Summary
Long noncoding RNAs (lncRNAs) are often expressed in a development-specific manner, yet little is known about their roles in lineage commitment. Here, we identified Braveheart (Bvht), a heart-associated lncRNA in mouse. Using multiple embryonic stem cell (ESC) differentiation strategies, we show that Bvht is required for progression of nascent mesoderm towards a cardiac fate. We find that Bvht is necessary for activation of a core cardiovascular gene network and functions upstream of MesP1 (mesoderm posterior 1), a master regulator of a common multipotent cardiovascular progenitor. We also show that Bvht interacts with SUZ12, a component of Polycomb Repressive Complex 2 (PRC2), during cardiomyocyte differentiation suggesting that Bvht mediates epigenetic regulation of cardiac commitment. Finally, we demonstrate a role for Bvht in maintaining cardiac fate in neonatal cardiomyocytes. Together, our work provides evidence for a long noncoding RNA with critical roles in the establishment of the cardiovascular lineage during mammalian development.
Keywords: ESCs, cardiomyocyte, cardiovascular progenitor, gene regulation, long noncoding RNA
Introduction
The heart is the first organ to function during vertebrate embryogenesis. In mammals, cardiogenesis involves the diversification of pluripotent cells in the embryo toward mesodermal and cardiac cell types including cardiomyocytes (Kattman et al., 2007). Coordination of this process depends on precise control of gene expression patterns, and disruption of transcriptional networks that govern cardiac commitment underlies congenital heart disease (CHD) (Bruneau, 2008; Srivastava, 2006). Notably, cardiovascular disease and ageing lead to a progressive loss of cardiomyocytes, yet the adult mammalian heart has limited regenerative capacity (Mercola et al., 2011; Steinhauser and Lee, 2011). The derivation of cardiomyocytes from pluripotent stem cells in vitro is a potentially promising strategy for regenerative therapy, but the inability to generate sufficient quantities of high quality cardiac cells has been a limitation to realizing this potential. Thus, knowledge of the molecular switches that control cardiac commitment is critical to achieve a better understanding of heart development and to design new approaches for treatment of cardiac-related diseases.
Development of the cardiovascular system including the heart is a multi-step process that is coordinated by a network of transcription factors (Murry and Keller, 2008; Olson, 2006). For example, the transcription factor mesoderm posterior 1 (MESP1) is critical for the establishment of a multipotent cardiovascular progenitor population during gastrulation (Bondue et al., 2008; Lindsley et al., 2008). MesP1 is transiently expressed in the nascent mesoderm and its expression marks those cells destined to give rise to the cardiovascular lineage (Saga et al., 1996; 1999; 2000). Consistent with this idea, forced expression of MesP1 during embryonic stem cell (ESCs) differentiation leads to an increase in the cardiogenic population (Bondue et al., 2008). MESP1 regulates a core network of transcription factors including many regulators of heart development as well as genes with roles in Epithelial-to-Mesenchymal transition (EMT) such as Snai and Twist (Bondue et al., 2008; Lindsley et al., 2008). EMT is critical for gastrulation and morphogenetic movements during organogenesis including heart formation (Lim and Thiery et al., 2012; von Gise and Pu, 2012). Thus, identifying additional factors that promote a mesoderm to cardiovascular transition may provide new insights into the regulation of heart development.
Long non-coding RNAs (lncRNAs) are broadly classified as transcripts longer than 200 nucleotides, that are 5′ capped and polyadenylated like most mRNAs, yet this class of transcripts has limited coding potential. LncRNAs function in a wide range of processes and can regulate gene expression by diverse mechanisms (Hu et al., 2012; Mercer et al., 2009; Ponting et al., 2009; Rinn and Chang, 2012). While thousands of lncRNAs have been identified across eukaryotes, many are species specific and appear less conserved than protein-coding genes (Cabili et al., 2011; Derrien et al., 2012; Ultisky et al., 2011). Importantly, lncRNAs are differentially expressed across tissues, suggesting that they regulate lineage commitment. Consistent with this idea, loss of function of two lncRNAs in Zebrafish embryos, cyrano and megamind, resulted in various developmental defects (Ulitsky et al., 2011). Moreover, HOTTIP plays a role in limb formation (Wang et al., 2011), whereas other lncRNAs function to promote or suppress somatic differentiation (Hu et al., 2011; Kretz et al., 2012a; 2012b). Furthermore, depletion of a subset of lncRNAs in mouse ESCs led to up-regulation of global lineage programs (Guttman et al., 2011). Despite these promising findings, our knowledge of lncRNAs that function in lineage commitment is limited to only a few examples, and a detailed understanding of the genetic pathways they regulate is lacking.
Here, we report the identification of AK143260, which we named Braveheart (Bvht), a lncRNA in mouse that is necessary for cardiovascular lineage commitment. Using multiple ESC differentiation strategies, we found that Bvht was necessary for activation of a core gene regulatory network that included key cardiac transcription factors (e.g. MesP1, Gata4, Hand1, Hand2, Nkx2.5, and Tbx5) and EMT genes (e.g. Snai and Twist). Further analysis revealed a significant overlap between the genes regulated by Bvht and MESP1, a master regulator of cardiovascular potential. Moreover, forced expression of MesP1 rescued the Bvht-depletion phenotype indicating that these two factors function in a similar genetic pathway. We show that Bvht interacts with SUZ12, a core component of Polycomb Repressive Complex 2 (PRC2), suggesting that this interaction may be critical for epigenetic regulation of network genes. We also demonstrate that Bvht is necessary for maintenance of cardiac fate in ex vivo neonatal cardiomyocytes. Together, these results indicate that Bvht functions to regulate gene expression programs that promote commitment toward the cardiovascular lineage. More broadly, our work identifies a potential new pathway for regulation of heart development and suggests that lncRNAs may represent new targets that can be exploited for efficient generation of stem cell-derived cardiac cells.
Results
Identification of candidate lncRNAs in the cardiac lineage
LncRNAs exhibit distinct expression patterns across tissues (Derrien et al., 2012; Dinger et al., 2008), however we have limited knowledge of their precise molecular roles during mammalian development. We reasoned that lncRNAs whose expression becomes restricted to specific cell types during ESC differentiation represent candidate regulators of lineage commitment. We analyzed lncRNA expression as measured by RNA-Seq in mouse ESCs as well as in differentiated tissues representing the three germ layers (Mortazavi et al., 2008). We identified 47 candidates that displayed high expression in ESCs and lower median expression across the tissues examined (Figure 1A and Table S1). To identify lncRNAs with potential roles in cardiac commitment, we next evaluated the expression of individual candidates in each tissue including the heart (Figure 1B). We focused on AK143260 located on mouse chromosome 18:61,799,307-61,807,126 (+ strand, mm9) because it displayed higher expression in the heart relative to the other tissues examined in this study. Thus, we named the candidate Braveheart (Bvht).
Figure 1. Identification AK143260 as a candidate heart-associated lncRNA.
A. Differential expression enrichment score calculated from RNA-Seq data from biological replicates of two independent mESC lines and adult tissues (brain, liver, and skeletal muscle) representing the three germ layers. Stars represent the position of 3 example candidates and their respective Genbank accession numbers. The y-axis represents the number of lncRNAs with a particular differential expression enrichment, as denoted by each bar. B. Transcriptome analysis in a broader range of tissues showed that AK143260 was predominantly expressed in ESCs and in heart. C. Depiction of transcript structure as defined by 5′ and 3′ RACE (lower panel) was consistent with RNA sequencing reads in ESCs (upper panel). D. Northern analysis using a full-length probe for AK143260 confirms the predicted size of the transcript (~590 nucleotides) and shows reduced levels upon shRNA-mediated depletion in mESCs. E. RNA sequencing reads from rat adult heart and human adult heart samples showed no evidence of a transcript expressed in a location syntenic to AK143260. Arrows mark direction of transcription. Related to Figure S1 and Table S1.
LncRNAs are a newly emerging class of RNA and their status as non-coding transcripts from genome annotations requires independent validation. We determined by RACE that Bvht is a ~590 nucleotide transcript comprising 3 exons, consistent with our RNA-Seq and Northern analysis in ESCs (Figure 1C,D). We also detected an isoform that lacked the first exon (~50 bases), which represented only a minor fraction of the total transcript as determined by RACE. Cell fractionation followed by qRT-PCR showed that ~33% of the spliced Bvht transcript resides in the nucleus (Figure S1A). Notably, HOTAIR, a lncRNA with nuclear function, is also distributed across both cell compartments in human fibroblasts (Khalil et al., 2009). While Bvht harbors short open reading frames (ORFs), the two largest are 48aa and 74aa, ORFs of this size would be expected by chance in a transcript of this length (Figure S1B). Consistent with the recent finding that lncRNAs are rarely translated in human cells (Banfai et al., 2012), we found no evidence of active translation or production of a protein product from Bvht (Figure S1C–F and Supplemental Information). Moreover, sequence homology searches revealed no clear orthologous Bvht sequence or transcripts in syntenic regions of the human or rat genomes, consistent with the idea that lncRNAs are less conserved than protein-coding genes (Cabili et al., 2011; Derrien et al., 2012). For example, analysis of RNA-Seq data from human and rat heart, a tissue in mouse that displays strong expression of both Bvht and the conserved neighboring miR143/145 cluster, revealed robust expression of the pri-miRNA in human and rat, whereas transcripts from the Bvht region were not detected (Figure 1E). Thus, Bvht appears to be a mouse-specific lncRNA and is an example of a newly evolved rapid gain of function non-coding transcript.
Braveheart is necessary for proper ESC differentiation
To dissect the function of Bvht in lineage commitment, we generated mESC lines that stably expressed short hairpin RNAs (shRNA) directed against either exon 2 or 3. We observed significant depletion (~80%) of the transcript in ESCs as measured by qRT-PCR and Northern analysis (Figure 2A and Figure 1D). Notably, Bvht depletion in ESCs did not affect colony morphology, expression of pluripotency markers, the fraction of apoptotic cells, or cell cycle kinetics compared to control cells harboring a non-targeting hairpin (Figure 2B and Figure S2A–C). Thus, Bvht is not required to maintain ESC self-renewal.
Figure 2. Bvht is required for proper ESC differentiation.
A. Targeting of AK143260 (Bvht) in mESCs by two independent hairpins directed to either exon 2 or 3 led to significant depletion of the transcript as measured by qRT-PCR. Experiments were performed in triplicate and error bars represent standard deviation within one representative experiment. B. Bvht depletion does not affect colony morphology and OCT4 staining in ESCs. Scale bar=100 μm. C. Percentage of spontaneously contracting EBs determined at Day 8 and 11 of differentiation (n>100 per time point). Bvht-depleted EBs showed a significant decrease in contracting EBs compared to controls. The number of resulting EBs was similar in each experiment. Experiments were performed in triplicate and error bars represent standard deviation. D. Expression of cardiac Troponin T (cTnT) measured by qRT-PCR at representative time points shows decreased levels in Bvht-depleted EBs. Error bars represent standard deviation of triplicate set of experiments. E. Antibody staining of paraffin embedded sections of Day 12 EBs showed decreased cTNT abundance in Bvht-depleted compared to controls. Scale bar=300 μm. F. Bvht-depleted EBs can form tissues representative of all three germ layers including gut-like epithelium (endoderm), cartilage (mesoderm), and neuroepithelial rosettes (ectoderm) as represented in H&E stained EB sections. Scale bar=100 μm. ** p<0.01 by two-tailed Student’s t-test. Related to Figure S2 and Table S2.
ESCs have the capacity to differentiate into derivatives of the three germ layers including cardiac cell types. We next induced Bvht-depleted ESCs to differentiate by allowing cells to aggregate into embryoid bodies (EBs). Given its expression in the heart, we investigated whether Bvht-depleted cells could form cardiac tissue, which can be visualized as spontaneously contracting EBs during differentiation. In control cells more than 25% of EBs beat at Day 11 of differentiation, compared to ~5% of the Bvht-depleted EBs (Figure 2C and Figure S2D,E). Consistent with this observation, cardiac troponin T (cTnT), an integral component of the contraction machinery in cardiac muscle, was expressed at significantly lower levels in Bvht-depleted EBs (Figure 2D,E). In contrast, cells depleted of AK085506, an ESC-specific lncRNA not expressed in the heart, displayed a similar proportion of contracting EBs (Figure 1B and Figure S2F). Examination of EB sections at Day 12 of differentiation showed a range of tissues, such as neural rosettes, gut endoderm, and cartilage (Figure 2F). Together, these data suggest that Bvht is not required for global ESC differentiation and may have specific functions in cardiac commitment.
Braveheart regulates the core cardiovascular gene network
LncRNAs can act in cis to regulate expression of neighboring genes or in trans by diverse mechanisms (Orom and Shiekhattar, 2011; Rinn and Chang, 2012). We first analyzed the expression of neighboring genes within a 100kb window, which included the convergent and overlapping miR143/145 cluster on the opposite strand (exons do not overlap) (Figure S3A). Expression of neighboring protein-coding genes was unaffected in Bvht-depleted cells during ESC differentiation compared to control cells; however, the pri-miRNA was not properly expressed during the time course (Figure S3B). It is likely that lack of expression of the pri-miRNA is due to depletion of cardiac cells in the Bvht-depleted EBs rather than direct regulation by Bvht. For example, miR143 and miR145 are typically transcribed in cardiovascular cell types (Cordes et al., 2009; Xin et al., 2009). Moreover, miR143/145 knock-out ESCs formed contracting, cTNT positive EBs and showed clear differences in gene expression patterns compared to Bvht-depleted cells (Figure S3C–E, S4 and Table S2). Thus, these data indicate that the Bvht phenotype is likely not a consequence of failure to regulate closely neighboring genes in cis including the mir143/145 cluster.
We next reasoned that analysis of global gene expression patterns during EB differentiation might reveal further clues toward elucidating Bvht function. To this end, we analyzed gene expression patterns at various time points by RNA-Seq. We found ~548 differentially expressed genes in the Bvht-depleted cells compared to controls (Figure 3A and Table S3). These genes were clustered by expression pattern, yielding several distinct clusters. Strikingly, genes in clusters that displayed lower expression at later stages (i.e. clusters G–I) showed enrichment for genes that function in heart and blood vessel morphogenesis, heart and muscle system process, and myofibril assembly (Figure 3B). In contrast, genes in clusters that displayed higher expression in Bvht-depleted cells (i.e. clusters A–F) were not enriched for specific gene ontology (GO) terms.
Figure 3. Bvht regulates a core network of genes that drive cardiac differentiation.
A. Heat map representing hierarchical clustering of all genes that displayed a 3-fold or greater difference in transcript levels in Bvht-depleted EBs compared to controls at Day 0, 3, 6, and 9. B. Significant gene ontology (GO) terms retrieved by clusters G–I. C. Pearson correlation network of the EB time course generated using the Spring embedded algorithm in Cytoscape. Genes in enriched GO categories are represented in the network. A sequence of cardiac transcription factors and genes involved in myofibril organization were not induced during EB differentiation in Bvht-depleted cells. Nodes represent genes and connections represent correlation coefficient. Related to Figure S3 and S4 and Table S3.
To gain further insights, we created a network diagram to illustrate how genes in the enriched GO categories changed in expression over time by computing a Pearson correlation coefficient of expression between all pairs of genes (Figure 3C). Our analysis revealed a complex network of transcription factors that failed to activate during differentiation including key regulators of heart development that have been implicated in cardiovascular diseases such as MesP1, Hand1, Hand2, Nkx2.5, and Tbx20 as well as genes with roles in cardiomyocyte structure and function (Bruneau, 2008; McCulley and Black, 2012). While a caveat of the EB assay is that it is difficult to distinguish subtle effects among related lineages, we observed that while expression of genes representing cardiovascular cell types including cardiomyocytes, vascular endothelium, and smooth muscle were significantly reduced, markers of paraxial mesoderm, hematopoiesis, and endoderm were expressed at normal or slightly elevated levels in Bvht-depleted EBs (Figure S5A). Moreover, Bvht-depleted cells displayed similar morphology upon neuronal differentiation by addition of retinoic acid and expressed similar levels of neural markers such as Sox1 and Nestin compared to controls (Figure S5B and data not shown). Together, these data suggest that Bvht specifically promotes activation of a core gene regulatory network in trans to direct cardiovascular cell fate.
Braveheart functions upstream of MesP1
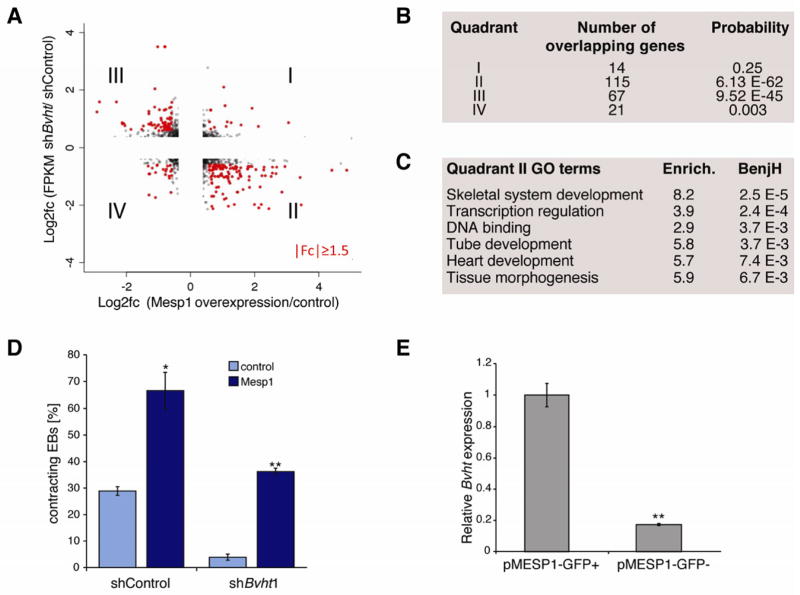
MESP1 was among the earliest transcription factors in the gene network that showed significantly lower expression in Bvht-depleted EBs. MesP1 marks a common multipotent cardiovascular progenitor that ultimately specifies all cell types of the heart (Bondue et al., 2008; 2011; David et al., 2008; Lindsley et al., 2008). To test if Bvht and MesP1 function in a similar pathway, we compared the differentially expressed genes in our EB time course with those changed upon MesP1 induction (Bondue et al., 2008). We observed a striking correlation among genes that showed lower expression in Bvht-depleted cells and higher expression upon MesP1 induction, denoted as Group II (P<6e-62). Genes that showed higher expression in Bvht-depleted cells and reduced expression upon MesP1 induction also overlapped significantly, denoted Group III (P<9e-45) (Figure 4A,B). Notably, Group II genes function in transcriptional regulation, heart development, and tissue morphogenesis, and included many of the transcription factors represented in the Bvht network (Figure 4C and Table S4). These results suggest that Bvht-depletion leads to failure to activate a MESP1-driven gene expression program.
Figure 4. Bvht functions upstream of MesP1.
A. Gene expression comparison between Bvht depletion and Mesp1 induction. B. The overlap is highly significant for quadrants II and III as measured by hypergeometric test with Benjamini correction. Genes in quadrant II failed to activate in Bvht-depleted cells and are expressed at higher levels upon Mesp1 induction, whereas quadrant III genes displayed the opposite expression pattern. C. Enriched GO terms based on genes in quadrant II. D. Forced MesP1 expression can rescue the contraction defect in Bvht-depleted EBs. Contracting EBs were counted at Day 11 (n>100). Error bars represent standard deviation of a triplicate set of experiments. E. Bvht is enriched in cells expressing a GFP reporter under the control of the Mesp1 promoter (pMESP1-GFP) compared to GFP negative cells as analyzed by FACS at Day 5 (peak of MesP1 expression) of EB differentiation followed by qRT-PCR. Experiments were performed in triplicate and error bars represent standard deviation in all panels. ** p<0.01; * p<0.05 by two-tailed Student’s t-test.
We expected that if Bvht functions upstream of MesP1, then forced expression of MesP1 should rescue the depletion phenotype. We employed a doxycycline-inducible MesP1 mESC line (Bondue et al., 2008) and induced expression at Day 2 of EB differentiation in Bvht-depleted and control cells. MesP1 cardiovascular progenitors comprise a very small proportion of cell types in early stage EBs and induction at this time point leads to expansion of the cardiogenic population (Bondue et al., 2008; Lindsley et al., 2008). Notably, MesP1 induction led to an increase in contracting EBs in both control and Bvht-depleted cells (Figure 4D). We also found that Bvht levels were ~10-fold higher in MESP1 population compared to negative cells using a mESC line that harbored GFP under the control of the MesP1 promoter (Bondue et al., 2011) (Figure 4E). Together, these data suggest that Bvht functions upstream of MesP1 as a potential regulator of cardiovascular fate.
Braveheart promotes cardiac cell fate from nascent mesoderm
To further dissect Bvht function in the cardiovascular lineage, we employed a directed in vitro cardiomyocyte (CM) differentiation assay that permits isolation of cell populations at well-defined stages (mESCs (ESCs), pre-cardiac mesoderm (MES), cardiac progenitors (CP), and cardiomyocytes (CM)) (Kattman et al., 2011; Wamstad et al., 2012) (Figure 5A). Using this assay, we observed that Bvht-depleted EBs were smaller and displayed an irregular shape at Day 4 (MES), and failed to form an adherent monolayer at Day 5.3 (CP) compared to controls (Figure 5B, middle and right panels). In contrast, no visible change in morphology was apparent at Day 2, a time point that precedes the addition of cardiac growth factors (Figure 5B, left panels). We also found an increase in apoptosis but no change in cell cycle kinetics at Day 4 in Bvht-depleted cells, whereas Day 2 cells showed no difference (Figure 5C and Figure S6A). Notably, shRNA-mediated depletion of MesP1 showed similar effects to Bvht-depletion and Bvht overexpression led to an increase in the levels of cardiac genes including MesP1 during CM differentiation (Figure S6B,C and data not shown).
Figure 5. Bvht is necessary for the transition from nascent to cardiac mesoderm in vitro.
A. ESCs were differentiated into beating cardiomyocytes (CM) and progress through precardiac mesoderm (MES) and cardiac progenitor (CP) stages by sequential addition of cytokines. B. Bvht-depleted EBs were smaller than controls at Day 4 and failed to attach and organize into a monolayer at Day 5.3. Scale bar=65 μm. C. Bvht-depleted cells showed increased levels of apoptosis at Day 4 as indicated by quantification of Annexin V-positive cells, whereas no significant difference was observed at Day 2. D. Brachyury and Eomes levels remained high at Day 5.3 compared to control as measured by qRT-PCR. E., F. Core cardiac transcription factors displayed lower expression at Day 4 (E) and Day 5.3 (F) in Bvht-depleted cells compared to control. G. Expression of cell surface markers, PdgfRa and Flk-1, was significantly decreased upon Bvht depletion at Day 4 and Day 5.3 compared to control. H. EMT genes displayed altered expression patterns in Bvht-depleted-cells at Day 5.3 as measured by qRT-PCR. Values for shControl samples were normalized to 1 for each individual time point. I. Equal numbers of mCherry-shControl and GFP-shBvht1 cells were mixed and subjected to cardiac differentiation. A dramatic decrease in Bvht-depleted cells was observed at Day 4 and Day 5.3, whereas cells cultured in ESC conditions maintain equal distribution. Error bars represent standard deviation of three replicates within one representative experiment. ** p<0.01; * p<0.05 by two-tailed Student’s t-test. Related to Figure S6.
Brachyury (T) and Eomesodermin (Eomes), transcription factors expressed in the primitive streak, have critical roles in mesoderm induction and regulate MesP1 expression by directly binding to its promoter (Costello et al., 2011; David et al., 2011). While T and Eomes were expressed normally or at elevated levels at Day 4 in Bvht-depleted and controls cells, both factors displayed higher levels in Day 5.3 Bvht-depleted cells (Figure 5D). Notably, expression of core cardiac transcription factors including MesP1 failed to induce at Day 4 and Day 5.3 in Bvht-depleted cells despite expression of T and Eomes (Figure 5E,F). Similarly, expression of cell surface markers of early cardiac identity (i.e. PdgfRa and Flk-1) was lower in Bvht-depleted cells (Figure 5G). Together, PdgfRa and Flk-1 define an early multi-potent cardiovascular progenitor population (marked by MesP1) and their co-expression enhances cardiogenic potential (Kattman et al., 2006; Yang et al., 2008). Conversely, Fgf8 levels were elevated in Bvht-depleted cells along with Mixl1 during CM differentiation, genes required for efficient induction of hematopoietic mesoderm, consistent with a reciprocal increase in hematopoietic markers upon loss of cardiac potential (Lim et al., 2009; Simoes et al., 2011) (Figure S6D).
MESP1 also regulates EMT genes whose expression is critical for migration of precardiac cells during gastrulation (Lindsley et al., 2008). We found that expression of EMT genes such as Snai1 and Snai2 as well as Twist1 and Twist2 (Yang et al., 2004) was not induced at Day 5.3 in Bvht-depleted cells compared to controls (Figure 5H). Snai1 and Snai2 repress E-cadherin and the switch from E-cadherin to N-cadherin expression is a hallmark of EMT (Lim and Thiery et al., 2012). Notably, E-cadherin levels remained high and N-cadherin was not expressed in Bvht-depleted cells (Figure 5H). Together, these results indicate that Bvht is required during a critical time during differentiation when cardiac cells are specified from nascent mesoderm.
Our observations suggest that Bvht functions in the same pathway as MesP1 in a cell autonomous manner to regulate a core cardiac gene network. To test this idea, we generated ESC lines that harbored either the shBvht or shControl vector additionally marked with GFP or mCherry to allow us to track each population. Equal numbers of shControl-mCherry and shBvht-GFP ESCs were combined and analyzed by FACS during CM differentiation. While the proportion of marked cells was similar in ESCs and at Day 2, we observed a dramatic over-representation of shControl cells at Day 4 and Day 5.3 indicating that Bvht-depleted cells cannot efficiently contribute to cardiac cell types (Figure 5I). Similar results were obtained using reciprocal markers (S6E). Notably, cardiac genes such Gata4, Nkx2.5, Tbx5, and Isl1 were expressed at normal levels at Day 5.3 (Figure S6F), indicating that control cells could differentiate properly in the presence of Bvht-depleted cells. Thus, our results support a cell autonomous role for Bvht in the transcriptional control of cardiac cell fate.
Braveheart interacts with SUZ12, a core component of PRC2
Some lncRNAs are thought to act through RNA-protein interactions to effect gene expression by diverse mechanisms (Rinn and Chang, 2012). Because T and Eomes have been shown to bind to the MesP1 promoter and to regulate its expression (David et al., 2011; Costello et al., 2011), we first tested whether Bvht interacted with T or EOMES to mediate their binding at the MesP1 promoter. We performed RNA immunoprecipitation (RIP) using antibodies against T or EOMES; however, we failed to detect a specific interaction with Bvht (Figure S7A). We also did not detect changes in the binding of T to the MesP1 promoter by chromatin immunoprecipitation (ChIP) followed by qPCR (Figure S7B and Supplemental Information). Thus, it is unlikely that Bvht directly regulates T or EOMES to control MesP1 activation.
LncRNAs can act as molecular scaffolds by interacting with chromatin modifying complexes (Guttman et al., 2011; Tsai et al., 2010). Using in vitro biotin-RNA pull down, we detected a specific interaction between Bvht and SUZ12 in ESCs; however, other factors thought to bind lncRNAs including hnRNPK, SNW1, RING1b, OCT4, BRG1, and MLL1 did not show a specific interaction (Figure 6A). These data were confirmed by RIP using SUZ12 specific antibodies (data not shown). SUZ12 is core subunit of PRC2, a complex that catalyzes tri-methylation of histone H3 lysine 27 (H3K27me3) and mediates transcriptional repression (Surface et al., 2010). A number of lncRNAs have been shown to interact with PRC2 to regulate target gene expression (Wang and Chang, 2011). We then constructed a series of Bvht deletions and found several regions of the transcript that were required for this interaction (Figure 6B) suggesting that Bvht directly interacts with PRC2.
Figure 6. Bvht directly interacts with SUZ12 during CM differentiation.
A. Biotin-RNA pull downs using full length Bvht transcript in nuclear ESC extracts showed specific binding to SUZ12. B. Deletion analysis of Bvht transcript showed that 5′ and 3′ regions were required for interaction with SUZ12. C. Bvht interacts with SUZ12 in Day 5.3 cells (CP stage) as determined by RIP using SUZ12 antibody. D. ChIP using SUZ12 antibody showed higher levels at promoter regions of cardiac and EMT genes in Bvht-depleted Day 5.3 cells compared to controls, whereas no significant changes were observed in genes expressed in other lineages (e.g. Sox1 and Sox17). Experiments were performed in triplicate and error bars represent standard deviation in all panels. ** p<0.01; * p<0.05 by two-tailed Student’s t-test. Related to Figure S7.
Next we tested whether Bvht interacted with SUZ12 during CM differentiation. Similar to ESCs, we found that SUZ12 interacted with Bvht by RIP at Day 5.3 cells (Figure 6C). We chose to analyze Day 5.3 cells because these cells comprise a largely homogenous population of >70% NKX2.5 positive cells and because Bvht is enriched in this population (Figure S7C). Many of the cardiac genes that failed to activate in Bvht-depleted cells at Day 5.3 are known Polycomb targets in ESCs (Boyer et al., 2006). These genes are poised for activation in ESCs as indicated by their bivalent chromatin marks (H3K27me3 and H3K4me3) (Bernstein et al., 2006) and differentiation towards specific lineages requires the selective loss of PRC2 at subsets of genes. We found that in Bvht-depleted cells SUZ12 levels remained high at the promoters of critical genes in the cardiovascular network including MesP1, Gata6, Hand1, Hand2, and Nkx2.5 compared to control cells as measured by ChIP-qPCR (Figure 6D). In contrast, SUZ12 levels were lower at T and Eomes, consistent with their higher expression at Day 5.3 in Bvht-depleted cells. SUZ12 levels were equivalent in both Bvht-depleted and control cells (Figure S7D). We also found that H3K27me3 levels paralleled SUZ12 enrichment in both Bvht-depleted and control cells. Notably, cardiac genes maintained their bivalent status at Day 5.3 (Figure S7E,F) consistent with a failure to activate the cardiac program. While our data do not distinguish between a direct or indirect mechanism, these results along with the demonstrated interaction between SUZ12 and Bvht suggest that Bvht functions, in part, with PRC2 to regulate cardiac commitment.
Braveheart is necessary for maintenance of cardiac fate in neonatal CMs
Our data show that Bvht is required for the commitment of ESCs towards a cardiac fate. Because we detected expression of Bvht in the adult mouse heart (Figure 1B), we investigated whether Bvht functions in later stages of differentiation. We analyzed the effect of Bvht depletion in primary murine neonatal ventricular cardiomyocyte (nCMs) cultures, a model that has been used extensively for investigating drug and injury responses (Louch et al., 2011). Briefly, nCMs were isolated from the newborn heart, a stage when myocytes display plasticity both in vivo and in vitro as evidenced by increased levels of early cardiac markers such as Gata4 and Nkx2.5 (Porrello et al., 2011; Zhang et al., 2010). Within days of isolation, nCMs are able to organize into myofibrils, forming sheets of beating cardiomyocytes in explant cultures. Thus, neonatal cardiomyocyte represent in vivo cell types in which to test the role of Bvht in modulating cardiac cell fate in the heart.
nCMs were infected one day after explant with Bvht or control shRNA lentiviruses harboring a GFP marker. We found that Bvht-depleted nCMs as marked by GFP displayed unevenly distributed and irregularly bundled myofibrils after 5 days in culture compared to control cells as evidenced by cTNT staining (Figure 7A). Consistent with this observation, the overall cell surface area of individual GFP-positive nCMs was significantly smaller (Figure 7B), indicating that nCM structure is disrupted in Bvht-depleted cells (Figure 7C). Moreover, structural genes that were not properly activated in Bvht-depleted EBs including alpha- and beta-MHC and Sma, displayed lower expression in the Bvht-depleted nCMs compared to controls (Figure 7D), whereas early cardiac markers Gata4 and Nkx2.5 displayed higher expression (Figure 7E). Notably, genes involved in EMT such as Snai and Twist members as well as Vimentin showed lower expression in Bvht-depleted nCMs (Figure 7F). These data suggest that Bvht plays a critical role in promoting cardiac gene expression programs and tissue remodeling in the developing heart.
Figure 7. Bvht is required to maintain cardiac fate in neonatal cardiomyocytes.
A. Bvht depleted or control nCMs were identified by the presence of a GFP reporter in the shRNA vector. Myofibrils clustered together in GFP positive control cells whereas Bvht depletion resulted in disorganized cardiac myofibril bundles. Myofibrils were visualized by IF using an antibody against cTnT (red), and GFP (green). Nuclei (blue) were stained with Hoechst. Scale bars=20 μm. B. Bvht-depleted nCMs were overall smaller than controls as assessed by the surface area of GFP positive versus control cells (n>100). C. Global levels of Bvht (qRT-PCR) in control and Bvht-depleted nCM cultures. D. Reduced expression of sarcomeric and myofibril components in Bvht-depleted nCMs as measured by qRT-PCR. E. Expression (qRT-PCR) of transcription factors involved in early cardiogenesis was increased in Bvht-depleted cells. F. Factors involved in EMT were expressed at lower levels in Bvht-depleted cells. ** p<0.01, * p<0.05 by ANOVA and Dunnett’s test.
Discussion
Dynamic regulation of gene expression programs is critical for cell fate transitions during lineage commitment and faulty regulation can lead to developmental failure and disease. Heart development is a multi-step process regulated by a network of transcription factors and disruption of transcriptional networks leads to congenital heart disease (Bruneau, 2008; McCulley and Black, 2012; Olson, 2006; Srivastava, 2006). While many of the genetic factors that govern heart development are known, we hypothesized that lncRNAs may represent an additional layer of regulation. Here, we identify Braveheart as critical regulator of cardiovascular commitment from nascent mesoderm, representing the first example of a lncRNA with potential implications in cardiovascular development.
The temporal activation of a core transcription factor network establishes the plan for a primitive heart. The genes in this network are highly conserved during evolution, whereas the formation of more complex and species-specific substructures is likely a consequence of evolution of species-specific regulators. Analysis of sequence conservation and potential transcripts in syntenic regions in the rat and human genomes did not identify a Bvht homolog. The lack of conservation is likely due to both weak selective pressure on the primary sequence of lncRNAs and rapid gain and loss of entire lncRNA genes. For example, non-coding genes such as Xist and Neat1 have undergone rapid sequence evolution while preserving their functional roles (Clemson et al., 2009; Nesterova et al., 2001). Increasing evidence also suggests that lncRNAs are largely species-specific with many transcripts arising from the primate lineage (Derrien et al., 2012). Thus, Bvht appears to represent a non-conserved lineage specific lncRNA, however, we expect that other species with a defined circulatory system may have evolved lncRNAs with similar functions.
Bvht is an upstream regulator of cardiovascular commitment
Our work suggests that Bvht functions upstream of and in the same genetic pathway as MesP1 as a permissive factor for cardiac commitment during ESC differentiation. MesP1 expression can specify all cell types of the cardiovascular lineage (Bondue et al., 2008; Lindsley et al., 2008) and its expression is sufficient to transdifferentiate dermal fibroblasts into cardiac progenitors along with the transcription factor ETS2 (Islas et al., 2012) indicating that Bvht may also be critical for forward programming and reprogramming toward a cardiovascular fate. EMT is critical for gastrulation and morphogenetic movements throughout development including during heart formation (Lim and Thiery et al., 2012; von Gise and Pu, 2012). EMT is equally critical for in vitro differentiation of stem cells and somatic cell reprogramming where cells transition through multiple stages (Esteban et al., 2012). Snai and Twist, well-characterized regulators of EMT, are downstream targets of MESP1 and have been implicated in various aspects of heart and vascular development (Lindsley et al., 2008). We found that Bvht was necessary for expression of an EMT gene network including Snai and Twist members in CM differentiation and in nCMs indicating that Bvht may also regulate cardiac commitment through organization of a functional tissue.
Bvht may function with PRC2 to regulate cardiac gene expression program
Epigenetic regulation plays an important role in development of the cardiovascular system (Chang and Bruneau, 2012). Recent studies have shown that Ezh2, the catalytic component of PRC2, is necessary for repression of the progenitor program during cardiac differentiation and for proper spatiotemporal regulation of cardiac gene expression (Delgado-Olguin et al., 2012; He et al., 2012). We found that Bvht directly interacted with PRC2 through SUZ12, a core component of the complex, at several stages during CM differentiation. Notably, SUZ12 and its associated modification H3K27me3 were enriched at promoters of cardiac genes including MesP1 in Bvht-depleted cells. Cardiac genes also remained bivalent in Bvht-depleted cells, similar to their configuration in ESCs, consistent with a failure to activate the cardiac program. These data suggest that Bvht binds SUZ12 and competes for PRC2 binding at target sites during the transition from nascent to cardiac mesoderm, leading to activation of the cardiac program. Alternatively, Bvht may recruit PRC2 to a repressor of the cardiac program where loss of Bvht would fail to silence the repressor. While further studies will be required to fully elucidate the mechanism of Bvht function, our work suggests that Bvht is a novel lncRNA that mediates cardiac commitment, in part, by epigenetic regulation of gene expression programs.
Implications of Bvht in cardiovascular development and disease
Cardiovascular disease is one of the most common causes of death, yet the mammalian heart has limited regenerative capacity. Thus, a detailed understanding of the pathways involved in cardiogenesis, and particularly factors dictating commitment, will be instrumental for recapitulating these processes for regenerative therapy. Stem cell models and the generation of specific cell types in vitro offer a potential avenue for studying diseases in a dish and ultimately for transplantation therapy (Burridge et al., 2010). Thus, our finding that Bvht is required for progression of cardiac commitment suggests that lncRNAs represent a new class of molecular modulators that can direct cell fate. Direct comparisons of cardiovascular-associated lncRNAs across species should contribute new insights into how these regulators are integrated into the gene regulatory networks that drive cardiogenesis, and may also provide important clues into the evolution of this complex organ. Moreover, our data suggest that lncRNAs may represent new disease loci that in some cases may be identifiable by genome-wide association studies (GWAS) (Mattick, 2009; Schonrock et al., 2012). For example, the human long non-coding RNA ANRIL has been associated with a locus implicated in cardiovascular disease (Congrains et al., 2012). In addition, human MIAT was identified as a novel long non-coding RNA associated with increased risk of myocardial infarction (Ishii et al., 2006). Thus, we anticipate that future studies to identify additional lncRNAs with roles in cardiac function will lead to a better understanding of heart development and will inform strategies for cardiac repair.
Methods
Detailed experimental and analysis methods can be found in Supplemental Information.
ESC lines and growth conditions
E14 (ola/129), V6.5 (SvJae129/C57BL/6), MesP1-GFP, A2lox-MesP1, NKX2.5-GFP, and miR-143/145 ESCs were cultured on irradiated MEFs using standard conditions.
Generation of ESC lines
shRNA-mediated depletion of lncRNAs
shRNAs directed against Bvht, Mesp1, and control hairpins were cloned into pLKO.1 vector. Production of Lentiviral particles and transduction of ESCs was performed according to protocols from The RNAi Consortium www.broadinstitute.org/rnai/trc.
Bvht overexpression
Full length Bvht cDNA was cloned into pSLIK vector and transgenic cells were generated as described above.
ESC differentiation
Embryoid body (EB) formation
ESCs were pre-plated to remove feeders, diluted to 100,000 cells/ml in complete growth medium lacking LIF and plated on low-attachment cell culture plates.
Cardiomyocyte differentiation
Directed differentiations and analysis were performed as described (Wamstad et al., 2012).
Identification of candidate lncRNAs
Expression data from ESCs (V6.5 and E14) (this study) and adult tissues (Mortazavi et al., 2008) were collated using two annotations (Flicek et al., 2011; Guttman et al., 2010) to define a set of murine lncRNAs. Candidate lncRNAs were identified by a stringent set of criteria outlined in the Supplemental Information.
Genomic analysis of Braveheart
5′ and 3′ Rapid amplification of cDNA ends (RACE) was performed using FirstChoice RLM-RACE kit (Ambion) on ESC RNA and products were cloned using TOPO TA cloning kit (Invitrogen). See Table S5 for primer sequences.
Preparation of RNA sequencing libraries and analysis
RNA-seq was performed as described (Wamstad et al., 2012) and reads were mapped using Tophat/Bowtie. Isoform identification and quantification were performed using Cufflinks. RNA-Seq quality control metrics are listed in Table S6.
Gene co-expression network
The cascade network of co-expression was generated based on the Pearson correlations of the RNA-Seq expression profiles, which are described by log2 fold change between Bvht and control knock-down cells. The layout of the network was created using the Spring-Embedded layout algorithm (Cytoscape plug-in) as described in Supplemental Information.
Competition assay/cell autonomy
GFP and mCherry coding sequences were cloned into plKO.1 vector containing control and Bvht hairpins (Table S5). Cells were isolated by FACS 4 days after infection to select for those expressing the fluorescent proteins. Equal numbers of ESCs were mixed, analyzed by FACS, and then plated under cardiomyocyte differentiation or ESC culture conditions. Cells were analyzed by FACS at various time points to determine the proportion of each fluorescently marked population.
RNA Immunoprecipitation
Native RIP was performed as described (Rinn et al., 2007). Antibodies are listed in Supplemental Information.
Chromatin Immunoprecipitation
The ChIP protocol has been adapted from previous studies (Creyghton et al., 2008). Antibodies are listed in Supplemental Information. ChIP-enriched DNA was quantified by qRT-PCR and analyzed as described in Milne et al., 2009. Primers are listed in Table S5.
Isolation and manipulation of primary mouse neonatal cardiomyocytes (nCM)
Murine nCMs were isolated from C57B6 mice within the first 24h after birth. A detailed protocol can be found in Supplemental Information.
Supplementary Material
Highlights.
Identification of Braveheart (Bvht), a heart-associated long noncoding RNA in mouse
Bvht is necessary for cardiovascular lineage commitment of embryonic stem cells
Bvht functions upstream of MesP1 to regulate a core cardiac gene network
Bvht interacts with PRC2 and may mediate epigenetic activation of cardiac genesto regulate cardiac gene expression program
Acknowledgments
We thank members of the Boyer lab especially Sera Thornton for helpful discussions and critical evaluation of the manuscript and members of the Burge lab especially Jason Merkin and Jessica Hurt for sharing unpublished data. We also thank members of the MIT BioMicro Center Fugen Li, Kevin K. Thai, and Stuart Levine for bioinformatics and technical support. We are especially grateful to Nadya Dimitrova and Tyler Jacks for miR-143/145 conditional KO ESC lines, Cedric Blanpain for inducible MesP1 and pMesP1-GFP reporter ESC lines, and for providing gene expression data. R.K.B was supported by the Damon Runyon Cancer Research Foundation (DRG 2032-09 and DFS 04-12). M.L.S was supported by NIH K08 (DK090147) and the Watkins Cardiovascular Leadership Award. C.A.K is a Novartis Scholar of the Life Sciences Research Foundation. J.C.S. is supported by an EMBO long-term fellowship. This work was supported by the NHLBI Bench to Bassinet Program (U01HL098179) (L.A.B), the Richard and Susan Smith Family Foundation, Chestnut Hill, MA (L.A.B.), and the Pew Scholar’s Program in the Biomedical Sciences (L.A.B.).
Footnotes
Supplemental Information includes Extended Experimental Procedures, seven figures, and six tables.
Accession Numbers
All RNA-sequencing data generated during the course of this study including raw reads and analysis files have been deposited to GEO under the accession ID GSE39656.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congrains A, Kamide K, Oguro R, Yasuda O, Miyata K, Yamamoto E, Kawai T, Kusunoki H, Yamamoto H, Takeya Y, Yamamoto K, et al. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis. 2012;220:449–455. doi: 10.1016/j.atherosclerosis.2011.11.017. [DOI] [PubMed] [Google Scholar]
- Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Muller-Hocker J, Kitajima S, Lickert H, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- David R, Jarsch VB, Schwarz F, Nathan P, Gegg M, Lickert H, Franz WM. Induction of MesP1 by Brachyury(T) generates the common multipotent cardiovascular stem cell. Cardiovasc Res. 2011;92:115–122. doi: 10.1093/cvr/cvr158. [DOI] [PubMed] [Google Scholar]
- Delgado-Olguín P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Bao X, Zhuang Q, Zhou T, Qin B, Pei D. The mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr Opin Genet Dev. 2012;22:423–428. doi: 10.1016/j.gde.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Nucleic Acids Res. 2011;39:D800–806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, Daley GQ, et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res. 2012;110:406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Alvarez-Dominguez JR, Lodish HF. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012;13:971–983. doi: 10.1038/embor.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng K, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. PNAS. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Adler ED, Keller GM. Specification of multipotential cardiovascular progenitor cells during embryonic stem cell differentiation and embryonic development. Trends Cardiovasc Med. 2007;17:240–246. doi: 10.1016/j.tcm.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al. Suppressor of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012a;15:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2012b Dec 2; doi: 10.1038/nature11661. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- Lim SM, Pereira L, Wong MS, Hirst CE, Van Vranken BE, Pick M, Trounson A, Elefanty AG, Stanley EG. Enforced expression of Mixl1 during mouse ES cell differentiation suppresses hematopoietic mesoderm and promotes endoderm formation. Stem Cells. 2009;27:363–374. doi: 10.1634/stemcells.2008-1008. [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louch WE, Sheehan KA, Wolska BM. Methods in cardiomyocyte isolation, culture, and gene transfer. J Mol Cell Cardiol. 2011;51:288–298. doi: 10.1016/j.yjmcc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci. 2009;1178:29–46. doi: 10.1111/j.1749-6632.2009.04991.x. [DOI] [PubMed] [Google Scholar]
- McCulley DJ, Black BL. Transcription factor pathways and congenital heart disease. Curr Top Dev Biol. 2012;100:253–277. doi: 10.1016/B978-0-12-387786-4.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mercola M, Ruiz-Lozano P, Schneider MD. Cardiac muscle regeneration: lessons from development. Genes Dev. 2011;25:299–309. doi: 10.1101/gad.2018411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Slobodyanyuk SY, Elisaphenko EA, Shevchenko AI, Johnston C, Pavlova ME, Rogozin IB, Kolesnikov NN, Brockdorff N, Zakian SM. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long non-coding RNAs and enhancers. Curr Opin Genet Dev. 2011;21:194–198. doi: 10.1016/j.gde.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10:345–352. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Harvey RP, Mattick JS. Long Noncoding RNAs in Cardiac Development and Pathophysiology. Circ Res. 2012;111:1349–1362. doi: 10.1161/CIRCRESAHA.112.268953. [DOI] [PubMed] [Google Scholar]
- Simoes FC, Peterkin T, Patient R. Fgf differentially controls cross-antagonism between cardiac and hemangioblast regulators. Development. 2011;138:3235–3245. doi: 10.1242/dev.059634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Steinhauser ML, Lee RT. Regeneration of the heart. EMBO Mol Med. 2011;3:701–712. doi: 10.1002/emmm.201100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res. 2012;110:1628–1645. doi: 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and Coordinated Epigenetic Regulation of Developmental Transitions in the Cardiac Lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li TS, Lee ST, Wawrowsky KA, Cheng K, Galang G, Malliaras K, Abraham MR, Wang C, Marban E. Dedifferentiation and proliferation of mammalian cardiomyocytes. PLoS One. 2010;5:e12559. doi: 10.1371/journal.pone.0012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.