Abstract
CD8+ T cells play a crucial role in the clearance of intracellular pathogens through the generation of cytotoxic effector cells that eliminate infected cells and long-lived memory cells that provide enhanced protection against reinfection. We have previously shown that the inhibitor of E protein transcription factors, Id2, is necessary for accumulation of effector and memory CD8+ T cells during infection. Here we show that CD8+ T cells lacking Id2 did not generate a robust terminally-differentiated KLRG1hi effector population, but displayed a cell-surface phenotype and cytokine profile consistent with memory precursors, raising the question as to whether loss of Id2 impairs the differentiation and/or survival of effector-memory cells. We found that deletion of Bim rescued Id2-deficient CD8+ cell survival during infection. However, the dramatic reduction in KLRG1hi cells caused by loss of Id2 remained in the absence of Bim, such that Id2/Bim double-deficient cells form an exclusively KLRG1lo CD127hi memory precursor population. Thus we describe a role for Id2 in both the survival and differentiation of normal CD8+ effector and memory populations.
Keywords: transcription factor, T cell memory, infection
INTRODUCTION
CD8+ T cells are an essential component of host resistance to viral and intracellular bacterial infections. Upon pathogen recognition in the context of appropriate co-stimulatory signals, naive CD8+ T cells expand and acquire effector functions, allowing them to kill infected cells and secrete cytokines. Upon resolution of infection, the majority of responding CD8+ T cells undergo apoptosis. However, a small number of antigen-specific effector cells remain and establish a heterogeneous memory T cell population that collectively serves to protect against reinfection. Exogenous signals received by responding CD8+ T cells early during infection including duration and levels of antigen and inflammatory cytokines are known to influence cell-fate decisions that dictate which cells will die soon after the resolution of infection or develop into memory-precursor cells from which the long-lived memory population is established (1–3). Phenotypic and functional analyses of CD8+ effector T cells have used levels of expression of KLRG1, CD127, CD27, CD43, CXCR3, and CD25 and production of IL-2 to distinguish cells that will go on to become terminally-differentiated, short-lived effector-memory cells versus long-lived memory cells (4–10). Numerous transcription factors have also been implicated in this cell-fate decision with T-bet, Blimp-1 and Id2 favoring shorter-lived, effector-memory cells expressing high levels of KLRG1, and with Eomesodermin, Bcl-6, TCF-1, STAT3 and Id3 expression supporting the formation of long-lived memory precursors with high levels of CD127 and CXCR3 (4, 11–22). Intimately intertwined in the formation of these distinct effector and memory populations is the relationship between surviving the contraction phase of the effector population and the initiation of a gene-expression program that supports the phenotypic and functional characteristics of protective memory.
Id2 is a member of the inhibitor-of-DNA-binding (Id) family consisting of transcriptional regulators that act antagonistically on E protein transcription factors to prevent their binding to DNA. Clear roles for E and Id proteins in lymphocyte development are well delineated (23). However, despite relatively high mRNA expression levels of Id2 in mature CD8+ effector and memory cells and high Id3 in naive and long-lived memory cells (19, 24, 25), the role of these transcriptional regulators in CD8+ effector T cell differentiation is still not fully understood. Previously, we showed loss of Id2 resulted in a diminished CD8+ effector response and led to fewer remaining memory CD8+ T cells which rapidly acquired a central memory (CD44hiCD62LhiKLRG1loCD127hi) phenotype (19, 25). This rapid loss of Id2-deficient CD8+ effector T cells was due to increased death by effector cells and correlated with high expression of the pro-apoptotic molecule Bim and low levels of the anti-apoptotic molecule Bcl-2. However, in these studies it was not clear if the severe loss of short-lived (CD44hiCD62LloKLRG1hiCD127lo) effector-memory cells was due solely to a failure of those cells to survive or if Id2 also regulated their differentiation. More recently, a role for Id3 expression in predicting memory potential and in supporting differentiation of long-lived memory cells has become clear as well, suggesting distinct functions for these two transcriptional regulators in CD8+ immunity (21, 24, 25).
One key molecule influencing the contraction phase of the CD8+ effector population is the proapoptotic molecule of the Bcl-2 family of proteins, Bim. Previous studies demonstrated a role for Bim in the induction of death of both CD4+ and CD8+ effector T cells after infection was resolved (26–28). Indeed, Bim-deficient CD8+ T cells failed to undergo apoptosis and continued to accumulate in the spleen indicating the importance of this protein in modulating the contraction phase of the CD8+ T cell immune response (26, 27). Surprisingly, Bim-deficiency appears to rescue both short-lived effector-memory and long-lived memory subsets from contraction (29–31). However, the memory cells rescued due to loss of Bim eventually wane, suggesting Bim-independent regulation of long-lived memory subsets (31). Ultimately, while it is understood that Bim is required for the contraction phase of the CD8+ T cell immune response, it is less clear if and how Bim is modulated to produce long-lived T cell memory.
In this study we investigated the role of Id2 in CD8+ T cells during infection. Here, we elaborate on the role of Id2 in formation of short-lived effector and long-lived memory CD8+ T cells, showing that Id2 influences the development of KLRG1hi effector T cells. We also show that high levels of Bim are responsible for the increased apoptosis observed in Id2-deficient cells responding to infection, likely due to unchecked E protein activity. Surprisingly, Bim-deficiency did not rescue the effector T cell differentiation defects observed in the absence of Id2, indicating Id2 controls the CD8+ T cell immune response at two levels: by influencing survival of effector cells and by regulating the effector and memory T cell differentiation program.
MATERIALS AND METHODS
Mice, Adoptive Transfers and Infection
Mice were bred and housed in specific pathogen-free conditions in accordance with the Institutional Animal Care and Use Guidelines of the University of California, San Diego. Mice with a mutated Id2 allele (Id2-knockout, Id2KO) were generated as previously described (19) and maintained on the C57/BL6 background. Mice with a mutated Bcl2l11 allele (Bim-knockout, BimKO) mice were purchased from Jackson Labs (32). For generation of fetal liver chimeras, 5 × 106 OT-I Id2+Bim+, BimKO, Id2KO, Id2KOBimKO E14-E15.5 fetal liver cells and 5 × 105 RAGKO bone marrow cells were injected i.v. into lethally irradiated (1000 RAD), congenically distinct hosts. Chimeras were rested for at least eight weeks to allow reconstitution of the host. Mixed Transfers: 2 × 104 OT-I wild type cells (CD45.1) were mixed with 2 × 104 OT-I Id2KO, BimKO or Id2KOBimKO cells (CD45.2) and were transferred into wild type CD45.1+CD45.2+ recipients. Single Transfers: 4 × 104 OT-I wild type, Id2KO, BimKO or Id2KOBimKO CD45.2 cells were transferred into wild type CD45.1 recipients. Infection: Mice were infected 1 day after mixed or single transfer with 5 × 103 colony-forming units of recombinant Listeria monocytogenes expressing chicken ovalbumin (Lm-OVA).
Flow Cytometry
Single-cell suspensions were prepared from the spleens of infected hosts. The following antibodies were used to phenotype the donor cells (all antibodies from eBioscience unless otherwise specified): CD8 (53-6.7), CD27 (LG-7F9), CD43 (1B11; Biolegend), CD44 (IM7), CD45.1 (A20-1.7), CD45.2 (104), CD62L (MEL-14), CD122 (TM-b1), CD127 (A7R34), CXCR3 (CXCR3-173), KLRG1 (2F1), Ly6C (AL-21; BD Biosciences). Antibodies were conjugated to fluorescing isothiocyanate, phycoerythrin, peridinin chlorophyll protein, peridinin chlorophyll protein-cyanin 5.5, allophycocyanin, Alexa Fluor 780, phycoerythrin-cyanin 7, Pacific Blue or biotin. Annexin V was purchased from Invitrogen. CXCL16-FC fusion protein was generously provided by Dr. M. Matloubian and used as previously described (33). For analysis of intracellular cytokine production, 4 × 106 splenocytes were incubated for a total of 6h at 37°C in RPMI (Mediatech) containing 10% (volume/volume) bovine growth serum (Hyclone) alone or with 10nM OVAp. After 3h 1μl/mL Golgi Stop (BD Biosciences) was added and cultures were incubated for an additional 3h. Cells were collected and stained with CD45.1, CD45.2, CD8 and KLRG1. Samples were fixed and made permeable in cytofix-cytoperm buffer (BD Biosciences). Intracellular staining for cytokines was preformed using IFNγ (XMG1.2), IL-2 (JES6-5H4) and TNFα (MP6-XT22). Samples were collected on a FACSCalibur, FACSAria or LSRIIFortessa (BD Biosciences) and were analyzed with Flowjo software (TreeStar).
Quantitative PCR and Chromatin Immunoprecipitation
KLRG1lo donor OT-I Id2+Bim+, BimKO, Id2KO, Id2KOBimKO splenocytes were sorted from host mice on day 6 of Lm-OVA infection. RNA was extracted with TRIzol reagent (Invitrogen) and treated with DNAse (Ambion), and cDNA was generated by RT-PCR with a Superscript III kit (Invitrogen). The abundance of mRNA was assessed by quantitative PCR with non-specific product detection (SYBR Green, Stratagene) with primers that amplify in a linear relationship for housekeeping genes. Results were normalized to levels of hprt mRNA. Chromatin immunoprecipitations were preformed as previously described (34) with 10 mg of DNA for each immunoprecipitation. A polyclonal antibody specific for HEB (A-20) was used to precipitate HEB-DNA complexes and rabbit Immunoglobulin G antibody (2729, Cell Signaling Technology) was used as a negative control. Immunocomplexes were bound to protein G sepharose beads (Cell Signaling Technology) and were washed four times. DNA was eluted and purified and was analyzed by quantitative PCR to detect E-box sites in the gene encoding Bim.
Microarray Analysis
KLRG1lo donor OT-I Id2+Bim+, BimKO, Id2KO, Id2KOBimKO splenocytes were sorted from host mice on day 6 of Lm-OVA infection. RNA was extracted with TRIzol reagent (Invitrogen) and was amplified twice with the MessageAmp RNA Amplicification kit (Ambion). RNA was labeled with biotin with the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics) and was purified with an RNeasy Mini Kit (Qiagen). The resulting cRNA was hybridized to GeneChip Mouse Gene 1.0 ST arrays and raw CEL files were obtained. Data were normalized and analyzed with the GenePattern software suite. The Gene Expression Omnibus (GEO) referenced accessions for these data is GSE41978 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41978&submit.x=0&submit.y=0).
Statistical Analysis
Differences between data sets were analyzed by an unpaired two-tailed student's t-test.
RESULTS
Loss of KLRG1hi short-lived effector cells in the absence of Id2
We previously showed that Id2 is necessary for CD8+ effector T cell survival during infection (19). Analysis of cell-surface markers showed early acquisition of a central memory phenotype by the few Id2-deficient effector CD8+ T cells recovered after infection. Thus, we were curious as to whether effector-memory populations were absent in the Id2-deficient response due to a failure of cells to survive or properly differentiate. Here, we investigated how loss of Id2 affected shorter-lived effector-memory (KLRG1hiCD127lo) and longer-lived memory precursor (KLRG1loCD127hi) CD8+ T cell populations during infection. We generated fetal liver or bone marrow chimeras reconstituted with either OT-I TCR transgenic Id2-sufficient (Id2+) or OT-I Id2-deficient (Id2KO) hematopoietic cells. Naive Id2+ or Id2KO OT-I cells (2 × 104) from chimeras were adoptively transferred into congenically distinct hosts, which were subsequently infected with Lm-OVA. Examination of cell-surface expression of KLRG1 and CD127 by transferred OT-I cells isolated on day 7 and 12 of infection showed a severe paucity of KLRG1hiCD127lo and KLRG1hiCD127hi effector cells in the Id2-deficient population (Fig. 1A). Analysis of absolute cell numbers of antigen-specific CD8+ KLRG1hi and KLRG1lo populations also showed a severe reduction (~17-fold on day 7 and 37-fold on day 12) in KLRG1hi cells in the Id2-deficient population compared to wild-type cells. However, the total numbers of KLRG1lo cells were similar between Id2+ and Id2KO cells (Fig. 1A). Id2KO CD8+ T cells also showed ~3-fold more cells with high CXCR3 expression, consistent with formation of a population of cells with a longer-lived memory phenotype (8). Further, we observed reduced (2–3-fold) CXCR6 expression by Id2-deficient CD8+ effector cells compared to Id2+ cells (Fig. 1B), a phenotype we previously showed was present in the Id2-deficient Natural Killer T (NKT) population as well (35). Thus, in the absence of Id2, at the peak of infection, CD8+ effectors do not accumulate as short-lived KLRG1hi effector-memory cells.
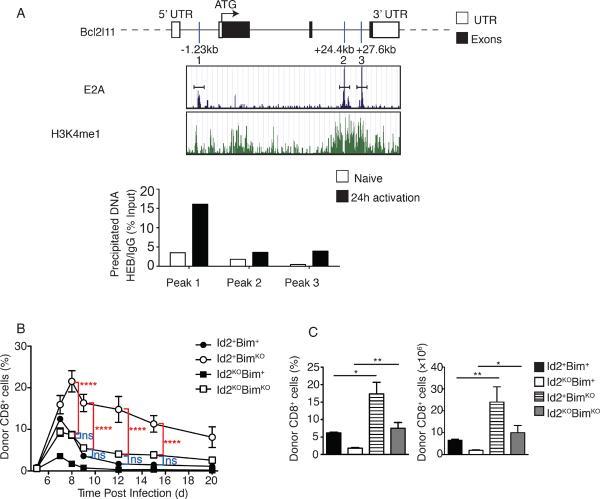
Figure 1. Id2-deficient CD8+ T cells display an altered phenotype during infection.
A 1:1 mixture of OT-I Id2-knockout cells (CD45.2) and OT-I Id2-wild type cells (CD45.1) were transferred into congenically distinct hosts 1 day before infection with Lm-OVA. (A) KLRG1 and CD127 expression by Id2+ and Id2KO OT-I cells recovered from spleen on days 7 and 12 post infection. Flow cytometry plots display surface phenotype of CD8+ and CD45.2+ or CD45.1+ gated cells. Numbers indicate percentage of cells in each quadrant (left). Average (±SEM) percentage donor CD8+ and total numbers of KLRG1hi and KLRG1lo cells summarizing flow cytometry data (bar graphs, right). Data are from two independent experiments, four mice per timepoint. (B) Histograms of CXCR6 and CD43 expression by donor cells, average (±SEM) of mean fluorescence intensity (right). Statistical significance was determined using unpaired two-tailed t test where ns, not significant, *, P < 0.05, **, P < 0.005, ***, P < 0.0005.
Knell et al.
Loss of Bim rescues survival defect in Id2KO CD8+ T cells during infection
We previously showed Bim mRNA levels were increased in Id2-deficient effector T cells (19) and Bim has previously been shown to be a direct target of E proteins (36, 37). The bcl2l11 (Bim) promoter and locus contains more than thirty conserved E box sites. To determine which E boxes potentially played a role in the regulation of Bim during the CD8+ T cell response, we examined previously published data in which E2A occupancy in A12 T cell lines was assessed by deep sequencing (ChIP-Seq) (36). We observed three E2A binding peaks containing the E2A binding motif CANNTG in the Bim locus and interestingly, H3K4 monomethylation (H3K4me1), a marker of active transcription, was also enriched at these three sites (Fig. 2A). To test whether E proteins bind these specific E box sites in the Bim locus upon CD8+ T cell activation, we isolated Id2+ OT-I cells directly ex vivo or after 24 hours of activation with OVA peptide in vitro and performed chromatin immunoprecipitation (ChIP). Using a HEB-specific antibody, we observed an increase in E protein binding upon activation at the potential E protein binding sites, with the first site (peak 1, in the promoter region of the Bim locus) showing the greatest increase (Fig. 2A). Together these data suggest that E proteins can bind DNA at the Bim locus and that Bim expression levels can be regulated directly by E protein transcription factors during CD8+ T cell activation.
Figure 2. Bim-deficiency rescues survival of Id2-deficient CD8+ T cells during infection.
(A) E2A occupancy (blue peaks) and H3K4me1 occupancy (green peaks) for the Bcl2l11 gene as reported by Lin et al. (17). E box binding sites are indicated relative to the transcriptional start site (ATG = 0kb). Immunoprecipitation of chromatin (with anti-HEB or immunologlobulin G, IgG) isolated from purified OT-I Id2+ cells directly ex vivo or 24 h after in vitro activation with OVAp followed by quantitative PCR analysis of input or precipitated DNA for indicated E-box sites in the Bcl2l11 gene. Data are representative of two independent experiments, n = 3. (B) Percentage of CD8+ OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO CD45.2 over the course of infection in PBL. CD45.1+ C57BL/6 mice received OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO (CD45.2+) cells (2 × 104) 1 day before infection with Lm-OVA, average (±SEM). Two-way ANOVA analysis indicated no significant difference between expansion and contraction of Id2+Bim+ and Id2KOBimKO cells throughout the course of infection. These analyses also revealed significant difference between Id2+BimKO and Id2KOBimKO cells throughout the course of infection. . (C) Bar graphs summarizing frequency of donor cells among CD8+ cells and total number of indicated donor cells recovered from spleen on day 8 of infection. Statistical significance was determined using unpaired two-tailed t test where ns, not significant, *, P < 0.05, **, P < 0.005, ***, P < 0.0005. Data are representative of three independent experiments with three mice per time point.
Knell et al.
To determine if loss of Bim expression could rescue Id2KO CD8+ T cell effector survival and phenotypic development, we crossed Bim-deficient mice to the Id2KO line. We generated fetal liver chimeras reconstituted with OT-I transgenic wild type (Id2+Bim+), Bim-deficient (Id2+BimKO), Id2-deficient (Id2KOBim+) or Id2-deficient Bim-deficient (Id2KOBimKO) hematopoietic cells. Following reconstitution, we transferred these OT-I cells into congenically distinct recipients and infected with Lm-OVA. We analyzed the expansion of the transferred CD8+ T cell populations in recipient mice during the course of infection (Fig. 2B). As expected, the Id2KOBim+ OT-I T cells did not accumulate during infection and Id2+BimKO CD8+ T cells failed to contract to the same extent as wild type effector cells following the peak of infection (27). However, Id2KOBimKO CD8+ T cells expanded and contracted to a similar degree as Id2+Bim+ CD8+ T cells, with total cell numbers comparable between Id2+Bim+ and Id2KOBimKO cells throughout the course of infection (Fig. 2B and C). Thus, loss of Bim resulted in rescue of the accumulation defect by Id2KO CD8+ cells observed during infection, supporting the idea that Id2 expression impacts effector cell survival.
Defects in differentiation of KLRG1hi effector-memory cells by Id2KOBimKO CD8+ cells remain in spite of rescued survival
Next, we examined the effector and memory phenotypes of Id2KOBimKO CD8+ T cells in order to determine if the reduction in the short-term effector population in the absence of Id2 was due simply to a failure of those cells to survive. As above, we transferred CD45.1+ OT-I Id2+Bim+, Id2+BimKO, Id2KOBim+ or Id2KOBimKO cells to CD45.2+ congenic hosts and infected with Lm-OVA. Splenocytes were isolated on day 8 (Supp. Fig. 1) and day 15 (Fig. 3) of infection and cell-surface expression of phenotypic markers associated with effector and memory populations were analyzed by flow cytometry. We noted that although the frequency of donor cells was restored to Id2+ levels (Fig. 2), Id2KOBimKO CD8+ T cells displayed a profound defect in the generation of a KLRG1hiCD127lo population (Fig. 3). Id2KO and Id2KOBimKO CD8+ T cells also showed increased frequency of CXCR3+CD43lo cells compared to Id2+ (Fig. 3, Supp. Fig. 1), a phenotype associated with long-lived memory cells capable of a strong recall response (8). As we previously reported, the frequency of effector memory cells (CD44hiCD62Llo) was lower and CD27 was higher in Id2KO populations, and this was sustained in the Id2KOBimKO CD8+ T cells at both time points (Fig. 3, Supp. Fig. 1) (19). CXCR6 expression remained lower in Id2KOBimKO CD8+ T cells compared to wild type cells (Fig. 3, Supp. Fig. 1). Recently it has been reported that CD25, the high affinity IL-2 receptor α chain is associated with terminal-effector cell differentiation (7, 10). Thus, we analyzed CD25 expression on Id2-deficient cells on day four after infection and observed that Id2KO cells had lower expression of IL-2Rα relative to wild type cells congruent with their long-term memory phenotype, yet this phenotype was not rescued by loss of Bim (Fig. 3).
Figure 3. Bim-deficiency does not rescue phenotypic defects in Id2KO CD8+ T cells during infection.
Flow cytometric analysis of cell-surface phenotype of donor CD8+CD45.2+ splenocytes on day 15 of infection KLRG1, CD127, CD62L, CD44, CD27, CXCR3, CD43, CD122 (gated on Ly6C+ population). CD45.1+ C57BL/6 mice received OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO (CD45.2+) cells (2 × 104) 1 day before infection with Lm-OVA. CXCR6 expression was examined on day 9 and CD25 expression was examined on day 4 of infection.
Knell et al.
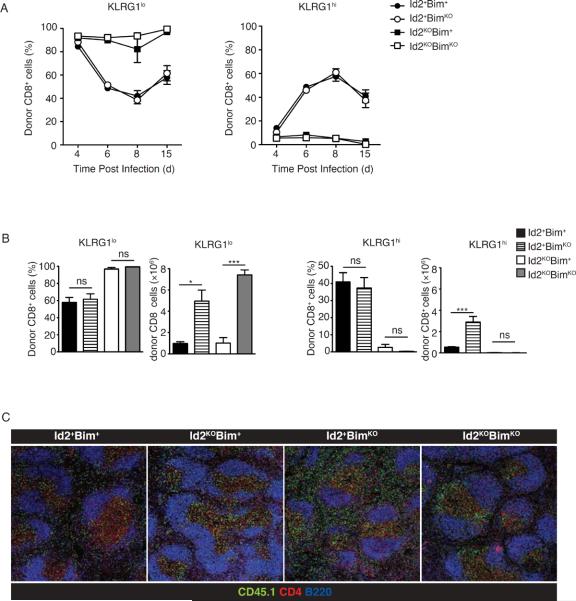
Examining peripheral blood on day 5 after infection, we noted that although Id2KOBim+ cells could upregulate KLRG1 expression early in the infection, the frequency of KLRG1hi Id2KOBim+ cells was reduced compared to wild type cells (Supp. Fig. 1A), suggesting that this population does not continue to differentiate or that these cells do not survive after their initial differentiation in the absence of Id2. Furthermore, the frequency of KLRG1hi cells in the absence of both Id2 and Bim was also reduced relative to wild type cells, indicating that although loss of Bim can rescue total OT-I cell frequency and number, it could not rescue the survival of KLRG1hi short-lived terminal effector cells (Supp. Fig. 1A). When we examined splenocytes throughout infection, we observed comparable expression of KLRG1 between Id2+Bim+ cells and Id2+BimKO cells or between Id2KOBim+ and Id2KOBimKO cells, once again indicating loss of Bim could rescue overall survival of Id2-deficient OT-I cells responding to infection, but Bim-deficiency did not restore the loss of KLRG1hi cell accumulation (Fig. 4A). Similarly, assessment of the total number of KLRG1hiCD127lo effector T cells revealed that loss of Bim expression, while rescuing cell survival, did not alter the loss of shorter-term effector-memory populations resulting from the absence of Id2 alone (Fig. 4B, Supp. Fig. 1).
Figure 4. Bim-deficiency does not restore KLRG1hi phenotype in Id2KO CD8+ T cells during infection.
(A) Percentage of KLRG1lo (left graph) or KLRG1hi (right graph) CD8+ OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO CD45.2 over the course of infection in spleen. CD45.1+ C57BL/6 mice received OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO (CD45.2+) cells (2 × 104) 1 day before infection with Lm-OVA, average (±SEM). Data are representative of one (day 4 and 6) to three (day 8 and 15) independent experiments, with three mice per timepoint. (B) Bar graphs summarizing average (±SEM) frequency of KLRG1hi or KLRG1lo donor cells among total donor CD8+ cells and total number of KLRG1hi and KLRG1lo donor OT-I donor cells recovered from spleen on day 15 of infection. (C) Confocal images of serial spleen sections of Id2+Bim+, Id2KOBim+, Id2+BimKO and Id2KOBimKO donor OT-I recipients on day 8 of Lm-OVA infection. Sections were stained for CD45.2, CD4 and B220. Images collected at 10× magnification. Data are representative of 2 independent experiments. Statistical significance was determined using unpaired two-tailed t test where ns, not significant, *, P < 0.05, **, P < 0.005, ***, P < 0.0005 relative to CD8+ OT-I Id2+Bim+ cells. Data are representative of three independent experiments, with three mice per timepoint.
Knell et al.
Due to the fact that Id2KO CD8+ cells lack a KLRG1hi short-lived effector population at the peak of infection, we also compared the phenotype between Id2+ and Id2KO cells gated only on the KLRG1lo population for days 8 and 15 of infection. The phenotypic differences observed in the entire CD8+ population (potentially including effector, effector-memory, and memory cells) were maintained in the KLRG1lo Id2+ and Id2KO populations, although these differences were less pronounced (Supp. Fig. 2A and B). Together, the data supported the more rapid emergence of the long-term memory phenotype among cells in the KLRG1lo subset when Id2 was absent, suggesting Id2 expression regulates gene expression and differentiation within this population.
Previous data show effector subsets, distinguished by KLRG1hi expression, localize to different areas of the spleen during infection (38). When we examined the CD8+ T cell localization in the spleens of recipient mice on day 15 of infection, we noticed that while wild type CD8+ T cells were diffusely located throughout the spleen, Id2-deficient CD8+ T cells were concentrated in the T cell zones (Fig. 4C), a phenotype associated with KLRG1lo longer-lived memory precursors (38). As expected, Id2+BimKO CD8+ T cells localized like wild type cells (Fig. 4C). Id2KOBimKO CD8+ T cells, however, were primarily found in the T cell zones, similar to Id2KOBim+ cells (Fig. 4C), again indicating that while loss of Bim rescues survival of Id2-deficient cells, it does not rescue their propensity for developing into shorter-lived effector-memory cells.
Id2-deficiency alters cytokine production by CD8+ T cells during infection
To assess the functional ability of Id2-deficient T cells in the presence or absence of Bim, we isolated Id2+Bim+, Id2+BimKO, Id2KOBim+ and Id2KOBimKO OT-I cells that had been transferred into recipient mice, which were then infected with Lm-OVA. As we previously observed, Id2KO cells produced IFNγ at equal levels compared to wild-type cells on both day 8 (Supp. Fig. 3B) and day 15 (Fig. 5B) of infection. Interestingly, Id2-deficient CD8+ T cells produced more IL-2 (Fig. 5A, Supp. 3A), a cytokine that has been associated with memory-precursor cells, consistent with their phenotypic alterations (9). CD8+ T cells lacking Id2 produced similar levels of TNFα compared to wild type cells (Fig. 5C, Supp. Fig. 3C). Loss of Bim alone had no effect on the cytokine profile of CD8+ T cells (Fig. 5). Cytokine production observed in the entire CD8+ population (potentially including effector, effector-memory, and memory cells) was maintained in the KLRG1lo Id2+ and Id2KO populations on both day 8 and 15 of infection (Supp. Fig. 4).
Figure 5. Id2-deficiency alters cytokine production by CD8+ T cells during infection.
Flow cytometric analysis of (A) IL-2 (B) IFNγ and (C) TNFα production by OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO (CD45.2+) splenocytes. CD45.1+ C57BL/6 mice received OT-I Id2+Bim+, BimKO, Id2KO or Id2KOBimKO (CD45.2+) cells (2 × 104) 1 day before infection with Lm-OVA. Splenocytes isolated on day 15 post infection were restimulated for 6 h in vitro with OVA peptide (solid line) or incubated in media alone (dotted line) followed by surface and intracellular staining. Bar graphs indicating average (±SEM) percentage of cytokine producing cells among donor cells. (D) E2A occupancy (blue peaks) and H3K4me1 occupancy (green peaks) in A12 cells overexpressing E47 as reported by Lin et al. (17). Immunoprecipitation of chromatin (with anti-HEB or IgG) isolated from purified OT-I Id2+ cells directly ex vivo or 24 h after in vitro activation with OVAp followed by quantitative PCR analysis of input or precipitated DNA for indicated E-box sites. Data are representative of three independent experiments.
Knell et al.
To examine if E proteins mediated a direct role in regulating cytokine production, we examined the IL-2, IFNγ and TNFα loci for enhanced E2A-binding occupancy using the previously described ChIP Seq profiles (36). Both IL-2 and TNFα contained an E box site capable of binding E proteins (Fig. 5D). There were no occupancy peaks present in the IFNγ locus. ChIP analysis showed minimal HEB binding in naïve CD8+ T cells. However, in CD8+ T cells activated for 24 hours in vitro with OVA peptide, we observed increased E protein binding at both the IL-2 and TNFα E box sites (Fig. 5D). These data indicate a possible role for direct E protein regulation of cytokines during T cell activation.
Altered gene expression by CD8+ effector cells in the absence of Id2
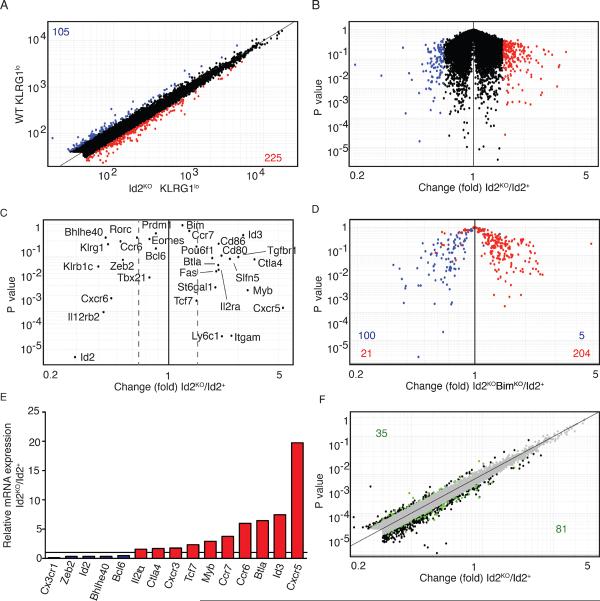
To understand further the role of Id2 during the CD8+ T cell immune response, we sorted KLRG1lo Id2+ and Id2KO OT-I CD8+ T cells on day 6 after Lm-OVA infection and compared their gene-expression profiles by microarray (Fig. 6). We observed that 225 genes were up-regulated ≥1.5-fold and 105 genes were down-regulated ≥1.5-fold, in the absence of Id2 compared to wild type cells (Fig. 6A and 6B). Interestingly, numerous genes associated with memory formation/function were up-regulated in Id2-deficient T cells, including Id3, Il2ra, tcf7, and cxcr3 (Fig. 6C), consistent with our phenotypic data. We also isolated Id2KOBimKO CD8+ T cells to compare the gene-expression profile of CD8+ T cells in the absence of Bim, with the goal of revealing a distinct profile that would implicate how Id2 promotes differentiation of shorter-lived effector-memory cells (Fig. 6D). We examined the expression of genes up- or down- regulated in Id2KOBimKO cells compared to Id2KO cells and observed that the expression pattern was remarkably similar (Fig. 6D). Genes down-regulated in Id2KO KLRG-1lo effectors (Fig. 6A, blue) were also down-regulated in Id2KOBimKO effectors compared to their Id2+ counterparts (Fig. 6D, blue) and vice versa (Fig. 6, red). Among the 5–10% of genes with expression that did not correlate between Id2KO and Id2KOBimKO effector cells, there was not significant inverse expression. Thus, a rescue of survival by Id2KO effectors did not reveal an altered expression pattern. We also examined the mRNA levels by qPCR analysis for Id2KO and Id2+ KLRG1lo cells separately isolated for a subset of genes of interest to CD8+ memory differentiation (Fig. 6E). These data confirmed the gene-expression profiles determined by microarray (Fig. 6E) and were consistent with a skewing of expression towards differentiation of cells with long-term memory potential. Last, we were interested if the genes that showed altered expression when Id2 was absent were potential targets of E protein transcription factors. Once again, we utilized published data that evaluated E2A occupancy in T cell lines by ChIP-Seq (36); those genes that we observed to be differentially regulated between Id2WT and Id2KO that were also identified as having significant E2A binding to putative regulatory elements are indicated in green (Fig. 6F). Notably, ~30–35% of the genes displaying altered expression due to loss of Id2 were also indicated as direct E protein targets. Together, the microarray analyses indicate that Id2 influences the gene-expression pattern in CD8+ T cells during infection and that this gene pattern is maintained irrespective of Bim expression and survival, further supporting the conclusion that Id2 plays a key role in differentiation of effector cells to short-term versus long-term effector and memory populations.
Figure 6. Gene-expression profile of Id2+ versus Id2KO KLRG1lo cells during infection.
Gene expression analysis of WT versus Id2KO effector cells. CD45.1.2+ mice received CD45.1 OT-I WT (4 × 104) cells mixed with CD45.2 OT-I BimKO, Id2KO or Id2KOBimKO (4 × 104) cells 1 day before infection with Lm-OVA. KLRG1hi and KLRG1lo cells were sorted on day 6. (A) Fold change plot of WT KLRG1lo versus Id2KO KLRG1lo CD8+ T cells. Numbers in corners indicate genes with a difference in expression of 1.7-fold or more. Genes upregulated in Id2KO KLRG1lo cells are indicated in red, genes downregulated are indicated in blue. Data are pooled from 3 independent experiments. (B) `Volcano' plot of expression data in (A). (C) Gene expression of key genes in WT KLRG1lo versus Id2KO KLRG1lo CD8+ T cells. Dotted lines indicate 1.5-fold difference. (D) Gene expression of genes 1.7 fold up- or downregulated in WT KLRG1lo versus Id2KOBimKO KLRG1lo CD8+ T cells plotted for wild-type versus Id2-knockout cells. Genes upregulated in Id2KOBimKO KLRG1lo CD8+ T cells are indicated in red, genes downregulated are indicated in blue. (E) mRNA expression of relevant genes in Id2KO compared to Id2+ KLRG1lo cells on day 6 of Lm-OVA infection. (F) Gene expression in WT KLRG1lo versus Id2KO KLRG1lo CD8+ T cells; green indicates E2A occupancy. Numbers in corners indicate number of genes with E2A occupancy.
Knell et al.
DISCUSSION
The factors controlling the survival of effector CD8+ T cells as they undergo contraction, as well as their differentiation to memory cells, has been the subject of intense scrutiny, with transcription factors, survival molecules and cytokines all being implicated in this process (1–3, 39). Of particular interest is the transcriptional regulation of differentiation leading to the eventual apoptosis of short-lived effector cells during the contraction phase versus the survival of memory-precursor cells that will seed the memory compartment. While the pro-apoptotic molecule Bim is a key regulator of contraction of the CD8+ effector population, the abundance of memory-precursor cells formed in the absence of Bim are eventually lost, implicating other factors in the regulation of CD8+ effector differentiation (31). In this study, we show the transcriptional regulator Id2 influences effector CD8+ T cells in two ways: by regulating KLRG1hi effector CD8+ T cell differentiation and survival of CD8+ effector T cells (Fig. 1 and 2). At all stages of infection, loss of Id2 resulted in impaired formation of the KLRG1hi short-lived effector cell population (Fig. 1 and Supp. Fig. 1A). We found that while loss of Bim led to rescue of the survival defect seen in the absence of Id2, Bim-deficiency failed to support formation of terminally-differentiated KLRG1hi effector cells by Id2KO CD8+ T cells (Fig. 3 and 4). Thus, by uncoupling Bim-mediated CD8+ effector contraction from differentiation and memory formation, we addressed the role of Id2 in these processes.
E protein transcription factors positively regulate expression of Bim; we previously showed upregulation of Bim in Id2KO effector CD8+ T cells, presumably due to unchecked E protein activity, led to defects in survival and subsequent memory formation (19). However, whether Id2 can also regulate CD8+ effector T cell differentiation, irrespective of cell survival, remained unaddressed. In the absence of Id2 alone, when increased E protein activity led to elevated Bim expression, both differentiation of short-lived KLRG1hi effector T cells and survival of effector cells were impacted (Fig. 1 and 2). Here, using mice genetically deficient for both Id2 and Bim, and thus removing the confounding effect of unchecked Bim expression, the CD8+ effector T cell survival phenotype was rescued (Fig. 2). Notably, loss of Bim alone rescued both the KLRG1hi and KLRGlo subsets when Id2 is expressed. However, differentiation of Id2-deficient CD8+ effector T cells into short-lived effector cells and long-lived memory-precursor cells was still perturbed when Bim was also absent, even thought the effector cells accumulated to wild type levels, indicating that E proteins together with Id proteins regulate genes necessary for CD8+ effector differentiation as well as genes necessary for survival. As loss of Bim does not favor KLRG1lo cells and gene expression of the Id2KOBim+ and Id2KOBimKO cells were the same, it does not appear likely that the rescued cells are developing aberrantly. We did, however, observe a small Id2KO KLRG1hi population at early time points (day 5, Supp. Fig. 1A), which was rapidly lost and not rescued to Id2WT levels by Bim deficiency. Thus, we suggest E proteins and Id proteins regulate not only the survival signals required for memory-precursor formation but also influence the gene-expression pattern required for the sustained differentiation of the terminal-effector population.
Aside from regulating Bim expression during CD8+ effector T cell contraction, how do E and Id proteins influence effector T cell differentiation? Our previous data indicate Id2 expression leads to development of short-lived effector cells, while Id3 expression marks cells that will contribute to the memory-precursor pool (25). Although microarray analysis revealed upregulation of Id3 expression in Id2-deficient T cells (Fig. 5C and E), Id3 was unable to compensate for either the differentiation or survival defect we observed with loss of Id2. These data support our hypothesis, that high Id3 expression marks cells for memory-precursor differentiation and is consistent with our previous data (25). Furthermore, these data imply Id2 and Id3 may have different binding affinities for E proteins or that certain E/Id proteins are expressed at different stages during effector T cell differentiation, thus providing a level of `fine-tuning' of the CD8+ effector immune response not previously appreciated. Increased expression of Id3 in the absence of Id2 also implies direct or indirect negative regulation of Id3 by Id2, a possibility requiring further investigation.
In our gene-expression analysis, we also observed upregulation of other genes previously associated with memory CD8+ T cell formation including Cxcr3, Tcf7 and Cxcr5 (Fig. 5C and E). These genes were upregulated in the absence of Id2 and irrespective of Bim expression, thus implicating unchecked E protein activity in directing CD8+ T cells toward a KLRG1lo CD127hi memory-precursor lineage during infection. As expected, Id2-deficient CD8+ effector T cells expressed high levels of Bim and Fas, both pro-apoptotic molecules (Fig. 5C). With loss of Bim in conjunction with loss of Id2, the T cell-differentiation gene-expression pattern was remarkably similar to that of Id2-deficient cells (Fig. 5D). Thus, unchecked E protein regulation of Bim expression, normally inhibited by Id2, does not appear to be intricately linked to regulation of genes associated with CD8+ effector T cell differentiation.
Recent publications have implicated various transcriptional regulators in the control of the effector T cell differentiation during infection. Here, we explored the role of Id2 in both surviving the contraction phase of the CD8+ T cell immune response and the initiation of the gene-expression profile required for short-lived effector and long-lived memory-precursor differentiation. We find E and Id proteins work together to regulate both the survival and differentiation of CD8+ T cells during the immune response.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Goldrath lab members for advice and critical review of the manuscript.
This work was supported by Cancer Research Institute, Pew Scholar Award, Leukemia and Lymphoma Society and grants from the NIH (AI067545 and AI072117) to A.W.G.; Leukemia and Lymphoma Society to L.M.D.
REFERENCES
- 1.Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 2.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joshi NS, Kaech SM. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J Immunol. 2008;180:1309–1315. doi: 10.4049/jimmunol.180.3.1309. [DOI] [PubMed] [Google Scholar]
- 4.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 2002;111:837–851. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 6.Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 2010;32:79–90. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hikono H, Kohlmeier JE, Takamura S, Wittmer ST, Roberts AD, Woodland DL. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J Exp Med. 2007;204:1625–1636. doi: 10.1084/jem.20070322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J Exp Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100:15818–15823. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 19.Cannarile MA, Lind NA, Rivera R, Sheridan AD, Camfield KA, Wu BB, Cheung KP, Ding Z, Goldrath AW. Transcriptional regulator Id2 mediates CD8+ T cell immunity. Nat Immunol. 2006;7:1317–1325. doi: 10.1038/ni1403. [DOI] [PubMed] [Google Scholar]
- 20.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, Wang E, Schrump DS, Marincola FM, Restifo NP, Gattinoni L. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegel AM, Heimall J, Freeman AF, Hsu AP, Brittain E, Brenchley JM, Douek DC, Fahle GH, Cohen JI, Holland SM, Milner JD. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 2011;35:806–818. doi: 10.1016/j.immuni.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 24.Ji Y, Pos Z, Rao M, Klebanoff CA, Yu Z, Sukumar M, Reger RN, Palmer DC, Borman ZA, Muranski P, Wang E, Schrump DS, Marincola FM, Restifo NP, Gattinoni L. Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8(+) T cells. Nat Immunol. 2011;12:1230–1237. doi: 10.1038/ni.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D'Cruz LM, Watowich SS, Murre C, Goldrath AW. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8(+) T cell subsets. Nat Immunol. 2011;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strasser A, Pellegrini M. T-lymphocyte death during shutdown of an immune response. Trends Immunol. 2004;25:610–615. doi: 10.1016/j.it.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Pellegrini M, Belz G, Bouillet P, Strasser A. Shutdown of an acute T cell immune response to viral infection is mediated by the proapoptotic Bcl-2 homology 3-only protein Bim. Proc Natl Acad Sci U S A. 2003;100:14175–14180. doi: 10.1073/pnas.2336198100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–545. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prlic M, Bevan MJ. Exploring regulatory mechanisms of CD8+ T cell contraction. Proc Natl Acad Sci U S A. 2008;105:16689–16694. doi: 10.1073/pnas.0808997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weant AE, Michalek RD, Crump KE, Liu C, Konopitski AP, Grayson JM. Defects in apoptosis increase memory CD8+ T cells following infection of Bim−/−Faslpr/lpr mice. Cell Immunol. 2011;271:256–266. doi: 10.1016/j.cellimm.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wojciechowski S, Jordan MB, Zhu Y, White J, Zajac AJ, Hildeman DA. Bim mediates apoptosis of CD127(lo) effector T cells and limits T cell memory. Eur J Immunol. 2006;36:1694–1706. doi: 10.1002/eji.200635897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 33.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 34.D'Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nat Immunol. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monticelli LA, Yang Y, Knell J, D'Cruz LM, Cannarile MA, Engel I, Kronenberg M, Goldrath AW. Transcriptional regulator Id2 controls survival of hepatic NKT cells. Proc Natl Acad Sci U S A. 2009;106:19461–19466. doi: 10.1073/pnas.0908249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung YW, Rutishauser RL, Joshi NS, Haberman AM, Kaech SM. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J Immunol. 2010;185:5315–5325. doi: 10.4049/jimmunol.1001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Cruz LM, Rubinstein MP, Goldrath AW. Surviving the crash: transitioning from effector to memory CD8+ T cell. Semin Immunol. 2009;21:92–98. doi: 10.1016/j.smim.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.