology) | http://bioontology.org/projects-/ontologies/birnlex | 1.3.1 |
| BRO | Biomedical Resource Ontology | http://bioontology.org/ontologies/BiomedicalResourceOntology.owl | 2.7.1 |
| BioPax | Biological Pathways Exchange Level 3 | http://www.biopax.org/release/biopaxlevel3.owl | 0.94 |
| NPO | Nano Particle Ontology | http://purl.bioontology.org/ontology/npo | 1.0 |
| OBI | The Ontology for Biomedical Investigations | http://purl.obolibrary.org/-obo/obi.owl | 1.0 |
| SAO | Subcellular Anatomy Ontology | http://ccdb.ucsd.edu/SAO/1.2 | 1.2 |
Guidelines L1 to L5 in Table 2 are proposed for describing meta-level information of an ontology or an ontology entity. Rows 1–3 in Table 3 show the exemplary evaluation for some sample representations in different OWL ontologies based on these guidelines. Row 1 in Table 3, for example, uses owl:versionInfo for version, copyright, and creators. It is difficult for computer programs and even for human readers to parse the information and interpret what it means. This representation does not demonstrate guidelines L1-L3. Our guidelines propose to represent the information as:
<owl:versionInfo>1.2</owl:versionInfo> <dc:rights>The University of Manchester</dc:rights> <dc:creator>Nick Drummond</dc:creator> <dc:creator>Georgina Moulton</dc:creator> <dc:creator>Robert Stevens</dc:creator> <dc:creator>Phil Lord</dc:creator>
The new proposed representation listed version, copyright, and creators separately using canonical tags recommended by the guidelines. This way the information can be easily queried and/or searched by computer programs without tedious configuration by source.
The examples in Rows 2 and 3 accord with the guidelines L4 and L5. Many biomedical terms can be represented in various ways. SKOS de-fined a tag skos:prefLabel for representing the preferred label of each term or concept. Guideline L6 recommends using skos:prefLabel whenever a preferred label needs to be presented. Note that SKOS allows only one value of skos:prefLabel per language, per context. For example, each concept in an ontology for clinical terms could have a preferred label for clinicians, and another preferred label for consumers. Rows 4–6 in Table 3 show examples of preferred labels. Both BIRNLex and NPO use a self-defined OWL property for representing preferred labels. BFO, on the other hand, uses skos:prefLabel as we recommended. Therefore, Guideline L6 is illustrated in BFO, but not in BIRNLex nor NPO.
In addition to preferred labels, we may also need to represent alternative labels for an ontology entity. Guideline L7 specifies how to use skos:altLabel for alternative labels. Synonyms, acronyms, abbreviations, etc, should also be considered as alternative labels. We recommend using skos:altLabel to represent this kind of information in addition to the original properties. W3C has not yet specified notations for annotation properties such as synonyms, acronyms, or abbreviations. These labels, however, quite commonly exist in the biomedical domain. Therefore, a standard way to represent these labels are highly desirable. We propose using the ISO TC37 Data Category Registry (http://www.isocat.org/) to represent these labels, which could potentially be declared as sub-properties of skos:altLabel.
For example, here is a way to represent the information in Row 7 in Table 3:
<ISOcat:abbreviation >cislt</ISOcat:abbreviation> <skos:altLabel>cislt</skos:altLabel>
Note that lexical labels such as acronyms or abbreviation correspond to a name (or a label) of a concept, not to the concept itself. For example, a concept might have a preferred label “Food and Agriculture Organization”, and an alternative label “FAO”. “FAO” is actually the abbreviation of “Food and Agriculture Organization”, but not of the concept itself. Therefore, theoretically the abbreviation should not annotate the concept directly. In SKOS, both the preferred labels and the alternative labels need to be attached to the concept. SKOS-XL, on the other hand, provides an approach where these representations can be linked to each other. In sections 5.3 and 5.4, we explore alternative ways to represent such information.
Guideline L8 specifies how to define languages used in an ontology. When specifying the language used on the ontology meta-level, we recommend using dc:language. For example, BFO uses dc:language with the value as the language tag “en” for English as shown below, which fulfils our guideline.
<owl:Ontology rdf:about=““> … <dc:language>en</dc:language> … </owl:Ontology>
Guideline L8 also specifies how to use the language tag [27] to identify languages used in a specific annotation property. For example, one can specify two preferred labels for an OWL class, one for English, one for German, like this:
<skos:prefLabel>Organisms transmitting pathogens@en</skos:prefLabel> <skos:prefLabel>Parasiten uebertragende Organismen@de</skos:prefLabel>
We also recommend using skos:note and its sub-properties to describe plain definitions and comments as guidelines L9 and L10 specify. SKOS provides a set of tags to define specific types of notes, comments, definitions, and examples. We decided to choose skos:note over rdfs:comment because using rdfs:comment as a general tag for multiple purposes could introduce ambiguity in many situations. Rows 9–10 in Table 3 show two examples. BIRNLex (Row 9) defined their own annotation property called definition, whereas, BRO uses a “definition” notation defined by another OWL ontology (biositemap.owl). Our guidelines recommend using skos:definition for representing the definition information of a concept. Therefore, Guideline L9 is not demonstrated in either examples. BioPax (Row 11 in Table 3) uses rdfs:comment for both definition and examples. This introduces ambiguity for both human readers and computer systems. Instead, we recommend using skos:definition for definitions, and skos:example for examples.
3.3. Semantic-information Guidelines
In this section, we discuss the guidelines for representing semantic information, especially when transforming terminologies and ontologies from other formats to OWL. Table 5 shows the detailed descriptions of these guidelines. We provide two options to fulfil different needs for representing the information in the biomedical domain.
Table 5.
Guidelines for Semantic Information: Column 1 shows the guideline numbers; Column 2 shows the guideline descriptions; and Column 3 gives some examples of how to represent information according to the guidelines.
| No. | Guideline | Example |
|---|---|---|
| SKOS Route | ||
| S1a | Use skos:Concept for concepts that are not from OWL | Use skos:Concept to represent OBO terms |
| S2a | Defining any relationship or association between two concepts as an instance of owl:ObjectProperty and a sub-property of skos:semanticRelation | representing OBO relationships |
| S3a | Use rdfs:subClassOf for parent-child hierarchical relationships | Use rdfs:subClassOf to represent relationships such as OBO is_a and UMLS hasSubtype |
| S3a | Use skos:broader and skos:narrower to assert any kind of direct hierarchical link between two SKOS concepts [31] | Defining OBO part_of as a sub-property of skos:broader |
| S4a | Use skos:related to assert an associative link between two SKOS concepts [31] | Use skos:related to represent Mesh see_also |
| OWL Route | ||
| S1b | Use owl:Class for all the concepts | Use owl:Class to represent OBO terms |
| S2b | Use owl:ObjectProperty and assertions for relationships between two concepts | representing OBO relationships |
| S3b | Use rdfs:subClassOf for parent-child hierarchical relationships | Use rdfs:subClassOf to represent relationships such as OBO is_a and UMLS hasSubtype |
| S4b | Use owl:equivalentClass for stating the equivalence of two named classes | Use owl:equivalentClass to represent relationships such as UMLS same_as |
| S5b | Use owl:disjointWith to assert that the class extensions of the two class descriptions involved have no individuals in common | Use owl:disjointWith to represent relationships such as OBO disjoint_from |
| S6b | Use owl:intersectionOf to describe a class for which the class extension contains precisely those individuals that are members of the class extension of all class descriptions in the list. | Use owl:intersectionOf to represent relationships such as OBO intersection_of |
| S7b | Use owl:unionOf to describe an anonymous class for which the class extension contains those individuals that occur in at least one of the class extensions of the class descriptions in the list. | Use owl:unionOf to represent relationships such as OBO union_of |
| S8b | Use owl:complementOf to describe two classes that are complement to each other. | |
| S9b | Use owl:ObjectProperty, owl:DataTypeProperty, and owl:AnnotationProperty in a semantically correct way | e.g., owl:AnnotationProperty cannot be inherited by subclasses. |
The SKOS route (Guidelines S1a, S2a, and S3) targets thesauri or classification schemes, which “do not assert any axioms or facts, but rather identify and describe information through natural language and define information in informal means” [31]. They mainly define a set of concepts as well as associations and hierarchies among these concepts. We propose using skos:Concept to represent the “concepts” in this kind of thesauri or classification schemes as Guideline S1a specifies. SKOS defines a skos:Concept as “the unit of thought, ideas, meanings, or (categories of) objects and events” without giving any further semantic assertions. This could apply to the named entities in many knowledge organization systems: terminologies, thesauri or classification schemes. For any associations or relations, we propose defining them as an instance of owl:ObjectProperty and a sub-property of skos:semanticRelation as Guideline S2a specifies. SKOS defines relations such as broader, narrower, or related in the same way. Since both the domain and the range of skos:semanticRelation are skos:Concept, any of its sub-property inherits the same domain and range. Therefore, we can use these properties to describe the relations between two skos:Concepts. Here we show an example using a sample term from the adult mouse anatomy OBO ontology5.
[Term] id: MA:0000002 name: spinal cord grey matter is_a: MA:0001112 ! grey matter relationship: part_of MA:0000216 ! spinal cord
Since this OBO ontology only defines terms, simple associations between terms (e.g., part_of), and hierarchies (using is_a), we follow the guidelines in the SKOS Route. Below is the RDF triple representations for this OBO term.
MA:0000002 rdf:type skos:Concept;
skos:prefLabel spinal cord grey matter;
rdfs:subClassOf MA:0001112;
part_of MA:0000216;
part_of rdfs:subPropertyOf skos:broaderTransitive
rdf:type owl:ObjectProperty
If one needs to formally express the semantic definitions of the knowledge in an ontology, we recommend the OWL route. A formal ontology in OWL is expressed as sets of axioms and facts, and provides formal definitions of the knowledge embedded in the ontology. OWL reference [1] distinguishes six types of class descriptions:
a class identifier (a URI reference)
an exhaustive enumeration of individuals that together form the instances of a class
a property restriction
the intersection of two or more class descriptions
the union of two or more class descriptions
the complement of a class description
The OWL Route allows us to use the above class descriptions to define a concept in an ontology or terminology. As specified by Guideline S1b, every concept needs to be defined as an OWL class. Guideline S2b specifies how to define relations between two classes. Each specific relationship between two OWL classes could be defined as an instance of owl:ObjectProperty and then we use assertions with restrictions to define the relations between two classes. To invoke such assertion axioms, however, we need to ensure the semantic definition is correct as we have discussed in Section 2. OWL also specifies how to represent hierarchical, equivalent, and disjoint relations between two classes as Guidelines S3b–S5b specify. Guidelines S6b and S7b specify how to assert the intersection and the union of a set of classes respectively. Guideline S8b specifies how to describe two classes that complement each other. Ontology designers should also ensure that OWL object properties, data type properties, and annotation properties are used in a semantically correct way as Guideline S9b specifies. In OWL DL, the sets of object properties, data type properties, and annotation properties must be mutually disjoint. In addition, an annotation property simply provides human readable annotations on classes, properties, individuals and ontology headers. Annotation properties do not provide any semantics, and therefore cannot support inferencing.
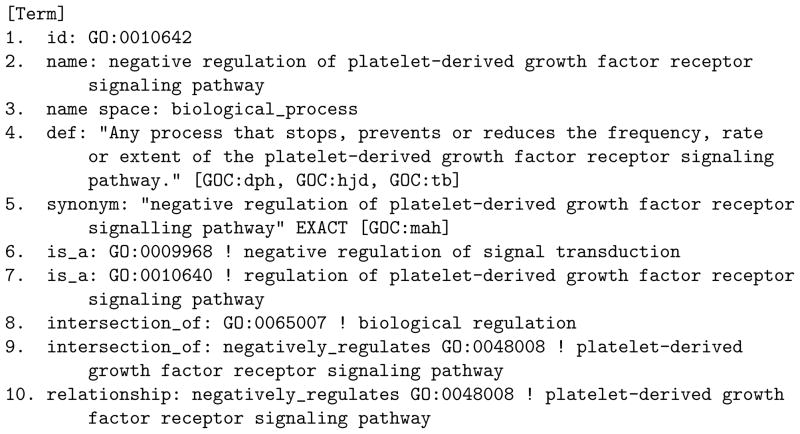
Figure 1 shows a sample OBO term from the Gene Ontology6 and Table 6 shows the OWL representation of that as rendered by Protégé 4. Because this OBO ontology involves axioms like intersection of, we believe it requires the OWL Route for its representation. Table 6 shows the sample illustration of how to represent semantics correspond to the guidelines.
Figure 1.
A Sample OBO Term from the Gene Ontology (http://www.geneontology.org/ontology/obo_format_1_2/gene_ontology_ext.obo)
Table 6.
Exemplary Illustration of the OWL Semantic Representations Rendered by Protégé 4 for the Example OBO term in Figure 1. Please note that the lexical information was not included in this table.
| Example in Original Format | Source | Illustration (Semantic Guidelines) | |
|---|---|---|---|
| 1 | |||
| <owl:Class rdf:about=“#GO_0010642”> | the GO term in Figure 1 | S1b illustrated | |
|
| |||
| 2 | |||
| <rdfs:subClassOf rdf:resource=“#GO_0009968”/> | Lines 6–7 | S3b illustrated | |
| <rdfs:subClassOf rdf:resource=“#GO_0010640”/> | |||
|
| |||
| 3 | |||
| <owl:equivalentClass> | Lines 8–9 | S2b, S6b illustrated | |
| <owl:Class> | |||
| <owl:intersectionOf rdf:parseType=“Collection”> | |||
| <rdf:Description rdf:about=“#GO_0065007”/> | |||
| <owl:Restriction> | |||
| <owl:onProperty rdf:resource=“#negatively_regulates”/> | |||
| <owl:someValuesFrom rdf:resource=“#GO_0048008”/> | |||
| </owl:Restriction> | |||
| </owl:intersectionOf> | |||
| </owl:Class> | |||
| </owl:equivalentClass> | |||
|
| |||
| 4 | |||
| <rdfs:subClassOf> | Line 10 | S2a illustrated | |
| <owl:Restriction> | |||
| <owl:onProperty rdf:resource= “#negatively_regulates”/> | |||
| <owl:someValuesFrom rdf:resource= “#GO_0048008”/> | |||
| </owl:Restriction> | |||
| </rdfs:subClassOf> | |||
Users can choose to use either the SKOS or OWL guidelines depending upon how formal they would like to make their terminologies/ontologies to be by following our guidelines. Although there seems to be a division between the representation following the SKOS route and the OWL route, there are options where SKOS and OWL may interact. W3C has proposed several patterns that allow users to use SKOS and OWL together [2].
4. Guideline Evaluations
We evaluated the guidelines from three aspects: (1) how well the guidelines cover the information in existing ontologies; (2) how well the proposed guidelines can be assessed consistently among different human experts; and (3) how well the existing ontologies already represent corresponding information in accordance to the guidelines. We focus on lexical guidelines only in this evaluation since we believe that the interpretations of the semantic representations require domain knowledge of each ontology, as well as the correct understanding of the ontology designer’s original intentions, which are beyond the scope of this paper.
4.1. Ontology Selection
We evaluated the guidelines using a set of commonly used ontologies. These ontologies were chosen based on the most-viewed-ontology list provided by the NCBO BioPortal [7]. The top 15 ontologies in OWL were downloaded from BioPortal. Table 7 shows the list of OWL ontologies we included in the study with their acronyms, names, URIs, versions, and the BioPortal “most viewed” ranks7.
Table 7.
Detailed Information for the OWL Ontologies studied: Column 1 shows the acronyms; Column 2 shows their full names; Column 3 shows their URIs; Column 4 indicates the particular version of the ontology we used; and Column 5 provides ranks in BioPortal Most-Viewed-Ontology List.
| Acronym | Name | URI | Version | Rank |
|---|---|---|---|---|
| AA | Amino Acid Ontology | http://www.co-ode.org/ontologies/amino-acid/2006/05/18/amino-acid.owl | 1.3 | 39 |
| BFO | Basic Formal Ontology | http://www.ifomis.org/bfo/1.1 | 1.1 | 86 |
| BioPax | Biological Pathways Exchange Level 3 | http://www.biopax.org/release/biopax-level3.owl | 0.94 | 13 |
| BIRNLex | BIRNLex (Biomedical Informatics Research Network controlled terminology) | http://bioontology.org/projects-/ontologies/birnlex | 1.3.1 | 26 |
| BRO | Biomedical Resource Ontology | http://bioontology.org/ontologies/BiomedicalResourceOntology.owl | 2.7.1 | 26 |
| DermLex | The Dermatology Lexicon | http://purl.bioontology.org/ontology/-DermLex | 1.0 | 38 |
| EFO | The Experimental Factor Ontology | http://www.ebi.ac.uk/efo/ | 2.4 | 18 |
| FMA | Foundational Model of Anatomy | http://purl.bioontology.org/-ontology/FMA | 3.0 | 2 |
| GALEN | The GALEN Ontology | http://www.co-ode.org/galen/-fullgalen.owl | 1.1 | 29 |
| MGED | The Microarray Gene Expression Data Ontology | http://mged.sourceforge.net/-ontologies/MGEDOntology.owl | 1.3.1.1 | 40 |
| NCIt | NCI Thesaurus | http://ncicb.nci.nih.gov/xml/owl/-EVS/Thesaurus.owl | 10.03 | 1 |
| NIFSTD | The Neuroscience Informatics Framework Lexicon | http://ontology.neuinfo.org/-NIF/nif.owl | 1.8 | 9 |
| OBI | Ontology for Biomedical Investigations | http://purl.obolibrary.org/-obo/obi.owl | 1.0 | 5 |
| OCRe | Ontology of Clinical Research | http://purl.org/net/OCRe/-OCRe-Start-Here | 0.95 | 27 |
| RadLex | A Lexicon for Uniform Indexing and Retrieval of Radiology Information Resources | http://bioontology.org/projects-/ontologies/radlex/radlexOwl | 3.0 | 3 |
4.2. Evaluation on the Guideline Coverage
To evaluate the coverage of the guidelines, we studied the annotation properties and data properties defined in the selected ontologies and classified them to four disjoint categories: domain specific, covered by OWL specifications, covered by the guidelines, and not covered by the guidelines.
Domain specific properties refer to the properties that are specific for the ontology domain. For example, BioPax defined properties such as chemicalFormula, sequence, and molecularWeight, which are specific for describing molecular biology data. Because our guidelines are designed for features that exist commonly in biomedical ontologies, we do not plan to include the definition of domain-specific properties in the guidelines.
Properties covered by OWL specifications include those properties that can be represented by OWL synopsis or schema directly. For example, Birn-Lex has a property called class_or_indiv. Whether an OWL entity belongs to a class or an individual can be defined using rdf:type directly without using additional self-defined properties. For those properties that are not domain specific nor covered by OWL specifications, we determined if they can be covered by the guidelines. Table 8 shows the numbers of properties that have been classified to each category8. As we can see there are three properties that are not covered by the lexical guidelines. The properties external_id_urls and external_ids from BIRNLex describe the mappings between a local concept to an external concept. Although we do not cover these in the lexical guidelines, we believe that these mappings can be covered by the semantic guidelines S4a and S4b. The mappings between two concepts across ontologies could be specified by using owl:equivalentClass for the OWL route or skos:related for the SKOS route. Note that owl:equivalentClass indicates exact mappings between resources and should be used when the two classes are semantically equivalent to each other. In many cases, the mappings between concepts are not exact [20]. In that case, an annotation property is more appropriate to be used just to indicate the possible mappings between the current ontology resource to external resources. In Section 5.1, we further discuss how we propose to define properties for mappings between concepts. In the SKOS route, SKOS provides a list of constructs (skos:closeMatch, skos:exactMatch, skos:broadMatch, skos:narrowMatch, and skos:relatedMatch) as sub-properties of skos:related that users can further choose to specify mappings in different situations. The guideline recommends skos:related as the default to define the mappings because the quality of the mappings is often not available. More specific properties should be used when detailed information of the mappings is available. In addition, NCIt defined a property called ALT_DEFINITION, which is similar to skos:altLabel except it is used for defining alternative definitions. In Section 5.2, we proposed a new tag to the SKOS community for representing alternative definitions. The complete list of the properties and their categories can be found in Appendix A.
Table 8.
Examination of the Guideline Coverage with Selected Ontologies
| Ontology | Domain specific | Covered by OWL specification | Covered by the guidelines | Not covered by the guidelines |
|---|---|---|---|---|
| AA | 0 | 0 | 1 | 0 |
| BFO | 0 | 0 | 0 | 0 |
| BioPax | 29 | 5 | 7 | 0 |
| BIRNLex | 0 | 1 | 6 | 2 (external id urls, external ids) |
| BRO | 4 | 1 | 3 | 0 |
| DermLex | 6 | 0 | 9 | 0 |
| EFO | 7 | 0 | 11 | 0 |
| FMA | 5 | 0 | 13 | 0 |
| GALEN | 0 | 0 | 0 | 0 |
| MGED | 1 | 2 | 8 | 0 |
| NCIt | 61 | 0 | 13 | 1 (ALT DEFINITION) |
| NIFSTD | 0 | 0 | 0 | 0 |
| OBI | 2 | 0 | 0 | 0 |
| OCRe | 0 | 0 | 2 | 0 |
We also listed the corresponding guideline(s) for each property that is classified as “covered by the guidelines” in Appendix A. Most of the conversions between these properties and their corresponding guidelines are straightforward – i.e. require one to one mappings. Sometimes a property can be covered by multiple guidelines. EFO defined a property called source_definition without giving any further description of how to use this property. From the usage of the property, we saw that EFO either uses the property to describe a textual definition of the entity or uses it to record an external URL that links to more detailed information about the entity. Whether one should use guideline L5 (source) or guideline L9 (definition) depends on each individual usage of the property. EFO also defined two properties definition_citation and definition_editor. The property definition_citation is defined as “a document, ontology class, person or organization from which the definition of the class is derived.” This property can be covered by using the combination of guidelines L9 (definition) and L5 (source). Using OWL 2, we can add annotations to another annotation. Therefore, we can add a source annotation to any definitions to represent a definition_citation. Similarly, we use the combination of guidelines L9 (definition) and L2 (author) to cover the property definition_editor.
4.3. Evaluation on Inter-Rater Reliability
To evaluate the inter-rater reliability for accessing the guidelines, three experts (Tao, Pathak, and Wei) studied the ontologies using the guidelines. Each ontology was first studied by three human experts independently, based on the 10 guidelines for lexical representations. For each ontology with each guideline, each human expert needs to determine whether the ontology covers the information referred by the guideline. If yes, they then need to determine if the ontology represents the contents following the representation guideline or if not, why would following the guideline help semantic interoperability among different ontologies.
The examination results were then compared to identify any conflicts and disagreements among the three experts. Kappa coefficients [12] were measured for the inter-rater agreements of the results. There were 13 conflicts and disagreements in the 450 ratings and the kappa coefficient is 90%. The disagreements fall into the following categories:
Including imported ontologies. During the examination, one expert considered imported ontologies as part of the main ontology itself, while the other two only considered the main ontology.
Properties defined but not used. Some ontologies defined properties that were never used to annotate lexical information to the ontology or a concept in the ontology. One expert took all the defined properties into consideration. The other two evaluators did not take the unused properties into consideration. For example, DermLex defined “Definition”, “Synonym Name”, and “Source” as annotation properties, but these properties were never applied.
Different interpretations. There were also some debates due to different interpretations of the properties among evaluators. For example, there is a property called “curator” in OCRe9; two experts consider it as contributor or creator, but one did not. Another example is from FMA. The version information was actually embedded in the name space itself. Two experts took it into consideration, but one did not.
The three experts then had study sessions together to resolve the disagreements. We decided to take all the defined properties into consideration, and to not include the imported ontologies. For the disagreements due to different interpretations, we took the result with the most votes. Table 9 shows our findings for the selected OWL ontologies with guidelines for lexical information. Each row in Table 9 presents the result of one ontology with the first cell of each row indicating the ontology acronym. The rest of the columns show results for each ontology on the lexical guidelines. L1-L10 indicates which particular guideline. A “Y” indicates that the ontology contains the information referred by the guideline and uses the solution proposed by the guideline to represent it; an “NA” indicates that the ontology does not contain the information referred by the guideline; and an “N” indicates that the ontology does not use the representation method proposed by the guideline for the corresponding contents, where an “N1” indicates that the ontology uses an W3C recommendation tag to represent the content, but the tag was used either in an ambiguous or improper way; an “N2” indicates that the ontology uses a self-defined property; and an “N3” indicates that the ontology uses a property defined by another ontology. Note that some cells could be marked as “N” for multiple reasons. For example, an ontology could use rdfs:comment to represent an editorial note, while use a self-defined tag to represent an example. Since either one of them could indicate an “N” in the result, we only reported one reason in the table.
Table 9.
Examination of a Set of Commonly-Used Ontologies for the Representation of Contents Referred by the Lexical Information Guidelines: Column 1 indicates the ontologies by their acronyms; and the rest of columns show the results for each ontology on the lexical guidelines. L1-L10 indicate the particular guideline.
| Ontology | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | L9 | L10 |
|---|---|---|---|---|---|---|---|---|---|---|
| AA | N1 | N1 | N1 | N1 | NA | N2 | N2 | Y | NA | N1 |
| BFO | Y | Y | Y | Y | Y | N1 | N1 | Y | N1 | N1 |
| BioPax | N1 | N1 | N1 | N1 | N2 | N2 | N2 | NA | N1 | N1 |
| BIRNLex | NA | NA | NA | NA | NA | N1 | N2 | NA | N2 | N2 |
| BRO | NA | NA | NA | NA | N2 | Y | NA | N2 | N3 | N2 |
| DermLex | N2 | NA | NA | NA | N2 | N2 | N2 | NA | N2 | N2 |
| EFO | Y | N2 | NA | NA | N2 | NA | N2 | NA | N2 | N2 |
| FMA | N1 | N2 | NA | NA | N2 | N2 | N2 | N2 | N2 | N1 |
| GALEN | NA | NA | NA | NA | NA | NA | N1 | NA | NA | NA |
| MGED | Y | Y | NA | NA | N2 | NA | N2 | NA | NA | N1 |
| NCIt | Y | NA | NA | NA | N2 | N2 | N2 | N2 | N2 | N2 |
| NIFSTD | Y | Y | Y | NA | N3 | Y | N3 | N3 | Y | Y |
| OBI | Y | Y | Y | Y | N1 | N1 | N3 | N3 | N1 | N3 |
| OCRe | NA | N2 | NA | NA | NA | N1 | NA | NA | N2 | N1 |
| RedLex | NA | NA | NA | NA | NA | N2 | N2 | N2 | N2 | N2 |
A “Y” indicates that the ontology uses the representation method proposed by the guideline; an “NA” indicates that the ontology does not contain the information referred by the guideline; and an “N” indicates that the ontology does not use the representation method proposed by the guideline, where an “N1” indicates that the ontology uses an W3C recommendation tag to represent the content, but the tag was used either in an ambiguous or improper way; an “N2” indicates that the ontology uses a self-defined property; and an “N3” indicates that the ontology uses a property defined by another ontology.
4.4. Ontology Evaluation Result
As we can see from Table 9, all the ontologies contain some information covered by the guidelines as all of them have at least one “Y” or “N” in the table. Most of the studied ontologies (11 out of 15) cover contents referred by at least 5 guidelines. Therefore, we can see that the contents referred by our proposed guidelines commonly exist in biomedical ontologies. The ontology representation of these contents can be classified into 4 categories:
Represented following the guideline. In this case, semantic interoperability among different ontologies can be ensured.
Represented using a tag in a W3C recommendation (either in an ambiguous way or improperly). Some ontologies use a single tag for many different purposes. For example, we found that rdfs:comment has been used in many different situations: for representing an example, a definition, an editorial note, etc. In addition, some tags were not used as designed. For example, information such as author, version, and copyright was all represented using owl:versionInfo. These ambiguous representations make it difficult for automatic terminology services to locate the proper information during querying, updating, and integrating ontology elements.
Represented using a self-defined property. Some ontologies defined their own properties for representing contents such as definition, preferred label, etc. Since these contents are very common, we believe that using a unified way to represent them could ensure better interoperability among ontologies than using arbitrarily defined properties.
Represented using a property defined in another ontology. Similar to above, using a tag from an ontology that is not a W3C recommendation can hardly ensure semantic interoperability since different ontologies could choose to import and use properties arbitrarily.
5. Future Direction: Additional Meta-Level Information For Interoperability
During our evaluation process, we found some meta-level information shared by the biomedical ontologies that is important for terminology interoperability but could not yet be reasonably represented by using W3C recommendations. In this situation, new tags need to be introduced and proposed to W3C. Here we list a few relevant new tags we proposed. We have discussed detailed information about these tags in a semantic web conference [33].
5.1. Properties for Concept Mappings
There are a large number of biomedical ontologies covering overlapping contents [14]. Many research efforts have been focusing on identifying mappings between ontology resources [9, 16]. Different approaches or users, however, could have different definitions on mappings. In many cases, these mappings are not exact mappings with semantic equivalence between two resources as owl:equivalentClass indicates, but are rather partial, lexical, or with uncertainties. SKOS provides a list of constructs–(skos:closeMatch, skos:exactMatch, skos:broadMatch, skos:narrowMatch, and skos:relatedMatch)–using which users can further choose to specify mappings in different situations. These properties, however, can only be used for mappings between SKOS concepts. CTS2 allows different mapping resolutions between ontology resources: different versions of mappings between the same pair of resources can be done by different algorithms or users. There is no standard tags from W3c, however, to represent this kind of information. We believe it is necessary to specify standard tags for mapping types and mapping methods for OWL ontology resources.
5.2. Preferred Definition
Textual definitions are very common in biomedical ontologies. Many biomedical concepts have more than one textual definitions. SKOS has defined prefLabel and altLabel, but no such constructs are provided for “definitions”. Akin to prefLabel and altLabel, our objective is to propose prefDefinition and altDefinition to the SKOS committee to be included in the future specification.
5.3. Designation Type
Typical designation types include acronym, synonym, eponym, abbreviation, etc., which commonly exist in the biomedical domain. Here we propose a new construct called designationType, using which ontology designers can declare the type of a lexical presentation. There is no standard recommendations to define designation types by W3C yet. To make it connect to standards, we propose adopting the ISO TC37 Data Category Registry (http://www.isocat.org/) to be the primary resource for designation types. Figure 2 shows an example using designationType. Here we use SKOS-XL data model to represent that a concept <A1> has an alternative label “FAO”. The first three lines in Figure 2 entail the expression <A1> skos:altLabel \FAO”. Using SKOS-XL, we can further specify that the its designation type is ISOcat:acronymFor. The skos plus:designationType10 itself is an OWL annotation property. We can define its range as the collection of a set of pre-defined OWL annotation properties that represent different designation types such as ISOcat:acronymFor, ISOcat:acronym, ISOcat:abbreviationFor, ISOcat:synonym, etc.
Figure 2.

An Example of Designation Type
5.4. Relations between Lexical Properties
Relations between two lexical properties (e.g., definitions, synonyms, etc.) are very common in biomedical ontologies. We propose a new property called noteRelation, which can be used to identify an association between two lexical properties. It can be viewed as a super-property of skosxl:labelRelation. In SKOS-XL, The object property skosxl:labelRelation is designed for representing binary links between instances of the class skosxl:Label. The new proposed noteRelation is designed for representing relations between not only two labels, but also any two lexical properties, such as definitions, notes, and examples. As with designationType, the types of these links could be adopted from the ISO TC37 Data Category Registry. Figure 3 shows an example for using skosxl:labelRelation to represent the property link between two labels. Similarly, we can represent the relations between any two notes using noteRelation.
Figure 3.

An Example of Property Link
5.5. Association Qualification
In many cases in the clinical domain, we need to modify a relation between two concepts or instances. For example, one can define an association, Polandanomaly Dextrocardia, where HAS_CLINICAL_SIGN is the association (relation) name, Poland anomaly is the association source, and Dextrocardia is the association target. This association instance also has an association qualification indicates how frequently the disease has the symptom. The association qualification has a name Frequency and a value Very frequent. Figure 4b shows how we represent this example using an N-ary relation definition proposed by the W3C [36]. We first declare a new node HAS_CLINICAL_SIGN relation_1 for the N-ary relation. For associationQualification, We define a new OWL annotation property, ctm:associationQualification11. Every actual association qualifier is defined as a sub-property of ctm:associationQualification, and therefore is also an instance of OWL annotation property.
Figure 4.
RDF Triples for an Example of AssociationQualifier.
6. Conclusion
In this research, we propose a set of guidelines using constructs in W3C recommendations such as RDF, OWL, and SKOS for representing common lexical and semantic information in the biomedical domain. The guidelines provide a unified semantic-web compatible model for representing biomedical ontologies and terminologies. Based on them, heterogeneous terminological and ontological information can be translated to or represented in semantic web notations with a well-defined interoperability. The biomedical informatics community can greatly benefit by applying semantic-web’s combination of formal semantics, rich expressiveness, and shared software base to biomedical and clinical terminologies. We illustrated the benefit of using the guidelines to enhance semantic interoperability with a set of popular ontologies in the biomedical domain. In addition, we have also identified several limitations of the existing W3C specifications and proposed new tags that warrant broader community engagement.
We propose terminology guidelines for representing biomedical ontologies in W3C notations.
The guidelines have been evaluated using popular biomedical ontologies.
These guidelines provide a unified representation for common elements in the biomedical domain.
They will synergistically tighten the Semantic Web and biomedical domain knowledge.
Semantic interoperability can be achieved by bringing a semantic harmonization over biomedical ontologies and terminologies.
Acknowledgments
This research is partially supported by the National Center for Biomedical Ontologies (NCBO) under the NIH Grant #N01-HG04028 and the NSF under Grant #0937060 to the Computing Research Association for the CIFellows Project. The authors would like to thank Drs Mark Musen, Natasha Noy, and Nigam Shah for their valuable suggestions during the preparation of the paper.
Footnotes
In this paper, we are using the tag and property interchangeably
Here we choose to use OWL DL because it provides maximum expressiveness, computational completeness, and decidability for reasoners; whereas OWL lite has limited expressiveness and OWL full does not have computational guarantees [35].
Please note that the BioPortal also includes ontologies in formats other than OWL, therefore the ranks of the top 15 ontologies in OWL are not necessary the top 15 overall.
RadLex was not included in this evaluation because it is in Protégé XML format, which does not define annotation or data properties.
In general a “curator” is a “contributor” with a more specific role. A set of tags for different contribution roles could be specified as sub-properties of dc:contributor to support work flows in different projects. However, it is out of scope of the current paper.
We use ”skos plus” as the name space for the tags we would like to add to SKOS
Here we use a ctm (common terminology model) to represent the name space for associationQualification temporarily.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.OWL Web Ontology Language Reference. http://www.w3.org/TR/owl-ref/
- 2.Using OWL and SKOS. http://www.w3.org/2006/07/SWD/SKOS/skos-and-owl/master.html.
- 3.OBO2OWL: Lossless transformation between OBO and OWL. 2008 http://www.cs.utexas.edu/~hamid/research/obo2owl.cgi.
- 4.LexGrid: The Lexical Grid. 2009 https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/LexGrid.
- 5.Barry Smith A, Werner Ceusters B. Computing, Philosophy, and Cognitive Science, chapter Ontology as the Core Discipline of Biomedical Informatics. Cambridge Scholars Press; 2006. [Google Scholar]
- 6.Alexandru Adriana, Filip Florin Gheorghe. Intelligent Medical Technologies and Biomedical Engineering: Tools and Applications, chapter Using Ontologies in eHealth and Biomedicine. IGI Global; 2010. [Google Scholar]
- 7.NCBO Bioportal. http://bioportal.bioontology.org/
- 8.Chute Christopher G. Clinical Classification and Terminology: Some History and Current Observations. Journal of American Medical Informatics Association. 2000;7(3):298–303. doi: 10.1136/jamia.2000.0070298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuttur MY. Challenges faced by ontology matching techniques: Case study of the oaei datasets. Research Journal of Information Technology. 2011;3(1):33–42. [Google Scholar]
- 10.Common terminology services 2. https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/Common\_Terminology\_Services_2.
- 11.DCMI namespace for the Dublin Core metadata element set. http://dublincore.org/documents/dces/
- 12.Fleiss JL, Cohen J, Everitt BS. Large sample standard errors of kappa and weighted kappa. Psychological Bulletin. 1969;72:323327. [Google Scholar]
- 13.Geller J, Perl Y, Halper M, Cornet R. Special issue on auditing of terminologies. Journal of Biomedical Informatics. 42(3):407. doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Ghazvinian Amir, Noy Natalya F, Jonquet Clement, Shah Nigam, Musen Mark A. What four million mappings can tell you about two hundred ontologies. In: Bernstein Abraham, Karger David R, Heath Tom, Feigenbaum Lee, Maynard Diana, Motta Enrico, Thirunarayan Krishnaprasad., editors. The Semantic Web - ISWC 2009, volume 5823 of Lecture Notes in Computer Science. Springer; 2009. pp. 229–242. [Google Scholar]
- 15.Horrocks Ian, Patel-Schneider Peter F, McGuinness Deborah L, Welty Christopher A. OWL: a Description Logic Based Ontology Language for the Semantic Web. In: Baader Franz, Calvanese Diego, McGuinness Deborah, Nardi Daniele, Patel-Schneider Peter F., editors. The Description Logic Handbook: Theory, Implementation, and Applications. 2. chapter 14. Cambridge University Press; 2007. [Google Scholar]
- 16.Kalfoglou Yannis, Schorlemmer Marco. Ontology mapping: the state of the art. Knowl Eng Rev. 2003 Jan;18(1):1–31. [Google Scholar]
- 17.Kashyap Vipul, Borgida Alexander. Representing the UMLS semantic network using OWL: (or “what’s in a Semantic Web link?”). International Semantic Web Conference; Sanibel Island, FL. 2003. pp. 1–16. [Google Scholar]
- 18.McCarthy JL, Kendall E, Warzel D, Bargmeyer B, Solbrig H, Keck K, Gey F. Data modeling and harmonization with owl: Opportunities and lessons learned. The Fifth International Workshop on Semantic Web Enabled Software Engineering; 2009. [Google Scholar]
- 19.Mirhaji P, Zhu M, Vagnoni M, Bernstam EV, Zhang J, Smith JW. Ontology driven integration platform for clinical and translational research. BMC Bioinformatics. 2009;10(S-2) doi: 10.1186/1471-2105-10-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitra Prasenjit, Noy Natalya F, Jaiswal Anuj R. Omen: A probabilistic ontology mapping tool. In Workshop on Meaning Coordination and Negotiation at the Third International Conference on the Semantic Web (ISWC-2004); Hisroshima. 2004. pp. 537–547. [Google Scholar]
- 21.Moreira DA, Musen MA. OBO to OWL: a protégé OWL tab to read/save OBO ontologies. Bioinformatics. 2007;23(14):1868–1870. doi: 10.1093/bioinformatics/btm258. [DOI] [PubMed] [Google Scholar]
- 22.Noy NF, de Coronado S, Solbrig HR, Fragoso G, Hartel FW, Musen MA. Representing the NCI Thesaurus in OWL DL: Modeling tools help modeling languages. Journal of Applied Ontology. 2008;3(3):173–190. doi: 10.3233/AO-2008-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The open biomedical ontologies. http://www.obofoundry.org/
- 24.Ontology metadata vocabulary for the semantic web. http://softlayer.dl.sourceforge.net/project/omv2/OMV.
- 25.OWL 2 web ontology language structural specification and functional-style syntax. http://www.w3.org/TR/owl2-syntax/
- 26.Pastor JA, Martinez FJ, Rodriguez JV. Advantages of thesaurus representation using the simple knowledge organization system (SKOS) compared with proposed alternatives. Information Research. 2009;14(4) [Google Scholar]
- 27.Phillips A. Davis M, editor. Tags for identifying languages. 2006 Sep; http://www.rfc-editor.org/rfc/bcp/bcp47.txt.
- 28.The RDF vocabulary. http://www.w3.org/1999/02/22-rdf-syntax-ns.
- 29.The RDF schema vocabulary (RDFS) http://www.w3.org/2000/01/rdf-schema.
- 30.Schulz S, Schober D, Tudose I, Stenzhorn H. The pitfalls of thesaurus ontologization the case of the nci thesaurus. Proceedings of the American Medical Informatics Association (AMIA) 2010 Annual Symposium; Washington DC. November 2010; pp. 787–791. [PMC free article] [PubMed] [Google Scholar]
- 31.SKOS vocabulary. http://www.w3.org/2006/07/SWD/SKOS/reference/20090315/skos.rdf.
- 32.SKOS XL vocabulary. http://www.w3.org/2006/07/SWD/SKOS/reference/20090315/skos-xl.rdf.
- 33.Tao C, Noy NF, Solbrig HR, Shah NH, Musen MA, Chute CG. Proposed skos extensions for bioportal terminology services. Proceedings of Joint International Semantic Technology Conference (JIST 2011); Hangzhou, China. December 2011. [Google Scholar]
- 34.van der Haring Egbert J, Bronhorst S, ten Napel H, Weber S, Schopen M, Zanstra Pieter E. Studies in Health Technology and Informatics. IOS Press; ClaML: A standard for the electronic publication of classification coding schemes; pp. 801–806. [PubMed] [Google Scholar]
- 35.OWL Web Ontology Language Reference Manual. W3C (World Wide Web Consortium); www.w3.org/TR/owl-ref. [Google Scholar]
- 36.Defining n-ary relations on the semantic web. http://www.w3.org/TR/swbp-n-aryRelations/