Abstract
Aims/hypothesis
Following on from the emerging importance of the pancreas circadian clock on islet function and the development of type 2 diabetes in rodent models, we aimed to examine circadian gene expression in human islets. The oscillator properties were assessed in intact islets as well as in beta cells.
Methods
We established a system for long-term bioluminescence recording in cultured human islets, employing lentivector gene delivery of the core clock gene Bmal1 (also known as Arntl)-luciferase reporter. Beta cells were stably labelled using a rat insulin2 promoter fluorescent construct. Single-islet/cell oscillation profiles were measured by combined bioluminescence–fluorescence time-lapse microscopy.
Results
Human islets synchronised in vitro exhibited self-sustained circadian oscillations of Bmal1-luciferase expression at both the population and single-islet levels, with period lengths of 23.6 and 23.9 h, respectively. Endogenous BMAL1 and CRY1 transcript expression was circadian in synchronised islets over 48 h, and antiphasic to REV-ERBα (also known as NR1D1), PER1, PER2, PER3 and DBP transcript circadian profiles. HNF1A and PDX1 exhibited weak circadian oscillations, in phase with the REV-ERBα transcript. Dispersed islet cells were strongly oscillating as well, at population and single-cell levels. Importantly, beta and non-beta cells revealed oscillatory profiles that were well synchronised with each other.
Conclusions/interpretation
We provide for the first time compelling evidence for high-amplitude cell-autonomous circadian oscillators displayed in human pancreatic islets and in dispersed human islet cells. Moreover, these clocks are synchronised between beta and non-beta cells in primary human islet cell cultures.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-012-2779-7) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Keywords: Beta cells, Circadian clocks, Human pancreatic islets, Time-lapse microscopy
Introduction
Circadian oscillations of biological processes have been described in virtually all light-sensitive organisms. They reflect the existence of underlying intrinsic clocks with near 24 h oscillation periods. Circadian control of physiology and behaviour is driven by a master pacemaker located in the suprachiasmatic nucleus (SCN) of the hypothalamus, which orchestrates subsidiary oscillators in peripheral organs. Rhythmicity in the SCN is entrained by external Zeitgeber (time-giver) cues, such as daily changes in light intensity [1]. In mammals, the CLOCK (circadian locomotor output cycles kaput) and BMAL1 (brain and muscle ARNT-like protein 1) transcription factors activate the expression of Per and Cry genes. Once the period (PER) and cryptochrome (CRY) proteins have reached a critical threshold, they attenuate the CLOCK/BMAL1-mediated activation of their own genes in a negative feedback loop [1, 2]. This rhythm-generating circuitry is functional in most cell types, including primary and immortalised cell lines [3, 4].
There is growing evidence for connections between metabolic syndromes, including obesity and diabetes, and the circadian clockwork [5]. Rhythmically expressed liver genes code for enzymes implicated in the metabolism of fatty acids, cholesterol, bile acids and xenobiotics, and the disruption of circadian oscillator function causes alterations in metabolism [6]. Animals with a liver-specific Bmal1 (also known as Arntl) disruption suffer from hypoglycaemia specifically during the resting phase [7]. Mice with disrupted circadian clock function (homozygous mutants of Clock) develop hyperphagia, obesity and features of the metabolic syndrome [8]. Further evidence of the interconnection between circadian oscillators and metabolism stems from several recent studies on the core clock gene Rev-erbα (also known as Nr1d1), which has been found to be indispensable for proper lipid and carbohydrate metabolism [9–11]. Moreover, CRY1 and CRY2 modulate fasting glucose levels through the inhibition of glucagon-induced gluconeogenesis [12]. Recently, evidence for self-sustained oscillators in mouse pancreatic islets has been provided by Bass and colleagues [13]. This work indicated that Clock∆19 mutant mice exhibited hyperglycaemia, elevated fasting glucose levels and impaired glucose tolerance. A similar phenotype was observed in Bmal1 knockout mice, suggesting that these metabolic alterations are not dependent on CLOCK function alone, but are general for core clock genes. Most importantly, this study revealed that ablation of the pancreatic clock might directly trigger onset of diabetes, representing an important milestone in our understanding of the molecular link between the clock, islet function and type 2 diabetes aetiology [13]. In line with these findings, pancreas-specific Bmal1 knockout in a different mouse model led to impaired insulin secretion [14].
In humans, glucose homeostasis is tightly controlled by the circadian system [15]. Genetic linkage analysis has shown that CRY2 and PER2 might be associated with blood glucose levels [16, 17]. Moreover, patients with circadian misalignments show profound perturbations of plasma glucose and insulin levels [6]. Finally, core clock gene expression analysis in human islets demonstrated that mRNA levels of PER2, PER3 and CRY2 might be downregulated in isolated islets from individuals with type 2 diabetes in comparison with islets from healthy counterparts, suggesting the link between the human islet clock and type 2 diabetes [18]. It is therefore of scientific and clinical importance to provide further insight into the emerging connection between circadian oscillator function, metabolic regulation and type 2 diabetes in humans, with a specific focus on the human islet circadian oscillator molecular makeup.
Methods
Human islet preparation
Human pancreases were obtained from brain-dead multi-organ donors. Details of islet donors are summarised in Table 1. Human islet isolations were performed at the Islet Transplantation Centre at the University Hospital of Geneva (Switzerland) as previously described [19, 20]. The use of human islets for research was approved by the local ethical committee. Islet purity was estimated by dithizone staining; islet viability was assessed by propidium iodide and fluorescein diacetate staining. After purification islets were cultured in Connaught Medical Research Laboratories (CMRL) medium for 24–72 h before the experiments. Whole islets were attached to dish surfaces covered with a laminin-5-rich extracellular matrix derived from 804G cells [21]. To dissociate islets into single cells, islets were re-suspended in Accutase (Innovative Cell Technologies, San Diego, CA, USA), incubated at 37°C until completely dissociated and then diluted in CMRL medium.
Table 1.
Islet donor characteristics
| Donor no. | Sex | Age (years) | BMI (kg/m2) | Purity (%) | Viability (%) |
|---|---|---|---|---|---|
| 1 | M | 58 | 29.3 | 90 | 79 |
| 2 | F | 56 | 23.8 | 90 | 85 |
| 3 | F | 56 | 19.7 | 90 | 87 |
| 4 | M | 56 | 29.1 | 90 | 70 |
| 5 | M | 48 | 20.3 | 95 | 94 |
| 6 | M | 52 | 23.1 | 95 | 96 |
| 7 | M | 61 | 27.8 | 85 | 47 |
| 8 | M | 56 | 28.6 | 85 | 60 |
| 1–8 | M = 6, F = 2 | 55 ± 4a | 25 ± 4a | 90 ± 4a | 77 ± 17a |
M, male sex; F, female sex
aData are means ± SD, n = 8
Lentivectors and lentiviral production
The Bmal1-luciferase (luc) lentivector was engineered by Liu and colleagues [22]. CMV-GFP lentivector (pLOX-CW-GFP) was previously described by us [23]. pRIP-GFP and pRIP-tomato were engineered using the rat insulin 2 (RIP) promoter [23], controlling the production of green fluorescent protein (GFP) or the tdTomato red living colour [24], respectively. Maps and sequences of lentivectors are available at our website (http://medweb2.unige.ch/salmon/lentilab/). Lentiviral particles were produced by transient transfection in 293T cells using the polyethylenimine method [25]. Lentiviral particles were harvested at 48 h post transfection, 100-fold concentrated, titred and used for the transduction of whole islets or islet cells. Multiplicity of infection (MOI) of three was found optimal in this system.
In vitro islet synchronisation and bioluminescence monitoring
To synchronise islets/islet cells, dexamethasone was added to the culture medium at a final concentration of 100 nmol/l. Following 30 min incubation at 37°C in a cell culture incubator, dexamethasone was replaced with normal culture medium, as previously described by us [26]. Bioluminescence patterns were monitored from human islets or islet cells 4 days following Bmal1-luc lentiviral transduction, as described previously [3, 26]. Briefly, synchronised islets/islet cells were transferred to a light-tight incubator (37°C) in the recording medium (CMRL containing 100 μmol/l luciferin). Bioluminescence from each dish was continuously monitored using a Hamamatsu photomultiplier tube detector assembly (Hamamatsu City, Japan). Photon counts were integrated over 1-min intervals. The Actimetrics LumiCycle Analysis program was used for analysis of rhythm variables (Actimetrics, Wilmette, IL, USA).
Temperature entrainment
Synchronisation with temperature cycles was performed according to the protocol described previously [27]. Briefly, simulated circadian body temperature cycles (38–34°C) were generated in a custom-built incubator equipped with photomultiplier tubes for simultaneous bioluminescence recording [27]. Luminescence traces are either shown as raw or detrended data. For detrended time series, luminescence signals were smoothened by a moving average with a window of 60 data points and detrended by an additional moving average with a window of 24 h.
Bioluminescence time-lapse microscopy and data analysis
Whole islets or dispersed islet cells were attached to glass bottom dishes and transduced with Bmal1-luc, or with Bmal1-luc and RIP-tomato lentivectors, respectively. Four days after lentiviral transduction islets/islet cells were synchronised with dexamethasone and subjected to combined bioluminescence–fluorescence imaging on an OlympusLV200 workstation (Bioimaging Platform, Geneva, Switzerland) equipped with a 63×UPLSAPO objective. Bioluminescence emission was detected for several consecutive days using an EM CCD camera (Image EM C9100-13, Hamamatsu) cooled to −90°C using exposure times of 30 min. The image series were analysed employing the ImageJ 1.32 software (http://imagej.nih.gov/ij/). The image analysis has been done by an adapted version of the software Circadian Gene Express (CGE, http://bigwww.epfl.ch/sage/soft/circadian/), a Java ImageJ’s plugin that we previously developed for tracking cells in the context of circadian studies [26, 28]. The measure of expression has been adapted to the islets considering that the labelled cells are mainly present at the periphery. The level of expression is obtained by subtracting the average of intensity inside a ring at the periphery of the islet and the average of intensity outside the islets, which represents the local background level (Fig. 1e). CGE now includes a module to fit a model to a profile of expression. The model that we have chosen is a sinusoid:
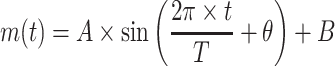 |
Fig. 1.
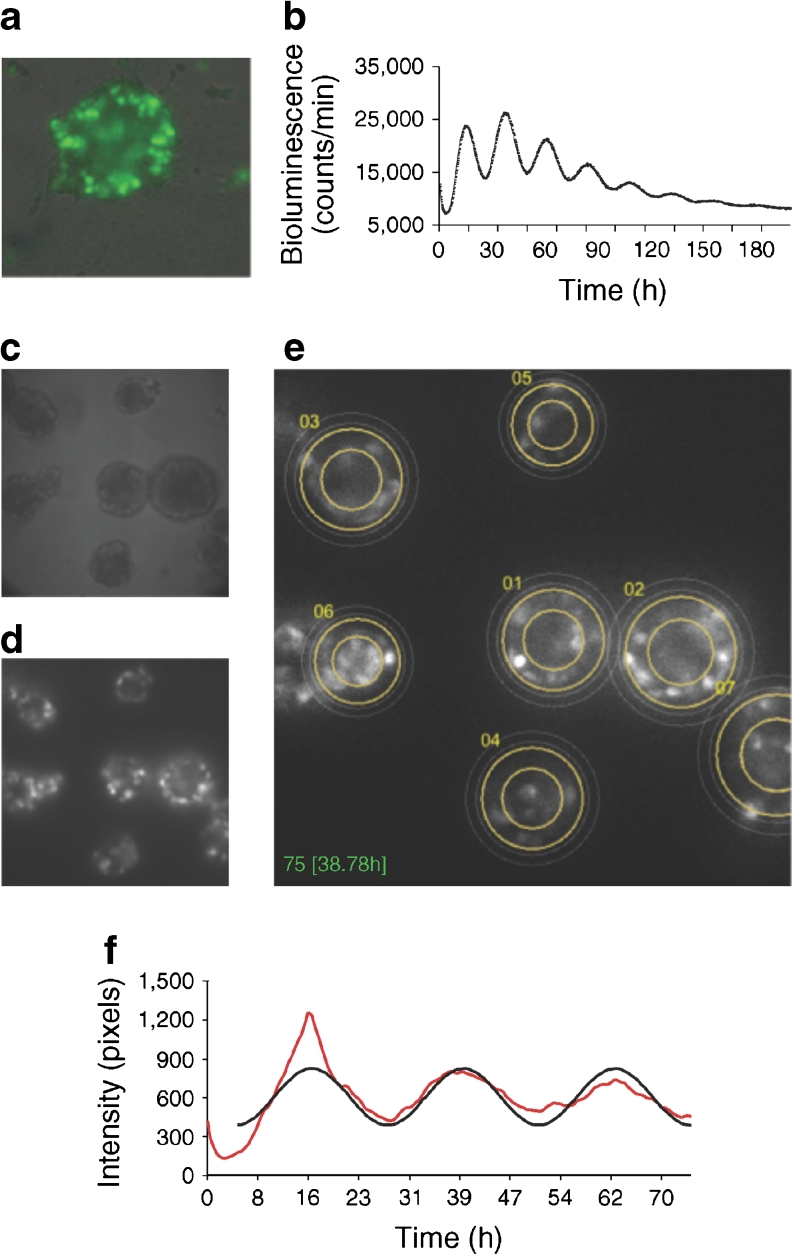
Cell-autonomous oscillators in human pancreatic islets. (a) Bmal1-luc and CMV-GFP co-expression in whole islets. Whole islets were co-transduced with lentiviral particles expressing Bmal1-luc and CMV-GFP constructs. Four days after infection, GFP expression evaluation was performed in n = 20 islets from two donors (ten islets from each donor). At least half of the cells were GFP positive in all visualised islets (representative CMV-GFP transduced islet is shown). (b) Bmal1-luc oscillation profile in synchronised islets. Islets were synchronised with dexamethasone and Bmal1-luc bioluminescence was recorded using an Actimetric Lumicycler for 200 h. Bmal1-luc oscillation profiles were recorded for three parallel dishes for each donor, total of five donors. (c, d) Individual islet circadian bioluminescence recording (ESM Video 1, left). Human islets were cultured on 804G matrix-covered dishes and transduced with Bmal1-luc viral particles. (c) Differential interference contrast (DIC) image. (d) Bioluminescence channel from the time-lapse microscopy images. (e) Trajectories of seven individual islets image by adapted CGE application (image corresponds to ESM Video 1, right). (f) Representative individual islet profile: the raw data (red curve) and the fitted sinusoid (black curve) are shown
The fitting is performed by a Levenberg–Marquardt optimisation procedure to recover the amplitude A and the period T. The bioluminescence image sequence has been de-noised using the PURE-LET method [29]. This method is optimised for images corrupted by mixed Poisson–Gaussian noise and it improves the signal-to-noise ratio of the bioluminescence signal.
RNA analysis by real-time quantitative PCR
Attached intact islets were synchronised by dexamethasone as described above, collected every 4 h for 48 h, deep-frozen in liquid nitrogen and kept at −80°C. Total RNA was prepared using RNeasy Plus Micro kit (Qiagen, Hombrechtikon, Switzerland). One μg of total RNA was reverse-transcribed using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and random hexamers, PCR amplified and quantified as described previously [11]. Mean values for each experiment were calculated from technical triplicates of PCR assays for each sample, and normalised to the mean of those obtained for GAPDH and 9S transcripts. GAPDH and 9S expression levels exhibited no significant variability throughout each experiment and therefore served as internal controls. Primers used for this study are listed in electronic supplementary material (ESM) Table 1.
Results
Recording circadian bioluminescence from human pancreatic islets
Bioluminescence profiles from cells producing luciferase driven from different circadian regulatory sequences can be recorded at the population level using photomultiplier tubes, or at the single-cell level using highly sensitive photon-counting microscopy [2]. We decided to employ both approaches to decipher the human islet clockwork. First, we set up the experimental system for long-term recordings of circadian reporter oscillations in cultured human islets. Whole islets were attached to dishes covered with a laminin-5-rich extracellular matrix derived from 804G cells [21] and transduced with lentivectors expressing the Bmal1-luc construct [30]. Transduction efficiency in whole islets was evaluated using a CMV-GFP lentivector [23] co-transduced with Bmal1-luc. At least half of the cells were GFP positive in all visualised islets, with intensive GFP staining observed on the islet surface, and significantly weaker penetration to the inner core (Fig. 1a). To access the impact of lentiviral transduction on islet function, glucose-stimulated insulin secretion (GSIS) was measured in the islet cells co-transduced with CMV-GFP and RIP-tomato lentivectors 5 days following the transduction, and compared with non-transduced counterparts (ESM Fig. 1, ESM Methods). No significant difference in GSIS was observed upon lentiviral transduction.
Cell-autonomous and self-sustained circadian clocks displayed in isolated human islets
To monitor circadian oscillation profiles in human islets, we first recorded circadian bioluminescence in the isolated human islets. Intact islets expressing Bmal1-luc lentivectors were synchronised by a dexamethasone pulse 4 days after the viral transduction, and circadian bioluminescence was continuously recorded for several days in an Actimetrics Lumicycler (Fig. 1b). We used the glucocorticoid hormone analogue dexamethasone for islet synchronisation since it was previously demonstrated to synchronise circadian oscillators in cultured rat-1 and NIH3T3 fibroblasts efficiently. In addition it transiently changes the phase of circadian gene expression in liver, kidney and heart [26, 31, 32], therefore representing a strong in vitro synchronisation stimulus. Our experiments revealed pronounced oscillations of Bmal1-luc expression with a period length of 23.6 ± 0.4 h (n = 5; Fig. 1b), indicating the presence of functional islet-autonomous circadian clocks in human islet population.
We next sought to visualise single-islet oscillations employing a bioluminescent time-lapse microscopy approach. Bmal1-luc transduced human islets were synchronised by a dexamethasone pulse and subjected to bioluminescence time-lapse microscopy recording for at least three consecutive days (ESM Video 1 [left] and Fig. 1c, d). The Bmal1-luc bioluminescence profiles of individual human islets were analysed by an updated version of the CGE software previously developed by us [28], specifically adapted to the islet analysis (ESM Video 1 [right] and Fig. 1e). A representative profile of bioluminescence oscillations in a human islet is shown in Fig. 1f. Of the analysed single islets, 76% (13 out of 17) were oscillating for 3 days of recording, with an oscillation period of 23.99 ± 1.20 h, closely matching the results obtained in the population of human islets (Fig. 1b). Our single-islet recording experiments unambiguously demonstrated the presence of high-amplitude cell-autonomous oscillators in human islets. Of note, this visualisation further ensures that Bmal1-luc oscillations were recorded from the islets themselves and not from adjacent cells or tissue fragments, which represent 5–15% contamination of human islet preparations (Table 1).
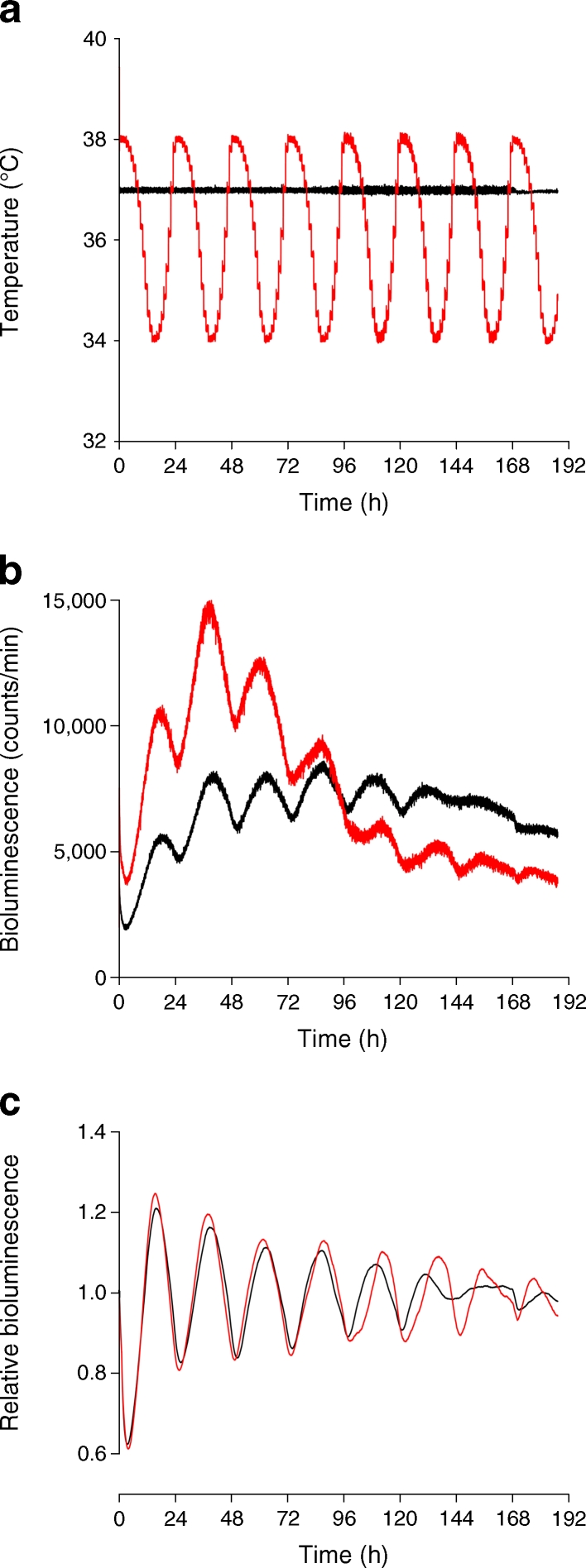
Multiple in vitro stimuli have previously been demonstrated to efficiently synchronise cultured cells [32]. Among them, forskolin has been used for in vitro synchronisation of isolated mouse islets [13]. As demonstrated in ESM Fig. 2, short pulses of dexamethasone, horse serum or forskolin were all able to strongly synchronise the islets isolated from mouse bearing the circadian Per2::luc reporter [33]. Furthermore, human islets expressing Bmal1-luc were subjected to simulated body temperature cycles oscillating between 38°C and 34°C (Fig. 2a). Similar to dexamethasone synchronisation, continuous temperature cycles resulted in high-amplitude circadian oscillations of Bmal1-luc bioluminescence in human islets (Fig. 2b, c).
Fig. 2.
In vitro entrainment of human islets by simulated body temperature cycles. Human islets transduced with Bmal1-luc reporters were synchronised by dexamethasone and maintained at constant 37°C temperature (black line), or continuously entrained by simulated body temperature cycles (red line). (a) Temperature measurements throughout bioluminescence recordings in dishes of dexamethasone-synchronised (black) and temperature-entrained (red) islets. (b) Raw Bmal1-luc traces of dexamethasone-synchronised (black) and temperature-entrained (red) islets. (c) Same data as in (b), which has been detrended as described in the Methods (y-axis shows detrended bioluminescence)
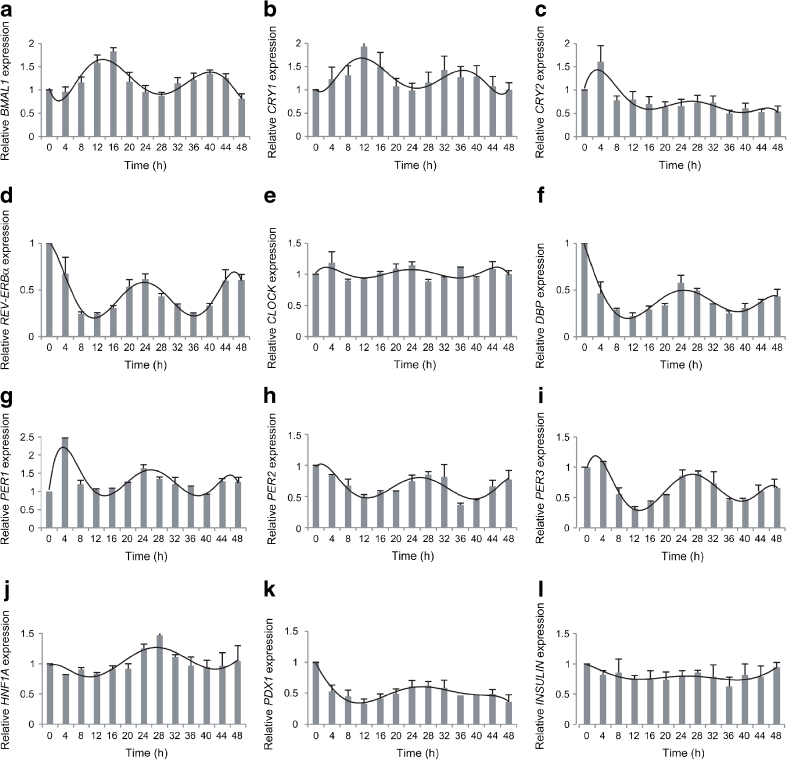
To validate our results obtained by circadian bioluminescence recording, we examined endogenous core clock gene expression profiles in the synchronised islets. To this end, intact islets were attached to laminin-5-rich extracellular-matrix-covered dishes (1,000 islets per dish) and subsequently synchronised by a dexamethasone pulse as described in Methods. The mRNA accumulation patterns from synchronised islets were monitored every 4 h for 48 h by quantitative RT-PCR, using amplicons for BMAL1, CRY1, CRY2, REV-ERBα, CLOCK, PER1, PER2, PER3 and DBP. The values obtained were normalised to the results determined for GAPDH and 9S mRNAs, which accumulated to similar levels throughout the day. In good agreement with our experiments using the Bmal1-luc reporter, endogenous BMAL1 transcript abundance exhibited pronounced circadian oscillations over 48 h in synchronised islets (Fig. 3a). CRY1 transcript oscillation was in phase with BMAL1 (Fig. 3b), while clearly antiphasic to those of REV-ERBα, DBP and PER1–3 transcripts (Fig. 3d, f–i), in good agreement with mouse islet and liver oscillator studies [13, 34]. CRY2 mRNA exhibited a weak oscillatory profile, in phase with REV-ERBα (Fig. 3c, d) and CLOCK expression was arrhythmic (Fig. 3e), as expected from previous studies ([35] and [36], respectively). Our experiments in in vitro synchronised human islets revealed the presence of core clock circadian genes, oscillating with a period length of about 24 h and circadian phases comparable with those described in mouse islets and other peripheral tissues [13, 33].
Fig. 3.
Oscillation of clock genes (a–i) and functional genes (j–l) in human islets. qPCR was performed in whole islets for core clock genes (a) BMAL1, (b) CRY1, (c) CRY2, (d) REV-ERBα, (e) CLOCK, (f) clock output gene DBP, (g) PER1, (h) PER2 and (i) PER3 and for islet functional genes (j) HNF1A, (k) PDX1 and (l) INSULIN transcripts, normalised to GAPDH and 9S house-keeping genes. Each time point represents 1,000 islets. Profiles are representative of three experiments (mean ± SEM), each using islets from one donor
Several key functional genes in the islet have previously been demonstrated to be expressed in a circadian fashion in mouse islets isolated around the clock [13]. We assayed human homologues of these potential clock-controlled genes in human islets synchronised in vitro. Expression pattern of HNF1A and PDX1 suggested a circadian oscillatory profile (Fig. 3j, k), in phase with those of REV-ERBα, PER1-3 and DBP. On the other hand, INSULIN (also known as INS) (Fig. 3l), GLUCAGON (also known as GCG), GLUT1 (also known as SLC2A1), GLUT2 (also known as SLC2A2) and CYCLIND1 (also known as CCND1) (not shown) transcripts exhibited weak or no measurable circadian amplitude.
Dispersed human pancreatic islet cells exhibit circadian oscillations
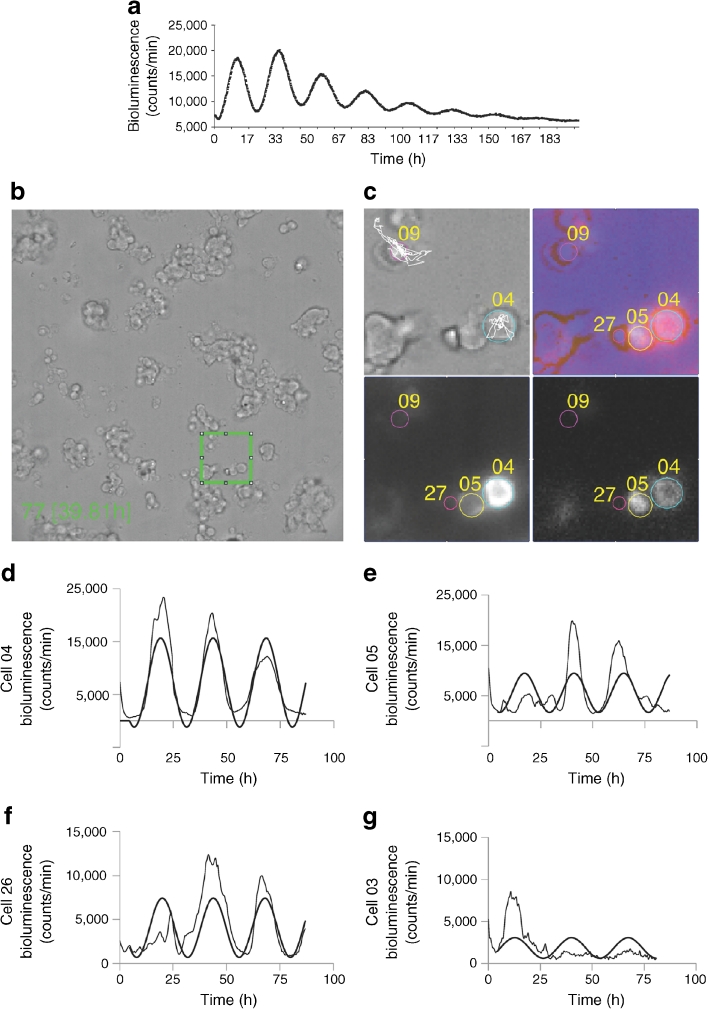
Our experiments in intact human islets revealed the presence of high-amplitude circadian oscillators. The next key question was whether the intact three-dimensional islet architecture is essential for the islet cellular clockwork. To address the impact of islet spatial organisation on cell oscillators, we compared the circadian oscillations of intact islets with those assayed in the dispersed islet cell population. To this end, intact islets were gently dispersed by Accutase and islet cells were then attached to matrix-coated dishes and synchronised by a dexamethasone pulse. Similarly to whole islets, dispersed human islet cells exhibited pronounced Bmal1-luc oscillations lasting for at least 8 days, with a period length of 24.3 ± 0.8 h (Fig. 4a). Oscillation period length, amplitude and dampening rate were comparable between synchronised islet and islet dispersed cell populations (Fig. 1b and Fig. 4a, respectively). Hence the islet architecture is not required for sustained daily oscillations.
Fig. 4.
Beta and non-beta cells possess cell-autonomous clocks. (a) Oscillation profiles of dexamethasone-synchronised human islet cells transduced with Bmal1-luc encoding lentiviral particles. Bmal1-luc oscillation profiles were recorded for three parallel dishes for each of the five donors. (b) Representative full DIC image (512 × 512 pixels) of human islet cells attached to a Wilco glass bottom dish and subjected to time-lapse bioluminescence/fluorescence microscopy (ESM Video 2). (c) Four analysed cells, No. 04, No. 05, No. 09 and No. 27, overlaid on a zoomed 76 × 76 pixel patch (corresponding to the green square in [b], ESM Video 3). Light-grey panel, DIC image; dark-grey panel, image taken with the red fluorescence channel (RIP-tomato expression); black panel, image taken with the bioluminescence channel (Bmal1-luc expression); lilac panel, composite image of all three channels. Representative trajectories (cell No. 04 and No. 09) are overlaid in white over the DIC image. (d, e) Bioluminescence expression profiles of two representative beta cells, No. 04 and No. 05, with average tomato expression intensity of 14,720 and 8,735 pixels/cell, respectively (threshold for beta cells: 8,000 pixels/cell). The zone between 6,000 and 8,000 pixels/cell was considered ‘unclassified’. The raw data and the fitted sinusoid curve (bold curve) are shown in the profiles of both cells. (f, g) Bioluminescence expression profiles of cell No. 26 and No. 03 (ESM Video 2). Both cells are considered to be non-beta cells with average tomato intensity below 6,000 pixels/cell. The raw data and the fitted sinusoid curve (bold curve) are shown in the profiles of both cells
Beta and non-beta cells possess cell-autonomous circadian clocks
Pancreatic islets harbour endocrine cells with different, or even opposite, physiological functions. We next aimed to compare beta and non-beta cell clock oscillations in the dispersed islet cells, employing combined bioluminescence–fluorescence time-lapse microscopy. RIP-GFP and tamoxifen inducible RIP-Cre/ER-GFP lentivectors [23] have been previously used to trace human beta cells. We avoided using GFP as a fluorescent reporter for combined bioluminescence–fluorescence time-lapse microscopy due to the strong background of luciferin in the GFP spectrum. We thus constructed a lentivector expressing the tdTomato gene [24] under the control of the RIP promoter (RIP-tomato; see Methods section for details), allowing the specific and stable labelling of human beta cells. Both RIP-GFP and RIP-tomato lentivectors exhibited about 90% transduction efficiency and about 95% specificity (shown for RIP-GFP in ESM Fig. 3) for human beta cells. To visualise the beta cell oscillation profiles, we conducted combined bioluminescence–fluorescence time-lapse microscopy of human islet cells transduced with the combination of RIP-tomato and Bmal1-luc lentivectors. ESM Videos 2 and 3 and Fig. 4b, c, provide time-lapse bioluminescence and fluorescence images recorded from human islet cells and illustrate the method used for the tracking of single islet cells. For each of the analysed 32 cells, bioluminescence and fluorescence profiles were obtained as described (legend to Fig. 4; Methods section), and the results are presented in Table 2. Cells were classified as beta and non-beta according to their RIP-tomato expression level (Fig. 4c). Among 32 analysed cells, 21 were classified as beta cells and seven as non-beta; four remained ‘unclassified’ as they expressed intermediate levels of RIP-tomato reporter (Table 2). Analysis of single beta cells (tomato positive) revealed that 17 out of 21 analysed cells were clearly oscillating, with a period length of 26.28 ± 2.26 h, while four cells were not cycling or exhibiting weak or uncertain oscillation profiles. For the non-beta cells, four out of seven were oscillating with a period of 26.01 ± 1.37 h. Representative beta and non-beta cell oscillatory profiles are shown in Fig. 4d, e and f, g, respectively. These experiments demonstrate circadian oscillations in human beta and non-beta cells, which were distinguished by using RIP-tomato labelling. The clocks of dispersed beta and non-beta cells seem to be well synchronised among themselves, and they exhibit oscillations with a similar period length to that observed in a mixed population of islet cells.
Table 2.
Summary of the oscillation analysis of beta and non-beta cells from time-lapse microscopy datasets
| Variable | Beta cells | Unclassified | Non-beta cells | Total number of cells |
|---|---|---|---|---|
| Tomato expression range | >8,000 | 6,000–8,000 | <6,000 | |
| No. of cells | 21 | 4 | 7 | 32 |
| Oscillating cell identification number | 01, 04, 05, 06, 07, 08, 10, 11, 12, 15, 17, 18, 20, 24, 28, 30, 31, 32 | 02, 22, 25 | 03, 14, 26, 29 | 25 |
| Non-oscillating cell identification number | 16, 19, 21 | 27 | 9, 13, 23 | 7 |
Discussion
We provide for the first time convincing evidence for cell-autonomous clocks displayed in cultured human islets. High-amplitude circadian oscillations were recorded at islet population level, at single-islet level, in the dispersed islet cells and in beta and non-beta cells separately.
It has been shown previously that circadian timekeepers in liver and lung explants can generate 20 or more daily cycles of PER2::luc expression [33]. Using the same reporter mouse, Marcheva and colleagues provided clear-cut evidence that isolated islets synchronised with forskolin possess self-sustained circadian oscillators [13]. Our study demonstrates unambiguously that living human pancreatic islets exhibit pronounced circadian oscillations and that we can assess these oscillations in in vitro synchronised human islets/dispersed islet cells for several days following the synchronisation. Human islets expressing Bmal1-luc circadian reporter were efficiently synchronised by dexamethasone and temperature cycles (Figs 1, 2). Dexamethasone, horse serum and forskolin pulses were able to synchronise cultured mouse islets expressing Per2::luc reporter (ESM Fig. 2). Thus high-amplitude circadian oscillations of in vitro synchronised pancreatic islets were observed irrespective of the synchronisation or entrainment pathway. It might be interesting to further explore whether the glucose, insulin or glucagon-like peptide 1 signalling pathways play a role in human islet synchronisation in vitro.
Pancreatic islets represent a unique model of endocrine cells tightly packed in a three-dimensional structure. We thus investigated whether islet architecture might influence oscillator properties in cells of intact islets, by comparing the oscillators of intact islets with those of dispersed single islet cells. Both intact islets and dispersed islet cells exhibited circadian oscillations, with a period length about half an hour longer for dispersed cells, and comparable amplitudes (Figs 1b, 4a; ESM Videos 1, 2). Previous study suggested that while intercellular oscillator coupling is indispensable for SCN neuron synchronisation, peripheral clocks do not show coupling within the organs [37]. In addition, the study by Nagoshi and colleagues provided elegant evidence that the clocks of cultured fibroblasts are not coupled, even if the cells are grown to confluence and hence establish physical contacts [3]. In line with these findings, our results suggest that three-dimensional islet architecture is not crucial for proper function of the cellular oscillator. We cannot formally exclude that islet cells are still coupled by paracrine signalling molecules, without the need for physical contact.
Taking advantage of recently developed combined bioluminescence–fluorescence time-lapse microscopy, we took one step further and analysed the oscillator profiles in synchronised beta and non-beta cells. Adaptation of the analysis software previously developed by us [28] for islet studies greatly facilitated time-lapse microscopy dataset analysis. These experiments suggest that the oscillators of different islet cell types are indeed synchronised in islets maintained in organotypic cultures (ESM Videos 2, 3; Fig. 4; Table 2).
In line with the outcome of our reporter experiments, endogenous clock gene expression measurements by real-time quantitative PCR (qPCR) suggested that the core clock genes BMAL1, CRY1, REV-ERBα, PER1, PER2 and PER3, as well as the clock output gene DBP, exhibit circadian oscillatory patterns in dexamethasone-synchronised human islets over 48 h (Fig. 3a–i). Moreover, expression of HNF1A and PDX1 genes involved in islet growth and development might be under circadian control in the human islet (Fig. 3j, k). Earlier studies demonstrated high DBP and HLF expression in human islets, all together suggesting a role of proper clockwork in human islet gene expression regulation [38]. Of note, genes encoding insulin and glucagon and other functional genes tested did not show clear oscillation patterns. However, to draw strong conclusions regarding the oscillation patterns of putative clock target genes, more accurate mRNA quantification methods, such as Nanostring counting [39], should be employed, since the transcript of these genes accumulate with rather low amplitudes in in vitro synchronised islets compared with mouse islets isolated around the clock (in vivo situation [13]).
This study was made possible by two major technical achievements. First, experimental conditions established in our laboratory for the continuous recording of circadian bioluminescence produced by a stably integrated Bmal1-luc reporter gene in islet cells provided a powerful and unique tool for studying circadian oscillators in living human pancreatic islets for several consecutive days with high temporal resolution. Second, using RIP-tomato-expressing lentiviral particles, we have developed an approach allowing study of specifically and stably labelled human beta cells. At present, the most commonly used method for human beta and non-beta cell sorting is based on beta cell labelling with Newport green [21]. Newport green labelling is non-toxic to the cells and does not interfere with gene expression and insulin secretion [21]. However, labelling with Newport green is transient and sorting thus would have to be performed before the experiments. Given that for the oscillator studies, as well as for many other purposes, cells have to be kept in culture for several days following sorting by FACS, and taking into account the high toxicity of the FACS procedure, avoiding the sorting represents a major advantage for human beta cell studies. Thus the RIP-tomato labelling we developed provides a valuable tool for studying human beta and non-beta cell function without sorting by FACS. This tool is particularly useful for fluorescence time-lapse microscopy but not restricted to it, as beta cells specifically labelled with RIP-tomato can be, for example, hand-picked under the fluorescent microscope.
Exploring the endocrine pancreas clockwork and its role in islet gene expression and function is likely to provide additional insight into the emerging connection between circadian oscillator function in peripheral tissues and metabolic regulation. In mouse models intact islet clocks have been demonstrated to play an important role in normal energy homeostasis [13, 14, 40]. This first detailed characterisation of human islet circadian oscillator opens the way for future studies that may perhaps link defects in these pathways with decreased beta cell function in type 2 diabetes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 7.05 kb)
Lentiviral transduction does not alter glucose induced insulin secretion in human islets. GSIS was measured in human islet cells transduced with RIP-tomato and CMV-GFP (RIP/CMV) lentivectors (MOI = 5 for each lentivirus) and compared to the non-transduced counterparts (NT). Insulin secretion (mean ± SDV) is shown for basal and stimulated conditions (left panel), normalised to total insulin content. Ratio (fold change) between stimulated and basal insulin secretion is shown in right panel. (PDF 984 kb)
Multiple stimuli strongly synchronise mouse islets in vitro. Bioluminescence profiles of Per2::luc mouse islets synchronised with 30 min pulse of 100 nmol/l dexamethasone (DEX), 50% horse serum (HS) or 10 μmol/l forskolin (FORS). (PDF 2,690 kb)
Specific expression of Rat Insulin 2 Promoter-GFP (RIP-GFP) construct in human beta cells. Human islet cells were transduced with a RIP-GFP lentivirus and stained with anti-insulin (INS) or anti-glucagon (GCG) antibodies. RIP-GFP exhibits close to 100% co-localisation with anti-insulin (upper panels), and less than 5% co-localisation with anti-glucagon staining (lower panels). (PDF 2,180 kb)
(PDF 10.8 kb)
Bioluminescence time-lapse microscopy of whole human islets. Human islets were transduced with Bmal1-luciferase lentivectors, synchronised by a dexamethasone pulse at time point 0, and subjected to bioluminescence time-lapse microscopy. Left video represents the recorded images; right video demonstrates the islet tracing performed by CGE plugin for ImageJ in order to obtain single islet profiles. If the file name obstructs the content of the video when viewed using VLC Media Player, users can remove it by disabling the On Screen Display (OSD) (Tools, Preferences, disable OSD). (MOV 18,900 kb)
Combined bioluminescence–fluorescence time-lapse microscopy of human islet cells. Human islet cells were transduced with Bmal1-luciferase and RIP-tomato lentivectors, synchronised by a dexamethasone pulse at time point 0, and subjected to bioluminescence–fluorescence time-lapse microscopy for at least 85 h. Upper left video shows differential interference contrast (DIC) image acquisition; lower left video–red fluorescence channel (RIP-tomato expression); lower right video–bioluminescence channel (Bmal1-luciferase expression); and upper right video–composite video of all three channels. Traces of 32 individual cells performed by adapted CGE application are delineated at DIC video. If the file name obstructs the content of the video when viewed using VLC Media Player, users can remove it by disabling the On Screen Display (OSD) (Tools, Preferences, disable OSD). (MOV 22,300 kb)
Combined bioluminescence–fluorescence time-lapse microscopy of human islet cells: close-up. This video represents the close-up (eightfold magnification) of the green square area delineated at the Fig. 4B in the manuscript body. If the file name obstructs the content of the video when viewed using VLC Media Player, users can remove it by disabling the On Screen Display (OSD) (Tools, Preferences, disable OSD). (MOV 14,600 kb)
Acknowledgements
We are grateful to: U. Schibler (Geneva University), G. Asher (Weizmann Institute of Science) and H. Reinke (Universitätsklinikum Düsseldorf) for constructive discussions and critical reading of the manuscript; K. Bouzakri (Geneva University) for generously sharing his expertise in human islet work and C. Barraclough and M. Docquier (NCCR Genomics Platform, Geneva University) for help in performing the qPCR experiments.
Funding
This work was funded by: the Swiss National Science Foundation Grant No. 31003A_130070/1; a grant from the EFSD/MSD Basic Research Programme; Ernst & Lucie Schmidheiny Foundation; Novartis Foundation for Medico-Biological Sciences Research; Swiss Life Foundation; Fondation Romande for the Diabetes Research to C. Dibner and Grant No. 31-2008-416 from the Juvenile Diabetes Research Foundation to Islet Transplantation Centre, HUG.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
PP, TM, DS and SL contributed to data acquisition, analysis and interpretation, LG, DB, MU, PS, MG-L, JM and CB facilitated experiments through the provision of reagents and data analysis, JP and PH contributed to the conception and design, CD designed the study, contributed to the data acquisition and analysis and drafted the manuscript. All authors took part in the revision of the manuscript and approved the final version.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Abbreviations
- BMAL1
Brain and muscle ARNT-like protein 1
- CGE
Circadian gene express
- CLOCK
Circadian locomotor output cycles kaput
- CMRL
Connaught Medical Research Laboratories medium
- CRY
Cryptochrome
- GFP
Green fluorescent protein
- GSIS
Glucose-stimulated insulin secretion
- MOI
Multiplicity of infection
- qPCR
Quantitative PCR
- PER
Period
- RIP
Rat insulin 2 promoter
- SCN
Suprachiasmatic nucleus
Footnotes
P. Pulimeno, T. Mannic and D. Sage contributed equally to this study.
References
- 1.Reppert SM. A colorful model of the circadian clock. Cell. 2006;124:233–236. doi: 10.1016/j.cell.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 3.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 4.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- 5.Kohsaka A, Laposky AD, Ramsey KM, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metabol. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delezie J, Dumont S, Dardente H, et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012;26:3321–3335. doi: 10.1096/fj.12-208751. [DOI] [PubMed] [Google Scholar]
- 11.Le Martelot G, Claudel T, Gatfield D, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biology. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang EE, Liu Y, Dentin R, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcheva B, Ramsey KM, Buhr ED, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadacca LA, Lamia KA, deLemos AS, Blum B, Weitz CJ. An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia. 2011;54:120–124. doi: 10.1007/s00125-010-1920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberhard D, Lammert E. The pancreatic beta-cell in the islet and organ community. Curr Opin Genet Dev. 2009;19:469–475. doi: 10.1016/j.gde.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Englund A, Kovanen L, Saarikoski ST, et al. NPAS2 and PER2 are linked to risk factors of the metabolic syndrome. J Circadian Rhythms. 2009;7:5. doi: 10.1186/1740-3391-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamenkovic JA, Olsson AH, Nagorny CL, et al. Regulation of core clock genes in human islets. Metabolism. 2012;61:978–985. doi: 10.1016/j.metabol.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Bucher P, Mathe Z, Morel P, et al. Assessment of a novel two-component enzyme preparation for human islet isolation and transplantation. Transplantation. 2005;79:91–97. doi: 10.1097/01.TP.0000147344.73915.C8. [DOI] [PubMed] [Google Scholar]
- 20.Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW. Automated method for isolation of human pancreatic islets. Diabetes. 1988;37:413–420. doi: 10.2337/diabetes.37.4.413. [DOI] [PubMed] [Google Scholar]
- 21.Parnaud G, Bosco D, Berney T, et al. Proliferation of sorted human and rat beta cells. Diabetologia. 2008;51:91–100. doi: 10.1007/s00125-007-0855-1. [DOI] [PubMed] [Google Scholar]
- 22.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genetics. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salmon P, Oberholzer J, Occhiodoro T, Morel P, Lou J, Trono D. Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol Ther. 2000;2:404–414. doi: 10.1006/mthe.2000.0141. [DOI] [PubMed] [Google Scholar]
- 24.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 25.Toledo JR, Prieto Y, Oramas N, Sanchez O. Polyethylenimine-based transfection method as a simple and effective way to produce recombinant lentiviral vectors. Appl Biochem Biotechnol. 2009;157:538–544. doi: 10.1007/s12010-008-8381-2. [DOI] [PubMed] [Google Scholar]
- 26.Dibner C, Sage D, Unser M, et al. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saini C, Morf J, Stratmann M, Gos P, Schibler U. Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev. 2012;26:567–580. doi: 10.1101/gad.183251.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sage D, Unser M, Salmon P, Dibner C. A software solution for recording circadian oscillator features in time-lapse live cell microscopy. Cell Div. 2010;5:17. doi: 10.1186/1747-1028-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luisier F, Blu T, Unser M. Image denoising in mixed Poisson–Gaussian noise. IEEE Trans Image Process. 2011;20:696–708. doi: 10.1109/TIP.2010.2073477. [DOI] [PubMed] [Google Scholar]
- 30.Brown SA, Fleury-Olela F, Nagoshi E, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biology. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balsalobre A, Brown SA, Marcacci L, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 32.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/S0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 33.Yoo SH, Yamazaki S, Lowrey PL, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 35.Miyamoto Y, Sancar A. Circadian regulation of cryptochrome genes in the mouse. Brain Res Mol Brain Res. 1999;71:238–243. doi: 10.1016/S0169-328X(99)00192-8. [DOI] [PubMed] [Google Scholar]
- 36.Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/S0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 37.Liu AC, Welsh DK, Ko CH, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allaman-Pillet N, Roduit R, Oberson A, et al. Circadian regulation of islet genes involved in insulin production and secretion. Mol Cell Endocrinol. 2004;226:59–66. doi: 10.1016/j.mce.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 40.Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythm. 2011;26:423–433. doi: 10.1177/0748730411416341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 7.05 kb)
Lentiviral transduction does not alter glucose induced insulin secretion in human islets. GSIS was measured in human islet cells transduced with RIP-tomato and CMV-GFP (RIP/CMV) lentivectors (MOI = 5 for each lentivirus) and compared to the non-transduced counterparts (NT). Insulin secretion (mean ± SDV) is shown for basal and stimulated conditions (left panel), normalised to total insulin content. Ratio (fold change) between stimulated and basal insulin secretion is shown in right panel. (PDF 984 kb)
Multiple stimuli strongly synchronise mouse islets in vitro. Bioluminescence profiles of Per2::luc mouse islets synchronised with 30 min pulse of 100 nmol/l dexamethasone (DEX), 50% horse serum (HS) or 10 μmol/l forskolin (FORS). (PDF 2,690 kb)
Specific expression of Rat Insulin 2 Promoter-GFP (RIP-GFP) construct in human beta cells. Human islet cells were transduced with a RIP-GFP lentivirus and stained with anti-insulin (INS) or anti-glucagon (GCG) antibodies. RIP-GFP exhibits close to 100% co-localisation with anti-insulin (upper panels), and less than 5% co-localisation with anti-glucagon staining (lower panels). (PDF 2,180 kb)
(PDF 10.8 kb)
Bioluminescence time-lapse microscopy of whole human islets. Human islets were transduced with Bmal1-luciferase lentivectors, synchronised by a dexamethasone pulse at time point 0, and subjected to bioluminescence time-lapse microscopy. Left video represents the recorded images; right video demonstrates the islet tracing performed by CGE plugin for ImageJ in order to obtain single islet profiles. If the file name obstructs the content of the video when viewed using VLC Media Player, users can remove it by disabling the On Screen Display (OSD) (Tools, Preferences, disable OSD). (MOV 18,900 kb)
Combined bioluminescence–fluorescence time-lapse microscopy of human islet cells. Human islet cells were transduced with Bmal1-luciferase and RIP-tomato lentivectors, synchronised by a dexamethasone pulse at time point 0, and subjected to bioluminescence–fluorescence time-lapse microscopy for at least 85 h. Upper left video shows differential interference contrast (DIC) image acquisition; lower left video–red fluorescence channel (RIP-tomato expression); lower right video–bioluminescence channel (Bmal1-luciferase expression); and upper right video–composite video of all three channels. Traces of 32 individual cells performed by adapted CGE application are delineated at DIC video. If the file name obstructs the content of the video when viewed using VLC Media Player, users can remove it by disabling the On Screen Display (OSD) (Tools, Preferences, disable OSD). (MOV 22,300 kb)
Combined bioluminescence–fluorescence time-lapse microscopy of human islet cells: close-up. This video represents the close-up (eightfold magnification) of the green square area delineated at the Fig. 4B in the manuscript body. If the file name obstructs the content of the video when viewed using VLC Media Player, users can remove it by disabling the On Screen Display (OSD) (Tools, Preferences, disable OSD). (MOV 14,600 kb)