Summary
Influenza A virus is unique as an RNA virus in that it replicates in the nucleus and undergoes splicing. With only ten major proteins, the virus must gain nuclear access, replicate, assemble progeny virions in the cytoplasm and then egress. In an effort to elucidate the coordination of these events, we manipulated the transcript levels from the bicistronic NS segment that encodes the spliced virus product responsible for genomic nuclear export. We find that utilization of an erroneous splice site ensures the slow accumulation of the viral Nuclear Export Protein (NEP) while generating excessive levels of an antagonist designed to inhibit the cellular response to infection. Modulation of this simple transcriptional event results in improperly timed export and loss of virus infection. Together, these data demonstrate that coordination of the influenza A virus life cycle is set by a ‘molecular timer’ that operates on the inefficient splicing of a virus transcript.
Introduction
Influenza A virus (IAV) is a negative sense, nuclear RNA virus comprised of eight viral segments encoding ten major proteins (Palese and Shaw, 2006). Despite its simplicity, IAV manages to usurp the necessary host components required for replication and coordinates both its nuclear import and export to coincide with virus replication and egress, respectively. Segment eight of IAV is one of two viral RNAs that undergoes splicing and thus encodes two proteins, non-structural protein 1 (NS1) and NS2 (herein referred to as the nuclear export protein (NEP)) (Lamb and Lai, 1980; O’Neill et al., 1998). IAV initiates transcription of the NS segment as a continuous product to generate NS1, whereas a weak 5′ splice site results in the second, less abundant splice product, NEP, which accounts for ~10–15% of NS-derived mRNA (Agris et al., 1989; Robb et al., 2010).
NS1 represents the dominant virus countermeasure to antagonize the cellular response to infection (Hale et al., 2008). NS1 has been described to inhibit a broad range of cellular functions as well as interfere with host mRNA export and splicing (Hale et al., 2008). The host antagonism function of NS1 was perhaps best illustrated by studies which found that IAV lacking NS1 was attenuated by more than four logs in vivo but maintained wild type virulence in mice lacking antiviral signaling capability (Egorov et al., 1998; Garcia-Sastre et al., 1998).
Unlike NS1, NEP performs a fundamental role directly pertaining to the virus life cycle. NEP is a 14kD protein, composed of 121 amino acids, and is necessary for the production of replication-competent virus (Neumann et al., 2000). The critical function of NEP is postulated to be in the nuclear export of nucleoprotein-associated (NP) vRNAs (vRNPs) (Boulo et al., 2007; Cros and Palese, 2003; O’Neill et al., 1998). NEP-mediated export of vRNPs is orchestrated by its association with the M1 matrix protein, which directly associates with the NP bound vRNA (Akarsu et al., 2003; Huang et al., 2001; Shimizu et al., 2011; Yasuda et al., 1993; Ye et al., 1999). Disruption of the nuclear export signal of NEP results in a complete loss of virus replication (Iwatsuki-Horimoto et al., 2004; Neumann et al., 2000). In addition to its role in vRNP export, NEP has also been implicated in controlling the level of complementary vRNA (cRNA), in determining host tropism, and in the synthesis of IAV-specific small virus RNA (svRNA) (Manz et al., 2012; Perez et al., 2010; Perez et al., 2012; Robb et al., 2009).
In this report we sought to define the interplay between the products of the non-structural (NS) segment to determine whether the balance of NS1 and NEP are critical for coordinating virus infection. To achieve this, we used microRNA- (miRNA) targeting to silence NS1 without impacting NEP and other modifications to generate viruses in which NEP levels were either elevated or reduced independent of NS1. Surprisingly, we found that silencing NS1 by greater than 90% does not impair the virus’s ability to replicate and incapacitate the cell’s antiviral response. In contrast, modulating the levels of NEP resulted in virus attenuation as a result of improperly timed vRNP export. Taken together, these data illustrate how IAV utilizes an inefficient splice site to slowly accumulate NEP as a by-product of transcribing NS1 and implicates this process as a ‘molecular timer’ that coordinates virus infection.
Results
Selective silencing of NS1 through miRNA-mediated targeting
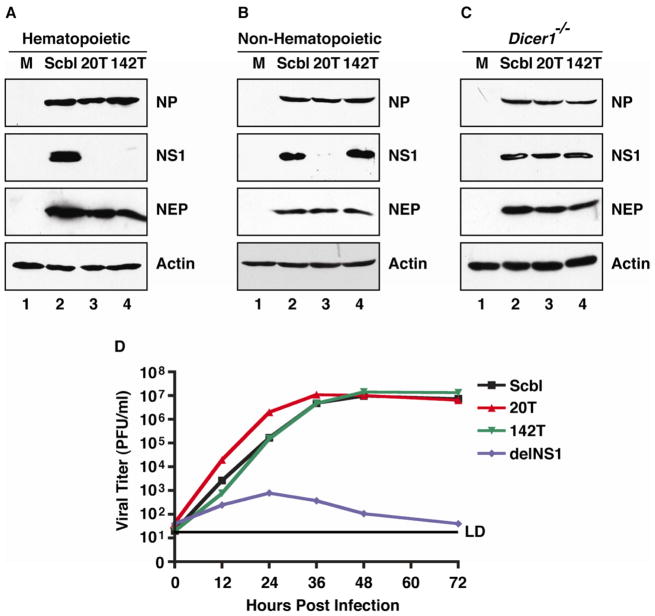
In an effort to selectively target NS1 without impacting the levels of NEP, we chose to harness the activity of endogenous miRNAs, a technique previously demonstrated to be successful for IAV (Langlois et al., 2012; Perez et al., 2009; Varble et al., 2010). For this purpose, we chose to exploit the ubiquitous expression of miR-20 to silence NS1 during virus infection. In addition, we engineered a second virus in which NS1 was targeted by the cell-specific expression of miR-142, which would limit expression only in hematopoietic cells (Langlois et al., 2012). Expression patterns of miR-20 and miR-142 have been described elsewhere but were independently verified for the relevant tissues and cells used in this study (Figure S1A) (Landgraf et al., 2007). To silence NS1, we inserted a scrambled (Scbl) sequence or tandem targets (T) of miR-20 or miR-142 into the 3′ untranslated region (UTR) of a modified NS viral vector (Figure S1B) (Varble et al., 2010). Perfect complementarity to either miRNA conferred sequence-specific silencing of NS1 at both a protein and mRNA level (Figure S1C). Plasmid-based rescue using each of these constructs resulted in the generation of three isogenic viruses (herein referred to as Scbl, 20T, and 142T) differing only in the NS1 3′ UTR. In order to confirm NS1 silencing of the recombinant IAV strains, we infected cells expressing different miRNA profiles. In hematopoietic cells, expressing high levels of both miR-20 and miR-142, silencing of NS1 was robust, generating no detectable NS1 at eight hours following infection with either 20T or 142T IAV strains (Figure 1A). In contrast to hematopoietic cells, infection of non-hematopoietic lineages demonstrated NS1 silencing only in response to the 20T strain (Figure 1B). Silencing of NS1 was confirmed to be miRNA-specific as virus infection in cells lacking Dicer1 yielded comparable levels of NS1 between Scbl, 20T, and 142T infections (Figure 1C). Moreover, these data demonstrate that independent of NS1 targeting, NP and NEP levels remain unaffected regardless of cell type or miRNA expression (Figure 1A–C). Taken together, we conclude that the engineered NS segment can be used to successfully target NS1 by host miRNAs without impacting NEP expression in the context of a replication-competent IAV.
Figure 1. Silencing of NS1 does not impact virus replication in vitro.
(A-C) Western blot analysis of whole cell extract (WCE) derived from (A) Jurkat T cells, (B) murine embryonic fibroblasts (MEFs), and (C) MEFs deficient in Dicer1 (Dicer1 −/−) infected with Scbl, 20T, and 142T viruses. Immunoblots depict NP, NS1, NEP, and Actin. (D) Multicycle growth curves performed in A549 cells in response to Scbl, 20T, 142T, and delNS1 IAV strains. Error bars represent average ± SD of the mean of samples from independent growth curves analyzed in triplicate. LD indicates limit of detection. See also Figure S1.
Silencing of NS1 does not impact virus replication in vitro
In an effort to characterize the consequences of NS1 silencing as it pertained to virus replication and the host response, we performed multicycle growth curves on the three recombinant viruses. Non-hematopoietic lung epithelial cells were treated with Scbl, 20T or 142T IAV strains or an IAV lacking NS1 (delNS1). Assaying for virus replication demonstrated that delNS1 infection was substantially inhibited whereas Scbl, 20T and 142T strains all accumulated to high levels of ~10E7 plaque forming units (pfu) per milliliter (Figure 1D). To ascertain the extent of NS1 silencing and the cellular response to virus infection, we analyzed both IAV expression and induction of the IFN-I-dependent transcript, Mx1 (Haller et al., 1979; Staeheli et al., 1986). In agreement with published reports, infection of the delNS1 virus demonstrated no NS1 and a dramatic decrease in both NP and NEP levels (Figure S1D) (Garcia-Sastre et al., 1998). Furthermore, complete deletion of NS1 resulted in elevated induction of Mx1. In contrast to delNS1, infection of the 20T IAV strain revealed a significant decrease in NS1 but no change in the expression of NP, NEP, or Mx1 suggesting that low levels of NS1 are sufficient to antagonize the cellular response in vitro.
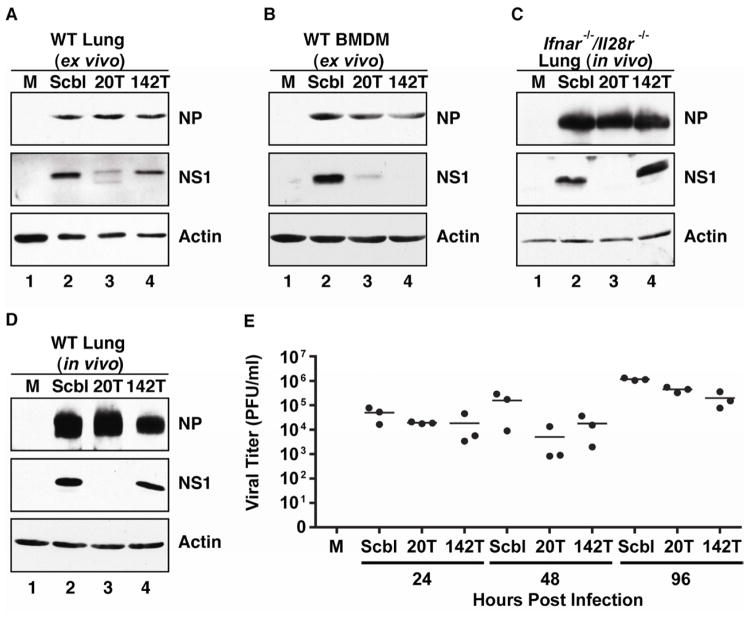
Given the efficient nature of viruses, we reasoned that there must be a biological requirement for sustained expression of NS1. One hypothesis for the lack of an in vitro phenotype is that low level production of NS1 was sufficient in immortalized cells whereas sustained and elevated levels may be necessary in primary cultures where interferon signaling is unperturbed (Stojdl et al., 2003). To test this hypothesis, we characterized Scbl, 20T, and 142T IAV strains in primary lung fibroblasts (pLFs) and bone marrow-derived macrophages (BMDMs) (Figure 2A, B). Infection of pLFs, which demonstrate robust levels of miR-20 and low levels of miR-142 (a possible result of resident macrophages), showed no substantial difference in the expression of NP amongst IAV strains (Figure 2A). However, in agreement with our in vitro data, NS1 levels in lung fibroblast cultures were inversely proportional to endogenous miRNA levels. Furthermore, comparable experiments in BMDMs also showed a substantial loss of NS1 without impacting NP in both 20T and 142T infections (Figure 2B). Together, these data corroborate our in vitro findings that low levels of NS1 expression are sufficient to allow for optimal replication.
Figure 2. Low levels of NS1 are sufficient for virus replication in vivo.
Western blot analysis of WCE from (A) primary lung fibroblasts (pLFs) or (B) bone marrow-derived macrophages (BMDMs) from wild type (WT) C57BL/6 mice infected with Scbl, 20T, or 142T IAV or mock (M) treated for 24 hours. Western blot analysis of (C) Ifnar1−/−/Il28r−/− or (D) WT mouse lungs infected intranasally with either Scbl, 20T, or 142T IAV or mock (M) treated at 48 hpi. Immunoblots depict NP, NS1, and Actin. (E) Viral titers from WT C57BL/6 mice infected with Scbl, 20T, 142T or mock (PBS) treated. Data points represent individual mice assayed in triplicate. See also Figure S2.
To ascertain whether the dramatic loss of NS1 impacted the course of an in vivo virus infection, we compared Scbl, 20T, and 142T IAV strains in the context of an animal model. To first ensure that NS1 targeting was robust in vivo, we infected mice devoid of interferon signaling, in which loss of NS1 function would be of little consequence, and analyzed whole lung for viral protein expression (Figure 2C). In agreement with both in vitro and ex vivo data, silencing of the 20T virus was robust, demonstrating a complete loss of detectable NS1 while having little, to no impact on NP expression. Successful silencing of NS1 in vivo could also be corroborated in wild type interferon competent mice (Figure 2D). Having confirmed in vivo targeting, we administered a sublethal dose of Scbl, 20T, and 142T IAV strains to wild type mice to determine the host response to infection. Surprisingly, the viruses demonstrated no substantial difference in overall virus titers, reaching levels between 10E5–10E6 pfu/mL (Figure 2E). Similarly, the host response, as measured by two interferon regulated genes, IFN Stimulated Gene 15 (ISG15) and IFN Regulatory Factor 7 (IRF7), demonstrated no substantial difference across viral cohorts (Figure S2A, B). These results suggest that low levels of NS1 are sufficient to antagonize the cellular response to infection and permit high levels of virus replication in vivo.
NEP accumulation defines virus replication levels
In an effort to understand the molecular basis for sustained NS transcription, we investigated the properties of the less abundant transcript of segment eight, NEP. To determine whether the intracellular concentration of NEP demonstrated comparable flexibility with regards to function as that observed for NS1, we monitored virus replication in model systems that either decreased or enhanced NEP levels. First, we utilized a recombinant virus in which NEP expression was controlled by a porcine teschovirus-1 2A (PTV-1 2A) ribosome ‘stop-carry on’ recoding site rather than being a spliced product of NS1 (Figure S3A) (Manicassamy et al., 2010). This virus, herein referred to as IAV 2A, while producing sufficient levels of NS1 (fused to GFP), demonstrated a significant decrease in NEP (Figure 3A). Consequently, IAV 2A virus, when compared to a wild type parental strain, resulted in a two log decrease of progeny virions, suggesting that insufficient NEP impaired replication despite normal protein synthesis of structural proteins such as NP (Figure 3A and 3B). To ensure the 2A phenotype was a result of decreased NEP expression, we also verified the impact on virus titers in the context of a wild type infection treated with short interfering RNAs (siRNAs) against NEP. To this end, we used two siRNAs with differing NEP silencing capacities (NEP1 and NEP2) to reduce, but not eliminate, NEP expression during IAV infection (Figure S3B). Comparable to the replication defect of IAV 2A, wild type IAV infections treated with siRNAs against NEP resulted in a significant decrease in viral titers, demonstrating a one log attenuation at 48 hpi (p=0.01) (Figure S3C).
Figure 3. NEP is essential in determining the levels of virus replication.
(A) Western blot analysis of WCE from fibroblasts infected with either WT or 2A viruses or mock treated for 3, 6, 9, or 12 hours post infection (HPI). (B) Multicycle growth curve for WT and 2A IAV. (C) Western blot analysis of WCE from fibroblasts infected with IAV-like vectors (VLVs) containing either an extragenic scrambled (Scbl) sequence or NEP on segment four. (D) Multicycle growth curve of WT IAV coinfected with VLVs characterized in (C). (A and C) Immunoblots depict NP, NS1, NEP, and Actin. (B and D) Error bars represent mean ± SD of the mean of samples from independent multicycle growth curve analyzed in triplicate. LD indicates limit of detection. See also Figure S3.
To ascertain whether excess NEP was also detrimental to overall virus replication, we designed a replication-incompetent IAV-like vector (VLV) that encoded an additional copy of NEP (VLV NEP) (Figure S3D). As a negative control, we also generated a VLV that expressed a scrambled non-coding RNA (VLV Scbl) (Figure S3D). VLVs expressing the extragenomic NEP and Scbl segments were grafted into segment four (which normally encodes HA) and propagated in HA expressing cells (Figure S3D). VLVs grew to high titers and could be used to transduce cells to express all of the IAV genes minus HA in addition to either the non coding RNA (VLV Scbl) or high levels of NEP (VLV NEP). Treatment of cells with the VLVs resulted in robust levels of NP and NS1 within 24 hrs and, in response to VLV NEP, high levels of NEP (Figure 3C). Given the nature of IAV to exchange segments through antigenic shift, we then performed an infection at a high multiplicity of infection mixing either VLV Scbl or NEP with wild type virus. Monitoring replication at 12hr intervals for 48hrs demonstrated that, while treatment with VLV Scbl permitted sustained levels of virus replication, an overabundance of NEP resulted in a two log attenuation (Figure 3D). Taken together, these results suggest that the levels of NEP are critical in determining the success of virus replication.
NEP splicing efficiency is essential in coordinating vRNP export
Given the critical nature of NEP to virus replication, we next investigated whether it was the NS splice site itself that was critical for achieving optimal NEP levels. To this end, we optimized the weak NEP 5′ splice site to create a transcript in which NS1 would be a minor ‘read-through’ product of the more dominant NEP transcript (herein referred to as NS optimal or NSo). As optimization resulted in a D12S amino acid substitution, we additionally produced a parental NS (NSp) IAV that carries this amino acid change while maintaining the endogenous, non-optimal, 5′ splice site. To confirm NSo had an inverse expression profile of NS1:NEP than that of NSp, we performed a single-cycle growth assay and determined protein levels by standard western blot (Figure 4A). As anticipated, NSo virus demonstrated a dramatic reduction in NS1 expression, with a corresponding increase in NEP, confirming that the quality of the NS splice site controls levels of NS1 and NEP. Proper splicing was also confirmed by RT-PCR (Figure S4A).
Figure 4. NEP accumulation is essential in coordinating virus replication.
(A) Western blot analysis of WCE from A549 cells infected with an NS encoding an optimal 5′ splice site (NSo), a parental splice NS (NSp) IAV, or mock treated for 3, 6, 9, or 12 hours post infection (HPI). Immunoblots depict NP, NS1, NEP, and Actin. (B) Multicycle growth curves for NSo and NSp IAV in A549 cells. (C) Same as (B) but performed in MDCK-NS1-GFP cells. Error bars represent mean ± SD of the mean of samples from independent multicycle growth curve analyzed in triplicate. LD indicates limit of detection. (D) Viral titers from WT C57BL/6 mouse lungs infected intranasally with NSo or NSp. (E) A549 cells stained for NP expression from NSo and NSp infection at 5 or 7 hours post infection and imaged by fluorescence microscopy. Hoechst dye used to visualize nuclei. See also Figure S4.
To ascertain whether the elevated NEP expression would result in a similar phenotype as the addition of the IAV NEP vector, we performed multicycle growth curves comparing NSo to NSp. As the levels of NS1 produced by NSo were comparable to the 20T virus in the presence of miR-20 (Figure S4B), we reasoned that any phenotypic differences between NSo and NSp could be attributed to NEP overproduction. In agreement with the NEP overexpression data, NSo replication was significantly attenuated, with virus titers demonstrating a two log reduction as compared to NSp virus (Figure 4B). To ensure this effect was indeed independent of NS1, we repeated the experiment in a NS1 complementing cell line, which also permits replication of delNS1 IAV (Figure S4C) (Kochs et al., 2009). Multicycle growth curves performed in the presence of NS1, demonstrated comparable results to unmodified cells, suggesting that the defect in NSo replication was directly the result of excess NEP (Figure 4C). Functional levels of NS1 from the NSo virus could also be confirmed through a lack of Mx1 induction (Figure S4D). Furthermore, NSo virus demonstrated a dramatic loss of replication in vivo, never accumulating to titers that exceeded 100pfu/mL. (Figure 4D). Infection was confirmed by qRT-PCR for viral NP levels (Figure S4E). Taken together, these data strongly suggest that the accumulation of NEP is dictated by the splice site of NS and is critical for proper IAV replication.
NEP is required for coordinating vRNP export
As NEP contributes to vRNP export during IAV infection (O’Neill et al., 1998), we hypothesized that the replication phenotypes of the NSo and IAV 2A viruses may be the result of improper timing of vRNP export. To test this hypothesis, localization of NP, an essential component of vRNP complexes, was monitored in lung epithelial cells. These results found that increasing NEP production through optimization of the 5′ splice site resulted in premature vRNP export, as NP staining could be observed as early as 5 hpi in response to NSo, whereas NSp NP staining, remained nuclear (Figure 4E). At 7 hpi, NSp infection began to show signs of vRNP export as ~50% of the infected cells displayed NP staining in the cytoplasm. In contrast, NSo at the 7 hpi time point demonstrated no cells in which IAV replication was exclusively nuclear (Figure 4E). In contrast, the decreased NEP production of the IAV 2A virus resulted in a significant delay in vRNP export when compared to wild type virus (Figure S4F). These results support the hypothesis that NEP accumulation alters the localization of vRNPs and implicates NEP in orchestrating the timing of IAV infection.
Discussion
Here we describe five complementing strategies to probe the molecular contributions of NS1 and NEP during virus infection. We exploited the endogenous expression of a ubiquitous miRNA to selectively target NS1, we used siRNAs to lower NEP expression, overexpressed NEP by providing it as an extragenic segment, decreased NEP expression by coupling it to an inefficient ribosomal re-initiation site, and modified the 5′ splice site to enhance expression of NEP at the expense of NS1. Surprisingly, these strategies all demonstrated that NEP accumulation plays a more substantial role during in vivo virus infection than the sustained expression of the IAV interferon antagonist protein, NS1. Given the limited coding capacity of IAV and the use of only a single promoter on each of the virus segments, usurping of the splicing machinery provides a clever method for the virus to control an aspect of its life cycle. Through the utilization of a poor 5′ splice site, the virus generates a ‘molecular timer’, which allows for the slow accumulation of NEP despite having comparable promoters on each of the eight segments. As the infection progresses, the virus must switch from transcription to replication; an event likely triggered by this accumulation (Perez et al., 2010; Robb et al., 2009). The data presented here on the manipulation of NEP levels certainly corroborates that deregulated NEP results in aberrant viral replication.
Lastly, it is noteworthy that splicing has also been implicated in maintaining the stoichiometric balance of retrovirus components during infection, although these examples do not extend to modulating the virus life cycle directly (Katz et al., 1988). Given that IAV proteins also need to be maintained in a stoichiometric balance, it was surprising to discover that NS1 is overproduced by an order of magnitude, at least late in infection, with regards to functionality. This misutilization of viral resources is not an error; rather, we show that this activity is essential for the accumulation of NEP to allow the proper timing of virus replication. In this regard, to achieve the slow accumulation of NEP, the virus needs to overproduce one of its own transcripts. Therefore, as overabundance of any of the polymerase, NP, or receptor components would alter replication, it is perhaps best suited that NEP accumulation be coupled to NS1 synthesis, where excess message during infection is inconsequential (or even beneficial). Taken together, the virus has exploited the predictable nature of the cell to establish a ‘molecular timer’ and successfully coordinate the processes of infection.
Experimental Procedures
Virus design
The modified NS segments (A/PR/8/34) have been described previously (Manicassamy et al., 2010; Varble et al., 2010). NS1/NEP-miR20T virus was generated by inserting four perfect complementary miR-20a target sites into the NS1/NEP intergenic region. NSo and NSP were generated by site directed mutagenesis (NSo mutations: G34A and A35G and NSp mutations: G34U and A35C). IAV-like vectors (VLVs) were also described previously (Marsh et al., 2007). In brief, fibroblasts were transfected with seven bi-directional plasmids (encoding-PB2, -PB1, -PA, -NP, -NA, -M, and –NS as well as the corresponding vRNA segments). In addition, a plasmid expressing the packaging sequence for segment four that encoded either a scrambled RNA or NEP were also included. This strategy has been described elsewhere (Gao et al. 2012). Transfected cells were co-cultured with HA-MDCK cells (Marsh et al., 2007). VLVs were propagated and titered on HA-MDCK cells.
Supplementary Material
Figure S1. miRNA-mediated targeting of NS1, Related to Figure 1. (A) Northern blot analysis of total RNA probed for miR-20 and miR-142 in indicated cell lines. U6 levels used as a loading control. 293, human embryonic kidney cells; 293 + miR-142, 293 cells transfected with a vector expressing miR-142; A549, human alveolar epithelial cells; Jurkat, human T lymphocyte; Dicer1−/−, Dicer1 knockout mouse embryonic fibroblasts; MEF, mouse embryonic fibroblasts; pLF, primary lung murine fibroblast; BMDM, bone marrow–derived murine macrophages. (B) Diagram of recombinant NS segment containing target sites in the 3′ untranslated region (UTR). Target sites are either a scrambled sequence (Scbl) or four identical target sites for miR-20a (20T) or miR-142 (142T). T indicates position of target sites. (C) (Top two panels) Western blots of WCE derived from fibroblasts expressing Scbl, 20T, or 142T vectors with either empty vector or vector producing miR-142 (miR-142). (Middle two panels) Northern blot of total RNA from samples above, probed for miR-142 and U6. (Bottom two panels) Semi-quantitative RT-PCR on total oligo-dT RNA using NS1- and beta tubulin specific primers. (D) Western blot analysis of WCE derived from samples in Figure 1D. Immunoblots depict Mx1, NP, NS1, NEP, or Actin.
Figure S2. Silencing of NS1 does not impact ISG15 or IRF7 levels in vivo, Related to Figure 2. qRT-PCR of (A) ISG15 and (B) IRF7 from mouse lungs from Figure 2E normalized to tubulin. Data points represent individual mice assayed in triplicate.
Figure S3. NS1/NEP 2A IAV, siRNAs, and VLVs, Related to Figure 3. (A) Diagram of segment 8 of NS1/NEP 2A IAV which is a non-splicing segment 8 expressing NS1-NEP as a fusion separated by a porcine teschovirus-1 (PTV-1) 2A sequence. (B) Western blot analysis of WCE from fibroblast transfected with different NEP siRNAs or scbl siRNA for 16 hours then infected with WT IAV for 8 hours. Immunoblots depict NP, NS1, NEP, or Actin. (C) Multicycle growth curves of WT IAV infection for 0, 12, 24, 36, and 48 hpi in fibroblast cells transfected with NEP1/2 siRNA or scbl siRNA for 16 hours prior to infection. P-values for 24, 36, and 48 hrs are 0.01, 0.001, and 0.01 respectively. (D) Diagram of how VLVs are constructed. In brief, wt segments 1,2,3,5,6,7, and 8 were combined with a recombinant segment 4 expressing either NEP or scrambled (Scbl) RNA. These eight segment are used to generate virus-like vectors (VLVs) in a MDCK cell line that complements the loss of wild type segment four by expressing exogenous HA protein.
Figure S4. Characterization of the NSo virus, Related to Figure 4. (A) Semi-quantitative RT-PCR on total oligo-dT RNA from fibroblast infected with WT, NSo, or NSp viruses for 3, 6, or 9 hpi. cDNA treated using primers to amplify NS1 and NEP as depicted. Tubulin was used as a loading control. (B) Western blot analysis of WCE from A549 cells infected with NSp, NSo, and 20T IAV strains or mock treated for 8 hours. (C) Western blot analysis of WCE from MDCK and MDCK-NS1-GFP cells infected with delNS1 virus for 24 hours. (D) Western blot analysis of WCE derived from samples in Figure 4B. Immunoblots depict Mx1, NP, NS1, or Actin. (E) qRT-PCR of viral NP levels from mouse lungs in Figure 4D normalized to tubulin. Data points represent individual mice assayed in triplicate. (F) A549 cells were fixed following infection with either WT or 2A IAV at 8 hpi. Cells were stained with the NP antibody prior to imaging by fluorescence microscopy. Hoechst dye used to visualize nuclei.
Highlights.
Demonstrates how a bicistronic gene can influence biological circuits
Characterizes potency of innate immune antagonism in vivo
Demonstrates how host factors can contribute to the timing of a virus life cycle
First example implicating NEP accumulation in the timing of IAV infection
Acknowledgments
This work is supported in part by the NIH, grant numbers A1093571 and A1080624 from the NIAID. J.T.P. and R.A.L. are supported by the MSSM Hematopoietic T32 training grant (T32HL094283). B.R.t. is supported in part by the Pew Charitable Trust and the Burroughs Wellcome Fund. We would like to thank Peter Staeheli and Georg Kochs (IMMH, Freiburg, Germany) for the Ifnar1−/−/Il28r−/− mice and for the Mx1 antibody, and Peter Palese (MSSM, NYU) for the recombinant segment four used for VLV construction and the MDCK-HA cells required for VLV construction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agris CH, Nemeroff ME, Krug RM. A block in mammalian splicing occurring after formation of large complexes containing U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins. Mol Cell Biol. 1989;9:259–267. doi: 10.1128/mcb.9.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akarsu H, Burmeister WP, Petosa C, Petit I, Muller CW, Ruigrok RW, Baudin F. Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2) EMBO J. 2003;22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulo S, Akarsu H, Ruigrok RW, Baudin F. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 2007;124:12–21. doi: 10.1016/j.virusres.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Cros JF, Palese P. Trafficking of viral genomic RNA into and out of the nucleus: influenza, Thogoto and Borna disease viruses. Virus Res. 2003;95:3–12. doi: 10.1016/s0168-1702(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Egorov A, Brandt S, Sereinig S, Romanova J, Ferko B, Katinger D, Grassauer A, Alexandrova G, Katinger H, Muster T. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J Virol. 1998;72:6437–6441. doi: 10.1128/jvi.72.8.6437-6441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chou YY, Doganay S, Vafabakhsh R, Ha T, Palese P. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J Virol. 2012;86:7043–7051. doi: 10.1128/JVI.00662-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- Haller O, Arnheiter H, Gresser I, Lindenmann J. Genetically determined, interferon-dependent resistance to influenza virus in mice. J Exp Med. 1979;149:601–612. doi: 10.1084/jem.149.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu T, Muller J, Levandowski RA, Ye Z. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology. 2001;287:405–416. doi: 10.1006/viro.2001.1067. [DOI] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K, Horimoto T, Fujii Y, Kawaoka Y. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J Virol. 2004;78:10149–10155. doi: 10.1128/JVI.78.18.10149-10155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Kotler M, Skalka AM. cis-acting intron mutations that affect the efficiency of avian retroviral RNA splicing: implication for mechanisms of control. J Virol. 1988;62:2686–2695. doi: 10.1128/jvi.62.8.2686-2695.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Martinez-Sobrido L, Lienenklaus S, Weiss S, Garcia-Sastre A, Staeheli P. Strong interferon-inducing capacity of a highly virulent variant of influenza A virus strain PR8 with deletions in the NS1 gene. J Gen Virol. 2009;90:2990–2994. doi: 10.1099/vir.0.015727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Lai CJ. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980;21:475–485. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois RA, Varble A, Chua MA, Garcia-Sastre A, tenOever BR. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc Natl Acad Sci U S A. 2012;109:12117–12122. doi: 10.1073/pnas.1206039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci U S A. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz B, Brunotte L, Reuther P, Schwemmle M. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat Commun. 2012;3:802. doi: 10.1038/ncomms1804. [DOI] [PubMed] [Google Scholar]
- Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Hughes MT, Kawaoka Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000;19:6751–6758. doi: 10.1093/emboj/19.24.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill RE, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Fields Virology. 5. Philadelphia, PA: Lippincott Williams & Wilkins; 2006. [Google Scholar]
- Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- Perez JT, Varble A, Sachidanandam R, Zlatev I, Manoharan M, Garcia-Sastre A, tenOever BR. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JT, Zlatev I, Aggarwal S, Subramanian S, Sachidanandam R, Kim B, Manoharan M, tenOever BR. A Small-RNA Enhancer of Viral Polymerase Activity. J Virol. 2012;86:13475–13485. doi: 10.1128/JVI.02295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb NC, Jackson D, Vreede FT, Fodor E. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J Gen Virol. 2010;91:2331–2340. doi: 10.1099/vir.0.022004-0. [DOI] [PubMed] [Google Scholar]
- Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J Gen Virol. 2009;90:1398–1407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Takizawa N, Watanabe K, Nagata K, Kobayashi N. Crucial role of the influenza virus NS2 (NEP) C-terminal domain in M1 binding and nuclear export of vRNP. FEBS Lett. 2011;585:41–46. doi: 10.1016/j.febslet.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell. 1986;44:147–158. doi: 10.1016/0092-8674(86)90493-9. [DOI] [PubMed] [Google Scholar]
- Stojdl DF, Lichty BD, tenOever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, et al. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell. 2003;4:263–275. doi: 10.1016/s1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Varble A, Chua MA, Perez JT, Manicassamy B, Garcia-Sastre A, tenOever BR. Engineered RNA viral synthesis of microRNAs. Proc Natl Acad Sci U S A. 2010;107:11519–11524. doi: 10.1073/pnas.1003115107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda J, Nakada S, Kato A, Toyoda T, Ishihama A. Molecular assembly of influenza virus: association of the NS2 protein with virion matrix. Virology. 1993;196:249–255. doi: 10.1006/viro.1993.1473. [DOI] [PubMed] [Google Scholar]
- Ye Z, Liu T, Offringa DP, McInnis J, Levandowski RA. Association of influenza virus matrix protein with ribonucleoproteins. J Virol. 1999;73:7467–7473. doi: 10.1128/jvi.73.9.7467-7473.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. miRNA-mediated targeting of NS1, Related to Figure 1. (A) Northern blot analysis of total RNA probed for miR-20 and miR-142 in indicated cell lines. U6 levels used as a loading control. 293, human embryonic kidney cells; 293 + miR-142, 293 cells transfected with a vector expressing miR-142; A549, human alveolar epithelial cells; Jurkat, human T lymphocyte; Dicer1−/−, Dicer1 knockout mouse embryonic fibroblasts; MEF, mouse embryonic fibroblasts; pLF, primary lung murine fibroblast; BMDM, bone marrow–derived murine macrophages. (B) Diagram of recombinant NS segment containing target sites in the 3′ untranslated region (UTR). Target sites are either a scrambled sequence (Scbl) or four identical target sites for miR-20a (20T) or miR-142 (142T). T indicates position of target sites. (C) (Top two panels) Western blots of WCE derived from fibroblasts expressing Scbl, 20T, or 142T vectors with either empty vector or vector producing miR-142 (miR-142). (Middle two panels) Northern blot of total RNA from samples above, probed for miR-142 and U6. (Bottom two panels) Semi-quantitative RT-PCR on total oligo-dT RNA using NS1- and beta tubulin specific primers. (D) Western blot analysis of WCE derived from samples in Figure 1D. Immunoblots depict Mx1, NP, NS1, NEP, or Actin.
Figure S2. Silencing of NS1 does not impact ISG15 or IRF7 levels in vivo, Related to Figure 2. qRT-PCR of (A) ISG15 and (B) IRF7 from mouse lungs from Figure 2E normalized to tubulin. Data points represent individual mice assayed in triplicate.
Figure S3. NS1/NEP 2A IAV, siRNAs, and VLVs, Related to Figure 3. (A) Diagram of segment 8 of NS1/NEP 2A IAV which is a non-splicing segment 8 expressing NS1-NEP as a fusion separated by a porcine teschovirus-1 (PTV-1) 2A sequence. (B) Western blot analysis of WCE from fibroblast transfected with different NEP siRNAs or scbl siRNA for 16 hours then infected with WT IAV for 8 hours. Immunoblots depict NP, NS1, NEP, or Actin. (C) Multicycle growth curves of WT IAV infection for 0, 12, 24, 36, and 48 hpi in fibroblast cells transfected with NEP1/2 siRNA or scbl siRNA for 16 hours prior to infection. P-values for 24, 36, and 48 hrs are 0.01, 0.001, and 0.01 respectively. (D) Diagram of how VLVs are constructed. In brief, wt segments 1,2,3,5,6,7, and 8 were combined with a recombinant segment 4 expressing either NEP or scrambled (Scbl) RNA. These eight segment are used to generate virus-like vectors (VLVs) in a MDCK cell line that complements the loss of wild type segment four by expressing exogenous HA protein.
Figure S4. Characterization of the NSo virus, Related to Figure 4. (A) Semi-quantitative RT-PCR on total oligo-dT RNA from fibroblast infected with WT, NSo, or NSp viruses for 3, 6, or 9 hpi. cDNA treated using primers to amplify NS1 and NEP as depicted. Tubulin was used as a loading control. (B) Western blot analysis of WCE from A549 cells infected with NSp, NSo, and 20T IAV strains or mock treated for 8 hours. (C) Western blot analysis of WCE from MDCK and MDCK-NS1-GFP cells infected with delNS1 virus for 24 hours. (D) Western blot analysis of WCE derived from samples in Figure 4B. Immunoblots depict Mx1, NP, NS1, or Actin. (E) qRT-PCR of viral NP levels from mouse lungs in Figure 4D normalized to tubulin. Data points represent individual mice assayed in triplicate. (F) A549 cells were fixed following infection with either WT or 2A IAV at 8 hpi. Cells were stained with the NP antibody prior to imaging by fluorescence microscopy. Hoechst dye used to visualize nuclei.