Abstract
Polymorphism at the GPSM3 gene locus is inversely associated with four systemic autoimmune diseases, including rheumatoid arthritis and ankylosing spondylitis. G-protein signaling modulator-3 (GPSM3) expression is most pronounced in myeloid cells, in which it targets heterotrimeric G-protein Gαi subunits of chemokine receptors, critical to immune function. To begin to explore the regulatory role of GPSM3 in monocytes, human THP-1 and primary mouse myeloid cells were cultured under stimulus conditions; GPSM3 was found by immunoblotting to be expressed at highest levels in the mature monocyte. To evaluate the effects of GPSM3 deficiency on a myeloid-dependent autoimmune disease, Collagen Antibody-Induced Arthritis (CAIA) was induced in Gpsm3−/− and control mice, which were then analyzed for clinical score, paw swelling, intra-articular proinflammatory markers, and histopathology. Mice lacking GPSM3 were protected from CAIA, and expression of monocyte-representative pro-inflammatory chemokine receptors and cytokines in paws of Gpsm3−/− mice were decreased. Flow cytometry, apoptosis, and transwell chemotaxis experiments were conducted to further characterize the effect of GPSM3 deficiency on survival and chemokine responsiveness of monocytes. GPSM3-deficient myeloid cells had reduced migration ex vivo to CCL2, CX3CL1, and chemerin and enhanced apoptosis in vitro. Our results suggest that GPSM3 is an important regulator of monocyte function involving mechanisms of differentiation, survival, and chemotaxis, and deficiency in GPSM3 expression is protective in acute inflammatory arthritis.
Keywords: cell signaling molecules, collagen induced arthritis, autoimmune diseases, macrophages, rheumatoid arthritis
1. Introduction
Myeloid derived cells such as monocytes and macrophages represent a major first line of defense against pathogens, but also contribute substantially to the pathogenesis of aseptic conditions such as chronic autoinflammatory and autoimmune disease (1, 2). Cytokines secreted by monocytes/macrophages comprise a diverse group of molecules including interleukin-1β (IL-1β), interleukin-6 (IL-6), tumor necrosis factor-α (TNF-α), interleukin-23 (IL-23), and interferon-γ (IFN-γ), all of which are involved in autoimmune pathogenesis (2, 3) and have been targeted effectively for the treatment of autoimmune disease (4).
G protein-coupled receptors (GPCRs) expressed on monocytes and macrophages regulate a variety of functions, including cell-to-cell contacts, cell activation and survival decisions, inflammatory mediator production, and chemotaxis (5). The intracellular signals transduced by cell-surface GPCRs are tightly controlled by a guanine nucleotide-binding complex of Gα, Gβ, and Gγ subunits, as well as a growing array of regulatory and accessory proteins (6). Consequently, this family of receptors has been extensively studied and exploited as drug discovery targets to identify valuable modulators of the immune response.
We and others have identified a novel set of regulators of G-protein signaling (6) involving association between inactive GDP-bound Gαi/o subunits and GoLoco motifs. Among the GoLoco class of proteins, one of the smallest called GPSM3 (‘G-protein signaling modulator-3’; a.k.a. AGS4 or G18) is known to bind not only to Gαi·GDP subunits but also to free Gβ subunits during their biosynthetic path towards Gβγ dimer formation (6). Gene atlas data of the human transcriptome demonstrates that GPSM3 mRNA has restricted tissue expression to hematopoietic organs with highest expression in myeloid cells (Fig. S1A), suggesting a potential role in immune regulatory responses. Additionally, GPSM3 mRNA was recently shown to decrease in the paw after anti-inflammatory treatment with dexamethasone in the Collagen Induced Arthritis (CIA) model (7), supporting a potential role for GPSM3 in inflammatory arthritis.
Recently, polymorphisms within the human GPSM3 gene locus (SNPs rs204989 and rs204991) were identified in GWAS studies (8, 9) as being significantly less prevalent in multiple autoimmune diseases, including rheumatoid arthritis (RA) and ankylosing spondylitis (AS). Based on the prominent expression of GPSM3 in myeloid cells and its regulatory implications in CIA, we chose to examine functional responses of wildtype and GPSM3-deficient mice in the Collagen Antibody-Induced Arthritis (CAIA) model, known to be highly dependent on monocyte function and innate immune responses (10). In vitro and ex vivo data suggest GPSM3 is an important regulator of monocyte function involving, at least in part, regulation of differentiation, chemotaxis, and survival, and its deficiency is protective in acute inflammatory arthritis.
2. MATERIALS AND METHODS
2.1. Reagents, cell culture, and microscopy
Monoclonal antibody against CD14 (clone rmC5-3) was from BD Pharmingen (San Diego). Anti-GPSM3 antibody was produced as described (6). Chemokines were from PeproTech (Rocky Hill, NJ). Cell lines were from American Type Culture Collection, grown in RPMI 1640 or DMEM (L-929) and supplemented with 10% FBS (Cellgro, Manassas, VA). THP-1 cells were transduced with control or GPSM3-silencing shRNA (sh19 or sh20), selected in 2.5 μg/ml puromycin, and confirmed for GPSM3 knockdown by immunoblot. Imaging of GPSM3 was performed as previously described (6).
2.2. CAIA arthritis model
All work was performed using IACUC-approved protocols in the AAALAC-accredited vivarium of UNC. Gpsm3-deficient (−/−) mice were obtained by breeding heterozygous C57Bl/6 mice purchased from the Texas Institute for Genomic Medicine. Parental mice were obtained by knocking out one copy of Gpsm3 via gene-trap insertion in C57Bl/6N ES cells (Fig. S2A). For CAIA, mice were injected on day 0 intraperitoneally with a cocktail of 5 anti-collagen monoclonals (5 mg/0.5 ml/mouse; Chondrex) and on day 3 with LPS (50 μg/0.1 ml/mouse). Mice were monitored daily for arthritis severity by a blinded observer as described (11, 12). Hind paws were isolated and fixed for 24h in Tellyesnickzky/Fekete fixative, decalcified using formic acid, and embedded in paraffin. Serial 5–7 μm sections were stained with H&E and analyzed by a blinded veterinary pathologist.
2.3. Quantitative real-time PCR
Joint tissues or cells were harvested at experiment termination. Expression of intra-articular mRNAs was measured by qRT-PCR using validated primers as previously described (11).
2.4. Chemotaxis
Chemotaxis experiments were performed as described (12) on Ly6C-enriched splenocytes. Briefly, mouse spleens were manually disrupted and filtered through 70 μm mesh, RBCs lysed, and enriched for Ly6C+ cells by magnetic separation using anti-Ly6C-biotin beads (Miltenyi Biotec, MA). Staining and flow cytometry analysis of migrated cells from the lower chambers was performed using anti-CD11b (eBioscience).
2.5. Flow cytometry
One million splenocytes were stained and analyzed using a Beckman-Coulter (Dako) CyAn ADP Analyzer. Anti-Ly6C-FITC and anti-Ly6G-PE were from eBioscience and anti-CCR2-APC was from R&D Systems.
2.6. Apoptosis
THP-1 cells were transduced with control or GPSM3-silencing shRNA as described (6). To induce apoptosis, 4–5 × 105 THP-1 cells were plated for 20h in culture media with or without 10 μM etoposide (Trevigen, Gaithersburg, MD). Cells were harvested, washed, stained with FITC-annexin V and propidium iodide using the TACS Annexin-V FITC kit (Trevigen), and analyzed by flow cytometry using a CyAn ADP Analyzer. Data is expressed as fold-increase in apoptotic cells between untreated and etoposide-treated matched samples.
2.7. Statistical analyses
For clinical disease and paw swelling, linear mixed models were used as described (11). The overall group effect was assessed using a likelihood ratio test (LRT) (α=.05) with SAS v9.2. For qRT-PCR, flow cytometry, and chemotaxis data, a two tailed Student’s t test was used and, for histopathology scores, a Mann-Whitney U test was used with Prism v4.
3. RESULTS
3.1. GPSM3 protein levels are highest in mature monocytes
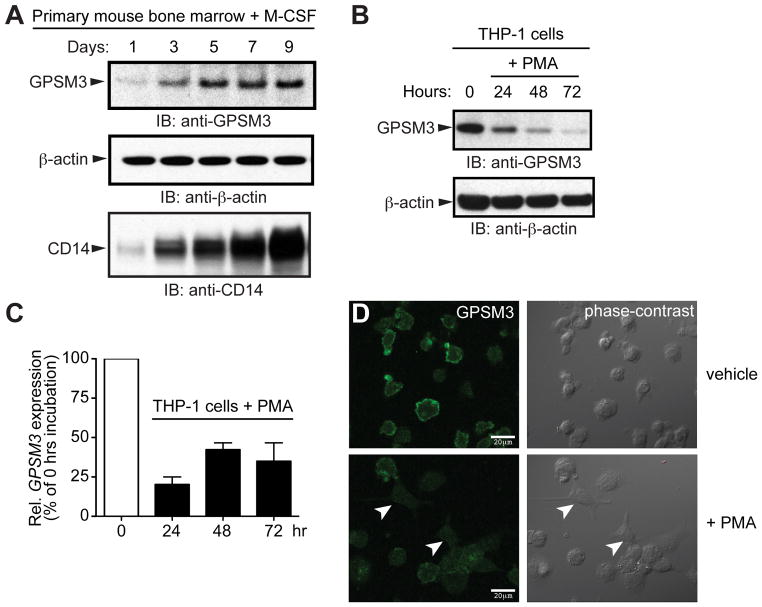
Based on the mRNA expression profile of GPSM3 within the human transcriptome (Fig. S1A), we hypothesized that GPSM3 protein expression may be restricted to the hematopoietic lineage with significant expression in monocytes. First, we screened immortalized human cell lines for endogenous GPSM3 expression using an antibody against human and mouse GPSM3 (6). A weak but specific signal at the expected GPSM3 molecular weight of 18 kDa was detected in lysates from human cell lines derived from acute lymphoblastic leukemia (Molt4), T cell leukemia (Jurkat), and Burkitt’s lymphoma (Ramos), but not in cells lines of non-hematopoietic origin (HEK293, BE2C, SH-SY5Y) (Fig. S1B). In contrast, strong expression was seen in the acute monocytic leukemia cell line THP-1 (Fig. S1B). To further confirm these data in primary cells, bone marrow from C57Bl/6 mice was differentiated using macrophage colony-stimulating factor (M-CSF) (13). Upregulation of CD14, a marker of the monocyte/macrophage lineage (14), increased coincident with levels of GPSM3 during differentiation (Fig. 1A).
Figure 1. GPSM3 is most highly expressed in mature monocytes.
(A) Bone marrow cells from C57Bl/6 mice were cultured on plastic; treatment over 9 days with M-CSF (L-929 conditioned medium) induced upregulation of CD14 expression and GPSM3 expression. (B) Differentiation of THP-1 cells into macrophages using PMA (50 ng/mL) decreased GPSM3 protein and (C) mRNA expression over 72 hours. GPSM3 transcript levels were normalized for relative expression levels using the 2-(ΔΔCt)method with 18S rRNA as the internal control. Data is presented as percent relative expression compared to the untreated (“zero hrs”) THP-1 cell line sample. (D) Endogenous GPSM3 expression was detected by indirect immunofluorescence and confocal microscopy in THP-1 cells cultured on poly-D-lysine-coated coverslips and treated with ethanol vehicle control (upper panels) or 50 ng/mL PMA for 72 hours (lower panels). Arrowheads denote THP-1 cells with macrophage-like morphology (less-rounded, extended processes). Scale-bar, 20 μm.
To determine whether GPSM3 expression might be altered following activation, we differentiated the monocytic THP-1 cell line into macrophage-like cells using phorbol 12-myristate 13-acetate (PMA) treatment (15). PMA treatment over 72 hours decreased GPSM3 protein levels (Fig. 1B) and GPSM3 mRNA levels (Fig. 1C). Decreased GPSM3 expression was also observed by indirect immunofluorescence and confocal microscopy. Untreated THP-1 cells exhibited robust expression of endogenous GPSM3 that localized to the plasma membrane; PMA treatment led to decreased GPSM3 immunostaining with more differentiated cells (identified by their macrophage-like, spreading morphology) showing the least GPSM3 expression (Fig. 1D).
3.2. Gpsm3−/− mice are protected from CAIA
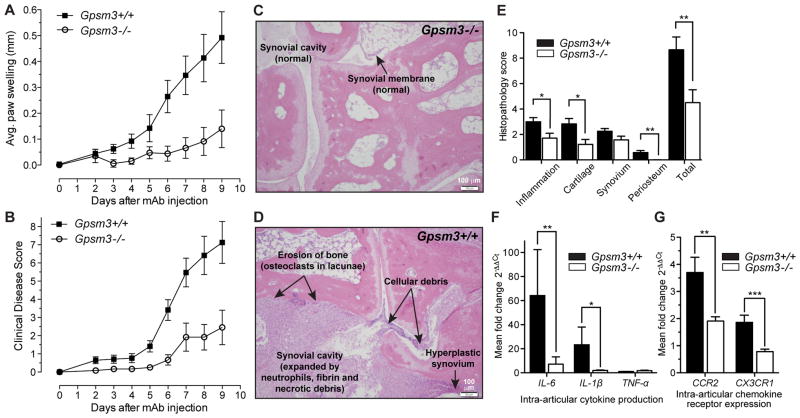
Because mature monocytes expressed the highest GPSM3 protein levels, we hypothesized that differential GPSM3 expression in monocytes might affect inflammatory disease in mice. Gpsm3−/− mice were obtained (Fig. S2A) and, when phenotyped, had normal hematopoiesis (Table S1), organogenesis, fecundity, and longevity. CAIA was induced in Gpsm3−/− mice and in age- and gender-matched wildtype controls. Gpsm3−/− mice had significant decreases in paw swelling and clinical disease scores (Fig. 2A, B). The incidence of disease was also reduced by GPSM3 deficiency to 34% compared to 95% in control mice (data not shown). At experiment termination, histological sections were scored by a blinded pathologist for the extent of inflammatory cell infiltration, cartilage erosion, synovial hyperplasia, and periosteal erosion (Fig. 2C, D); the severity of inflammation and erosion was reduced or absent in Gpsm3−/− mice compared to controls (Fig. 2E).
Figure 2. Inflammatory signs of arthritis are reduced in Gpsm3−/− mice.
All data are combined from two separate experiments (Gpsm3−/−, n=10; Gpsm3+/+, n=15). (A) Mean clinical index over time (+/− SEM) was significantly reduced in Gpsm3−/− mice (LRT = 6.5, df=2, p <0.05). (B) Mean difference in paw swelling from baseline thickness over time (+/− SEM) was also significantly different (LRT = 13.5, df=3, p<0.01). (C, D) Representative H&E histology of hind limbs at experiment termination (day 10) shows that late stage inflammation and erosion are decreased in Gpsm3−/− mice. Arrows highlight major inflammatory and erosive differences between genotypes. (E) Histological evaluation of both hind paws were examined (day 10) and scored on a 0–4 scale for each subcategory. Quantitative RT-PCR demonstrated a significant reduction in expression of transcripts for the pro-inflammatory cytokines IL-6 and IL-1β (F) as well as the chemokine receptors CCR2 and CX3CR1 (G) in Gpsm3−/− mice arthritic paws compared to wildtype controls. Transcript levels were normalized for relative expression levels using the 2-(ΔΔCt) method with IDUA rRNA as the internal control. Data is presented as percent relative expression compared to a mouse paw without arthritis.
3.3 Gpsm3−/− arthritic paws have reduced expression of proinflammatory cytokines and chemokine receptors
To analyze the expression of validated monocyte-derived proinflammatory mediators (11), RNA was extracted from paws of CAIA mice at experiment termination and transcripts for IL-1β, IL-6, and TNF-α were analyzed by qRT-PCR. IL-1β and IL-6 transcripts levels were significantly reduced in inflamed paws from Gpsm3−/− mice (Fig. 2F), whereas TNF-α transcripts were similar in both groups.
Two major subsets of monocytes have been identified in mice based on their functional properties and level of surface expression of the chemokine receptors CCR2 and CX3CR1 (14). Proinflammatory monocytes are recruited to the inflamed synovium by the action of inflammation-driven chemokines such as Monocyte Chemoattractant Protein-1 (MCP-1 or CCL2), the ligand for CCR2 (14, 16) and Fractalkine (FKN or CX3CL1), the ligand for CX3CR1 (11). Real-time PCR analysis of CCR2 and CX3CR1 transcripts in arthritic paws revealed that the expression levels of both were significantly reduced in Gpsm3−/− mice (Fig. 2G).
3.4. Reduced number, viability, and chemotactic function of GPSM3-deficient myeloid cells
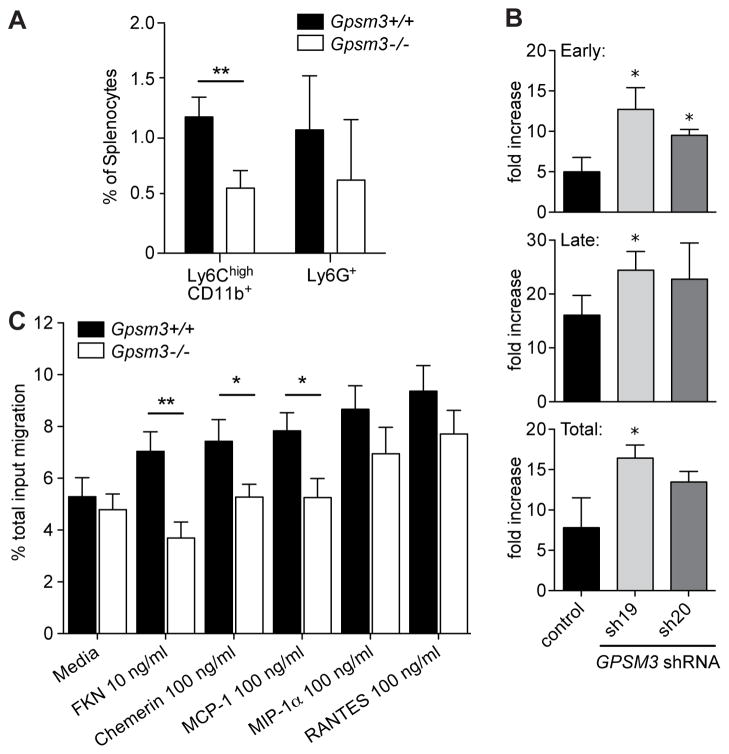
Monocytes characterized by the cell surface markers Ly6Chigh and CD11b+ are thought to be proinflammatory in that they mobilize rapidly in response to infection and inflammation (17). Flow cytometry of isolated splenocytes from Gpsm3−/− mice revealed that the abundance of inflammatory monocytes, characterized by Ly6ChighCD11b+ expression was significantly reduced in comparison to littermate control mice (Fig. 3A), whereas the granulocyte Ly6G+ population was not significantly altered. Furthermore, monocytic THP-1 cells deficient in GPSM3 (6) were more sensitive to death induced by apoptotic stimuli (Fig. 3B), suggesting effects on viability.
Figure 3. GPSM3-deficient splenocytes have decreased number and chemotactic function of the Ly6C+CD11b+ myeloid population, and GPSM3-deficient myeloid THP-1 cells exhibit enhanced apoptosis.
(A) Flow cytometry analysis of splenocytes revealed decreased numbers of Ly6ChighCD11b+ monocytes in Gpsm3−/− mice versus wildtype controls, while the numbers of Ly6G+ granulocytes were not altered (n = 3; **, p < 0.05). (B) GPSM3-deficient THP-1 cells (bearing stable expression of lentiviral-transduced shRNAs sh19 and sh20; (6)) have enhanced apoptosis in early (annexin V positive, propidium iodide negative) and late (annexin V positive, propidium iodide positive) stages following etoposide treatment (n=3, *p<0.05). (C) Migration in response to indicated chemokines was determined by anti-CD11b staining and flow cytometry of migrated Ly6C+ enriched cells from LPS-treated Gpsm3−/− mice vs littermate controls. Data are represented as percent migration of total input (mean +/− SEM, n=6).
Activation of the monocyte/macrophage system is a hallmark of rheumatoid arthritis (16); such activation is potentiated in the CAIA model by LPS injection in vivo (18), allowing for subsequent isolation of physiologically activated monocytes/macrophages (19). To analyze the chemotactic responsiveness of mature, proinflammatory myeloid cells, Gpsm3−/− and control mice were injected with LPS and resultant Ly6C-enriched/CD11b+ splenocytes were tested for their migration ex vivo towards a panel of proinflammatory monocytic chemokines. Ly6C-enriched/CD11b+ splenocytes from Gpsm3−/− mice had significantly reduced chemotaxis toward CCL2/MCP-1, CX3CL1/FKN, and chemerin, although no statistical differences were observed for CCL3/MIP-1α or CCL5/RANTES (Fig. 3C).
4. DISCUSSION
Monocytes are critical to autoimmune arthritis disease pathogenesis through their roles in antigen presentation and production of pro-inflammatory cytokines (1, 2); in particular, chemotaxis of circulating monocytes toward the inflamed joint, and their subsequent differentiation into proinflammatory macrophages, is critical to the development and exacerbation of RA (16). Our results support a key role of GPSM3 in monocyte function and in disease pathogenesis in an acute inflammatory arthritis disease model–a finding that, in turn, strengthens the association of human GPSM3 polymorphisms in autoimmune diseases including RA and AS (8, 9). Part of the protection to CAIA conferred by GPSM3 deficiency is believed to occur through altered chemotaxis toward multiple proinflammatory chemokines that target monocytes to the synovium, which suggests that GPSM3 may be a promiscuous regulator of multiple Gαi-coupled receptors. Additionally, impaired survival of GPSM3-deficient monocytes upon exposure to pro-apoptotic stimuli may also affect overall monocyte differentiation and function in vivo and, thus, the severity of disease sequelae. These data provide compelling rationale for future studies examining GPSM3 deficiency in chronic autoimmune models such as CIA, as well as an examination of GPSM3 gene products and monocyte function in humans with and without autoimmune inflammatory arthritis.
Supplementary Material
(A) Based on data obtained from the public GeneAtlas database of the human transcriptome (accessible at http://biogps.org/), expression of the human GPSM3 transcript is restricted to hematopoietic cells and is detected with the highest signal strength in whole blood and CD14+ monocytes. (B) GPSM3 was detected by immunoblotting total lysates from human hematopoietic cell lines Molt4, Jurkat, Ramos, and THP-1, U937, and non-hematopoietic neuroblastoma cell lines SK-N-BE(2)-C (“BE2C”) and SH-SY5Y, and human embryonic kidney cell line HEK293.
Gpsm3−/− mice were obtained by breeding heterozygous mice purchased from the Texas Institute for Genomic Medicine. Parental mice were obtained by knocking out one copy of the Gpsm3 locus via gene-trap insertion in mouse C57Bl/6N cells, generating the ES cell clone IST12408A1. (A) Illustration of the insertion site of the gene-trap into the 5′ UTR region of the Gpsm3 gene locus. (B) Mice were genotyped by PCR from mouse tail DNA. Primers use for genotyping as illustrated in (A) are: fwd1 primer 5′-cttcctgtgttgtagctagttcc-3′, rev1 5′-tcacctgttcaccatcttcttcc-3′, rev2 5′-ccaataaaccctcttgcagttgc-3′. (C) GPSM3 expression is ~50% reduced in bone marrow monocytes from Gpsm3+/− mice and absent in Gpsm3−/− mice. Mouse bone marrow cells were differentiated over 3 days with M-CSF prior to immunoblotting with anti-GPSM3 and α-actin mAbs as a loading control.
RESEARCH HIGHLIGHTS.
GPSM3 is highly expressed in mature monocytes.
GPSM3-deficient monocytes have decreased chemotaxis to CCL2, CX3CL1, and chemerin.
GPSM3-deficient THP-1 cells have enhanced apoptosis.
Gpsm3−/− mice have decreased arthritis in the Collagen Antibody Induced Arthritis (CAIA) model.
Acknowledgments
This study was supported by NIH R01 GM082892 (to D.P.S.), R03 AR059286 and Rheumatology Research Foundation funding (to T.K.T.), NIH CTSA UL1RR025747, and an NC TraCS pilot grant 50KR71116 (to T.K.T. and D.P.S.) via support from NIH CTSA UL1TR000083.
Footnotes
Authorship contributions. TT and DS conceived of and designed the studies. PG wrote the manuscript, and PG and GL performed the monocyte differentiation studies. TT, RG, and BB performed the arthritis studies, and OF evaluated the histopathology. DE performed and oversaw the statistical analysis. PL performed the flow cytometry studies. MB performed the apoptosis studies. MM performed the qRT-PCR studies and analysis. TT and MB performed the chemotaxis studies.
Conflict of interest. The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ambarus C, Yeremenko N, Tak PP, Baeten D. Pathogenesis of spondyloarthritis: autoimmune or autoinflammatory? Current opinion in rheumatology. 2012;24(4):351–358. doi: 10.1097/BOR.0b013e3283534df4. [DOI] [PubMed] [Google Scholar]
- 2.Rong C, Hu W, Wu FR, Cao XJ, Chen FH. Interleukin-23 as a potential therapeutic target for rheumatoid arthritis. Molecular and cellular biochemistry. 2012;361(1–2):243–248. doi: 10.1007/s11010-011-1109-6. [DOI] [PubMed] [Google Scholar]
- 3.Kagari T, Doi H, Shimozato T. The importance of IL-1 beta and TNF-alpha, and the noninvolvement of IL-6, in the development of monoclonal antibody-induced arthritis. J Immunol. 2002;169(3):1459–1466. doi: 10.4049/jimmunol.169.3.1459. [DOI] [PubMed] [Google Scholar]
- 4.Miller AV, Ranatunga SK. Immunotherapies in rheumatologic disorders. The Medical clinics of North America. 2012;96(3):475–496. ix–x. doi: 10.1016/j.mcna.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Lattin J, et al. G-protein-coupled receptor expression, function, and signaling in macrophages. J Leukoc Biol. 2007;82(1):16–32. doi: 10.1189/jlb.0107051. [DOI] [PubMed] [Google Scholar]
- 6.Giguere PM, Laroche G, Oestreich EA, Siderovski DP. G-protein signaling modulator-3 regulates heterotrimeric G-protein dynamics through dual association with Gbeta and Galphai protein subunits. J Biol Chem. 2012;287(7):4863–4874. doi: 10.1074/jbc.M111.311712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt N, et al. The anti-inflammatory fungal compound (S)-curvularin reduces proinflammatory gene expression in an in vivo model of rheumatoid arthritis. The Journal of pharmacology and experimental therapeutics. 2012;343(1):106–114. doi: 10.1124/jpet.112.192047. [DOI] [PubMed] [Google Scholar]
- 8.Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5(12):e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corona E, Dudley JT, Butte AJ. Extreme evolutionary disparities seen in positive selection across seven complex diseases. PloS one. 2010;5(8):e12236. doi: 10.1371/journal.pone.0012236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandakumar KS, Backlund J, Vestberg M, Holmdahl R. Collagen type II (CII)-specific antibodies induce arthritis in the absence of T or B cells but the arthritis progression is enhanced by CII-reactive T cells. Arthritis Res Ther. 2004;6(6):R544–550. doi: 10.1186/ar1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tarrant TK, et al. Decreased Th17 and antigen-specific humoral responses in CX(3) CR1-deficient mice in the collagen-induced arthritis model. Arthritis Rheum. 2012;64(5):1379–1387. doi: 10.1002/art.34320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarrant TK, et al. Granulocyte chemotaxis and disease expression are differentially regulated by GRK subtype in an acute inflammatory arthritis model (K/BxN) Clin Immunol. 2008;129(1):115–122. doi: 10.1016/j.clim.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. Cold Spring Harb Protoc. 2008;2008(12) doi: 10.1101/pdb.prot5080. pdb.prot5080- [DOI] [PubMed] [Google Scholar]
- 14.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 15.Cassol E, Alfano M, Biswas P, Poli G. Monocyte-derived macrophages and myeloid cell lines as targets of HIV-1 replication and persistence. J Leukoc Biol. 2006;80(5):1018–1030. doi: 10.1189/jlb.0306150. [DOI] [PubMed] [Google Scholar]
- 16.Tarrant TK, Patel DD. Chemokines and leukocyte trafficking in rheumatoid arthritis. Pathophysiology. 2006;13(1):1–14. doi: 10.1016/j.pathophys.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Sunderkotter C, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172(7):4410–4417. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- 18.Terato K, et al. Collagen-induced arthritis in mice: synergistic effect of E. coli lipopolysaccharide bypasses epitope specificity in the induction of arthritis with monoclonal antibodies to type II collagen. Autoimmunity. 1995;22(3):137–147. doi: 10.3109/08916939508995311. [DOI] [PubMed] [Google Scholar]
- 19.Saluk-Juszczak J, Wachowicz B. The proinflammatory activity of lipopolysaccharide. Postepy Biochem. 2005;51(3):280–287. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Based on data obtained from the public GeneAtlas database of the human transcriptome (accessible at http://biogps.org/), expression of the human GPSM3 transcript is restricted to hematopoietic cells and is detected with the highest signal strength in whole blood and CD14+ monocytes. (B) GPSM3 was detected by immunoblotting total lysates from human hematopoietic cell lines Molt4, Jurkat, Ramos, and THP-1, U937, and non-hematopoietic neuroblastoma cell lines SK-N-BE(2)-C (“BE2C”) and SH-SY5Y, and human embryonic kidney cell line HEK293.
Gpsm3−/− mice were obtained by breeding heterozygous mice purchased from the Texas Institute for Genomic Medicine. Parental mice were obtained by knocking out one copy of the Gpsm3 locus via gene-trap insertion in mouse C57Bl/6N cells, generating the ES cell clone IST12408A1. (A) Illustration of the insertion site of the gene-trap into the 5′ UTR region of the Gpsm3 gene locus. (B) Mice were genotyped by PCR from mouse tail DNA. Primers use for genotyping as illustrated in (A) are: fwd1 primer 5′-cttcctgtgttgtagctagttcc-3′, rev1 5′-tcacctgttcaccatcttcttcc-3′, rev2 5′-ccaataaaccctcttgcagttgc-3′. (C) GPSM3 expression is ~50% reduced in bone marrow monocytes from Gpsm3+/− mice and absent in Gpsm3−/− mice. Mouse bone marrow cells were differentiated over 3 days with M-CSF prior to immunoblotting with anti-GPSM3 and α-actin mAbs as a loading control.