Abstract
Protein phosphatases type 2C (PP2Cs) from group A, which includes the ABI1/HAB1 and PP2CA branches, are key negative regulators of ABA signaling. HAI-1 gene had been shown to affect both seed and vegetative responses to ABA, which is one of PP2Cs clade A in Arabidopsis thaliana. Transgenic plants containing pHAI-1::GUS (β-glucuronidase) displayed GUS activity existing in the vascular system of leave veins, stems and petioles. Green fluorescent protein fused HAI-1 (HAI-1-GFP) was found in the nucleus through transient transformation assays with onion epidermal cells. The water-loss assays indicated the loss-of-function mutants did not show symptoms of wilting and they had still turgid green rosette leaves. The assays of seed germination by exogenous ABA and NaCl manifested that the loss-of-function mutants displayed higher insensitivity than wild-type plants. Taken together, the final results suggest that the HAI-1 (AT5G59220) encoded a nuclear protein and it can be highly induced by ABA and wound in Arabidposis, the stress-tolerance phenotype showed a slightly improvement when HAI-1 gene was disrupted.
Keywords: Loss-of-function mutant, GFP, GUS, Stress tolerance, Water-loss assays, Seed germination
Introduction
Abiotic stresses adversely affect growth and productivity and trigger a series of morphological, physiological, biochemical and molecular changes in plants. ABA regulates diverse plant processes, ranging from adaptation to water stress to seed germination and dormancy [10, 18], and post-embryonic development, such as lateral root development [5, 9]. Proteins encoded by these genes, which are ABA-mediated processes are important for plant tolerance to several abiotic stresses including salt, drought and freezing [30, 43].
Although numerous factors related to ABA responses had been reported [19], however, the ABA signaling model was dramatically updated due to two findings. Recently, a core signaling pathway has been established by the discovery of PYR/PYL/RCAR as a new type of soluble ABA receptor [33, 40], and the identification of a protein phosphatase–kinase complex [type 2C protein phosphatase (PP2C)-SNF1-related protein kinase 2 (SnRK2)] as downstream components of PYR/PYL/RCARs [49]. SRK2E plays a key role in stomatal responses to ABA in Arabidopsis [52, 55], while SRK2D and SRK21 are probably involved in ABA signaling during seed germination and root growth [12].
Salt stress, drought and, to a lesser extent, cold stress elevate ABA levels [35]. ABA upregulates the expression of many, but not all, drought-response genes [25, 31], indicating that there are ABA-dependent and ABA-independent salt and drought stress responses in plants [54].
Sixty-nine PP2Cs are encoded in the Arabidopsis genome [22, 47], according to sequence alignment of the catalytic phosphatase core, the clade A of PP2Cs is arranged in two subgroups, one including ABI1, ABI2, HAB1 and HAB2, and a second one formed by PP2CA/AHG3, AHG1, At5g59220, At1g07430 and At2g29380 [47], have largely overlapping but different roles as negative regulators of ABA signaling, mainly during germination [19]. The recessive loss-of-function mutants hab1-1 shows ABA hypersensitive inhibition of seed germination and enhanced ABA-mediated stomatal closure [26]. Genetic evidence has largely supported the negative role of PP2Cs in ABA signaling, and certain triple loss-of-function pp2c mutants display partial constitutive response to ABA [44].
Among the ABI genes, ABI1 and ABI2 are unique in that they encode PP2Cs, which are ubiquitously found in all eukaryotes and involved in phosphorylation-mediated signaling; In addition, these genes function through seed maturation and germination to vegetative growth. The mutants show a broad range of ABA-related phenotypes, including reduced seed dormancy, ABA-resistant seed germination and seedling growth, abnormal stomatal regulation, and defects in various responses to drought [11, 24].
Both PP2CA/AHG3 and AHG1 appear to play an essential role for ABA signaling during seed development and germination [26, 36, 57], but in contrast to pp2ca-1, the ahg1-1 mutant has no ABA-related phenotype in adult plants and expression of AHG1 is restricted to seed [36].
HAB1 is broadly expressed in the plant and strongly induced by ABA [29, 45]. Constitutive expression of HAB1 under a 35S promoter led to reduced ABA sensitivity both in seeds and vegetative tissues, compared to wild-type plants [45].
We previously reported that group A PP2Cs is functionally redundant at the molecular level, but they have distinctive roles in different tissues and organs, as indicated by tissue-specific expression patterns. The plant PP2CA genes appear to be expressed ubiquitously in various organs, albeit at varying levels. ABI1 is expressed in various tissues, including seeds and guard cells, and AHG1 and AHG3 are specifically localized in the nucleus [49]. Therefore, we took advantage of transgenic plants containing pHAI-1::GUS (β-glucuronidase) to study the expression of HAI-1 gene. To test the role of HAI-1 gene in Arabidopsis, we had constructed over-expression transgenic plants and gained the homozygote mutants of hai-1, which the T-DNA insertion was in the HAI-1 gene.
In this study, we successfully isolated from the T-DNA insertion mutant of HAI-1, namely hai-1. Furthermore, the mutants of hai-1 showed greatly enhanced tolerance to drought stress and highly enhanced ABA or NaCl insensitivity. Moreover, HAI-1 gene (highly ABA-induced PP2C gene 1) was highly induced by wound and ABA through generating transgenic Arabidopsis plants, which were carrying the HAI-1 promoters fused to the GUS gene.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana L. Heynh. Ecotype columbia was used in this study unless otherwise indicated. Plant growth conditions have been described elsewhere [37].
Transgenic plants
Using genomic sequences from the TAIR database (http://www.arabidopsis.org), the full-length HAI1 (AT5G59220) open reading frame were amplified from cDNA by PCR with primers Pro-F (sense, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTGAATATCTTATAATTTTTGCCC-3′) and Pro-R (antisense, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTCTCTTCTCCTCCGCCTCTGTAA-3′) and were inserted into pDONR201 Entry vector (Promega) by Gateway cloning technology and sequenced then was inserted into 35S pleela vector by LB clonase (Invitrogen company). Agrobacterium GV3101 90RK were transformed with these plasmids and used for infection of flowering plants by the floral dip method [7].
Loss-of-function insertion lines
Loss-of-function lines were obtained from the Arabidopsis biological resource center (ABRC). 7-day-old homozygous plants were identified by the kanamycin tolerance test and a PCR-based method using loss-of-function left- or right- border primers [1].
Root growth and germination assays
The root growth assay for scoring ABA sensitivity was performed by measuring root growth after 3 days cold treatment and 6-day-old seedlings were growing onto MS plates. To measure ABA sensitivity, seeds were plated on solid medium, composed of MS basal salts, 3 % sucrose and increasing concentrations of ABA (0, 0.1 0.3, 0.6 or 1 μM) and of NaCl (0, 20, 50 or 70 mM) [22]. In order to score seed germination, the percentage of seeds which had germinated and developed fully green expanded cotyledons was determined.
Drought stress and water-loss assays
The two different water-loss assays were performed. The short-term water-loss assays were performed in detached leaves at the same developmental stage and size from 20-day-old plants. The short-term water-term assay was been described previously [2]. Four samples of five leaves per genotype were excised and fresh weight was determined by submitting the leaves to the drying atmosphere of greenhouse at 22 °C for 4 h. Data were averages ± SE from three independent experiments (n = 5). The difference in weight was considered as water loss.
Long-term water-loss assays were performed after removing watering in 20-day-old plants maintained under greenhouse conditions. To this end, plants (10 individuals per experiment, three independent experiments) were grown under normal watering conditions for 21 days and then subjected to drought stress by completely terminating irrigation under greenhouse conditions [44].
Construction of pHAI-1::GUS and histochemical staining
The promoter region was amplified from the genomic DNA of Columbia ecotype and fused to a GUS reporter gene in the binary vector pGKB5, which contained a promoterless GUS reporter gene fused to the right border, and the genes conferring kanamycin and Basta resistance as plant selection markers with primers as Table 1 using the gateway cloning system (invitrogen) [51]. The binary plasmid derived from a pBGS plasmid [48], harbors a bacterial kanamycin resistance gene, and is able to replicate both in Escherichia coli and in Agrobacterium. The GUS-nos3′ reporter cassette from pBI101.1 [21]. The constructed fusion expression vector pGKB5 was introduced into Columbia by vacuum infiltration, and T2 lines were used for analysis. GUS staining was performed for 2–3 h at 37 °C using X-Gluc (Sigma-Aldrich, St. Louis, MO, USA) as described previously [21]. Finally, the tissue was mounted in 70 % ethanol on a glass microscope slide and observed under an olympus BX51 microscope (Olympus Corporation, Japan), as described previously [50]. Histochemical GUS reporter assay was performed.
Table 1.
Primers used for PCR and qPCR analysis
| Genes | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| PP2CA2oxF | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCCGGAATTCATGGCTGAGATTTGTTACGAGAA | GGGGACCACTTTGTACAAGAAAGCTGGGTGCCGCCCGGGTTACTTATGATCGGAGGATAAAGCAA |
| PP2CA2oxB | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCCCGGAATTCATGCCAGACCGTC CGGACGA | GGGGACCACTTTGTACAAGAAAGCTGGGTGCCGCCCGGGCTACGTGTCTCGTCGTAGATCA |
| pOK | TGGTTCACGTAGTGGGCCATCG | |
| pLB | AGAAGTATTCACGCACCAAGG | AATCTCAGCCATGTGATCGTC |
| 59220Promoter | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTGAATATCTTATAATTTTTGCCC | GGGGACCACTTTGTACAAGAAAGCTGGGTGTCTCTTCTCCTCCGCCTCTGTAA |
| PP2CA2ox | ATGGCTGAGATTTGTTACG | CTACGTGTCTCGTCGTAGA |
| 59220pEDST17 | GGGGACAAGTTTGTACAAAAAAGCAGGCTTC ATGGCTGAGATTTGTTACG | GGGGACCACTTTGTACAAGAAAGCTGGGTG CTACGTGTCTCGTCGTAGA |
| Actin2 | CACTGTGCCAATCTACGAGGGT | CACAAACGAGGGCTGGAACAAG |
Bombardment experiments
Each plate was shot twice. Each shot contains 270 μg gold particles (1.0 μm in diameter), and particles were coated with 2 μl of p35S:HAI-1-GFP recombinant plasmid at 0.5 μg/μl. The gold-coated DNA particles were delivered into onion epidermal cells using the PDS-1000/He biolistic particle delivery system (BioRad Laboratories, Hercules, CA, USA), and the bombarded onion epidermal peels were maintained at 25 °C for at least 12 h until they were examined by fluorescence microscopy (Nikon, Tokyo, Japan) [51].
Results
Physiological characterization of hai-1 mutants
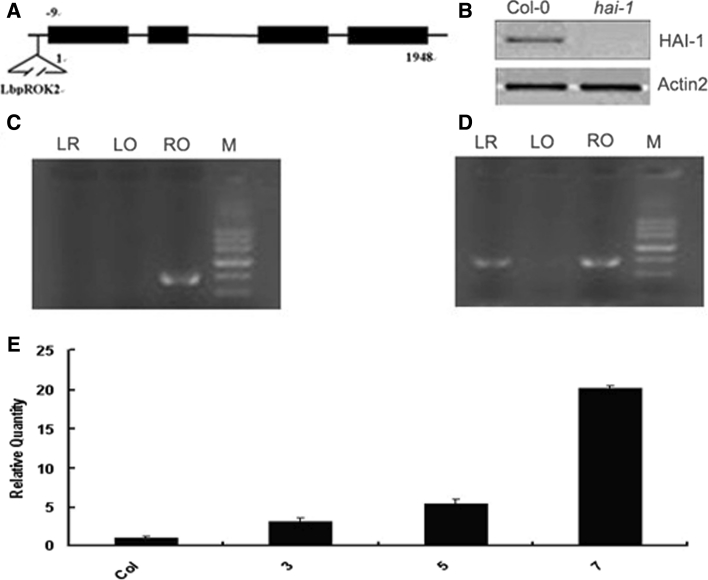
T-DNA insertion mutants of HAI-1 were identified in the Salk collection (Columbia background), corresponding to donor stock number Salk_039723, and it was named hai-1. Sequencing of the T-DNA flanking region in hai-1 showed that the insertion was localized nine nucleotides upstream of the ATG start codon (Fig. 1a). T-DNA insertions severely impaired HAI-1 expression, based on reverse transcription (RT) -PCR (Fig. 1b). Homozygous individuals were identified by three primers PCR method. The primers of sequences of T-DNA insertion loci of flanking region both sides and within T-DNA insertion fragment were designed, homozygous loss-of-function mutants were identified from genomic DNA with pOK/pLB/pRB primers (Table 1). PCR amplification results were shown that there was an electrophoretic band amplified from homozygous genomic DNA. Heterozygous genomic DNA can amplify two electrophoretic bands (Fig. 1c, d).
Fig. 1.
Physiological characterization of loss-of-function mutant hai-1 and HAI-1 over-expression transgenic plants a Diagrammatic representation of the T-DNA insertions in the At5g59220 gene. b RT-PCR analysis of mRNAs from wt and hai-1 mutant seedlings. c The loss-of-function mutant of HAI-1 gene was identified as homozygous mutant by gel electrophoresis. d The loss-of-function mutant of HAI-1 gene was identified as heterozygous mutant by gel electrophoresis. e Relative expression levels of HAI-1 gene in HAI-1 over-expression transgenic plants (3, 5 and 7) compared to wild type by Q-PCR analysis. The expression level of HAI-1 gene in Col-0 was defined as “1”
To further elucidate the role of HAI-1 in ABA signaling, we designed the primers according to its cDNA sequence of HAI-1 gene from 1 to 1,242 bp by high fidelity PCR polymerase. Making use of gateway system BP and LR reaction, the sequences were cloned into the entry vector pDONR201 and the expression vector pLeela, respectively. Homozygous mutant plants were identified by Q-PCR. The HAI-1 gene expression levels of over-expression transgenic lines 3, 5 and 7 were higher than that of Col-0 (Fig. 1e).
Phenotypic characteristics of over-expression mutants and wild type
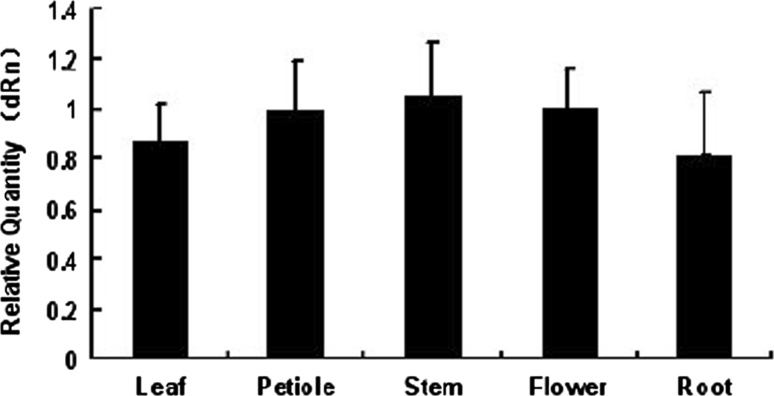
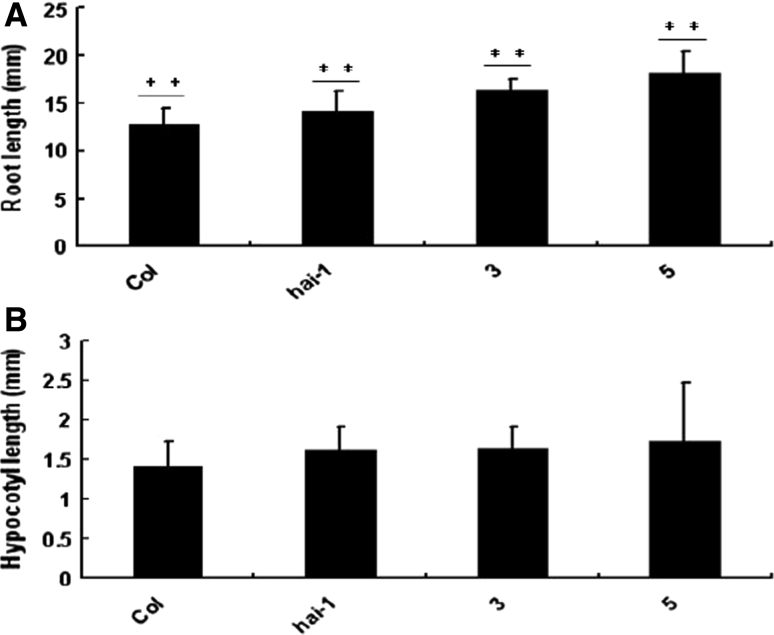
To examine the expression patterns in various young tissues, total RNA were extracted from different organs of 7-day-old Col-0 wild type plants. Q-PCR analyses showed that the higher levels of HAI-1 mRNA were observed in stems and flowers, and expressed at slightly lower levels in all of the organs (Fig. 2). 6-day-old seedlings of Col-0, loss-of-function mutants and HAI-1 over-expression transgenic plants were exposed to natural condition. There were a few differences in root length and hypocotyl length among these mutants. The root length and hypocotyl length of over-expression mutants were longer than those of loss-of-function mutants and Col-0 (Fig. 3).
Fig. 2.
The expression of HAI-1 gene in different tissues, including stem, root, leaf, petiole and flower of 15-day-old Col-0 seedlings by Q-PCR analyses
Fig. 3.
Phenotypes analysis of mutant plants a The means of hypocotyl lengths of seedlings of wild type, loss-of-function mutants and HAI-1 over-expression transgenic plants are shown. b The means of root lengths of seedlings of wild type, loss-of-function mutant and HAI-1 over-expression transgenic plants are shown. 6-day-old seedlings were scored after cold treatment. Values are averages ± SD (n = 30). *P ≤ 0.05, **P ≤ 0.01
Mutants of hai-1 confer NaCl insensitivity
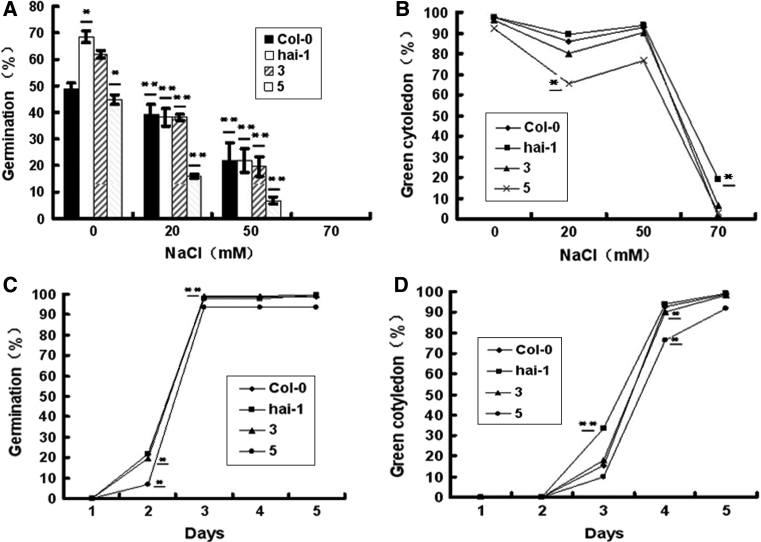
We then subjected the mutant seeds in the presence of various concentrations of NaCl (0, 20, 50 or 70 mM). A dose–response curve showed germination rates of mutant plants clearly decreased with increasing NaCl concentration at 2 days after stratification (Fig. 4a). The green cotyledon percentage of hai-1 is higher than that of others at 4 days after stratification in the presence of 20 mM NaCl. At 50 mM, the green cotyledon percentage of hai-1 mutants appear almost identical to that of wild-type plants (Fig. 4b).
Fig. 4.
NaCl-insensitive germination inhibition of hai-1 mutants as compared to wild-type and over-expression transgenic plant seeds a hai-1 mutants showed more NaCl-insensitive seed germination response at 2 days after cold treatment, b hai-1 mutants showed more green cotyledon percentage in the presence of various NaCl concentrations at 4 days after cold treatment, c seed germination kinetic profiles were determined in the presence of 50 mM NaCl, d green cotyledon percentage kinetic profiles were determined in the presence of 50 mM NaCl. Values are averages ± SD (n = 40–50), three replicates. *P ≤ 0.05, **P ≤ 0.01
The germination and green cotyledon kinetic profile showed hai-1 mutants had higher germination rate and green cotyledon percentage than the wild type and over-expression transgenic plants; especially over-expression line 5 was seen clearly significant (Fig. 4c, d). We can intuitively know about the germination rates and green cotyledon percentages were almost the same with 20 mM NaCl (Fig. 5a). However, the green cotyledon percentage of hai-1 mutants was higher than those of wild type and over-expression transgenic plants with 70 mM NaCl at 5 days (Fig. 5b). These experiments indicate that HAI-1 gene probably has sensitive function in NaCl-mediated seed germination inhibition response.
Fig. 5.
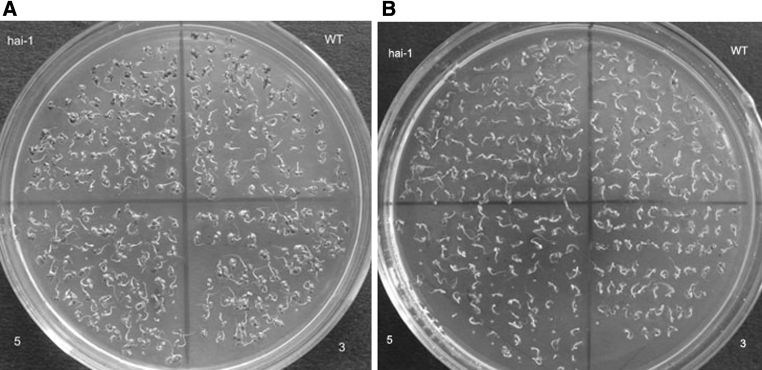
hai-1 mutants enhance NaCl-insensitive inhibition germination a The seed germination was determined in the presence of 20 mM NaCl. b The seed germination was determined in the presence of 70 mM NaCl. Photographs were taken at 5 days after cold treatment; 3, 5 represented HAI-1 over-expression transgenic plant lines
Mutants of hai-1 confer ABA insensitivity
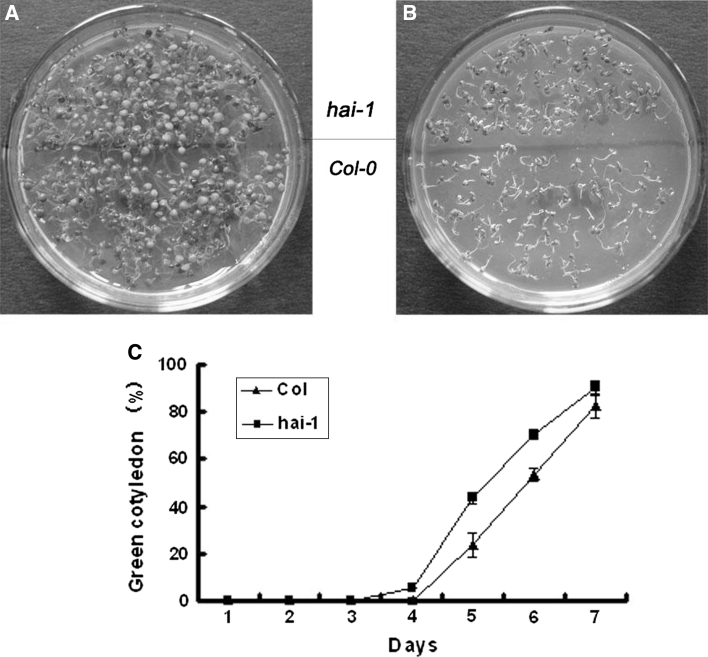
In order to investigate the relative contribution of At5g59220 to ABA signaling, we analyzed ABA-response of the At5g59220 loss-of-function mutant Salk_039723, which we have named hai-1. After 6 days culture on the medium containing 0 or 0.1 μM ABA, wild type (Col) and the loss-of-function mutant (hai-1) had the same germination inhibition (Data no shown). But the green cotyledon percentage of hai-1 was slightly higher than Col-0, the loss-of-function mutant hai-1 is insensitive to ABA response than Col-0 (Fig. 6).
Fig. 6.
Mutants of hai-1 slightly inhibit ABA response in seed germination. The comparison of seed germination sensitivity to the ABA stress between wild type and hai-1 mutant plants. Photographs were taken at 6 days after cold treatment. In MS culture without ABA (a) or with 0.1 μM ABA (b), c hai-1 mutants showed slightly higher green cotyledon percentage kinetic profiles with 0.1 μM ABA
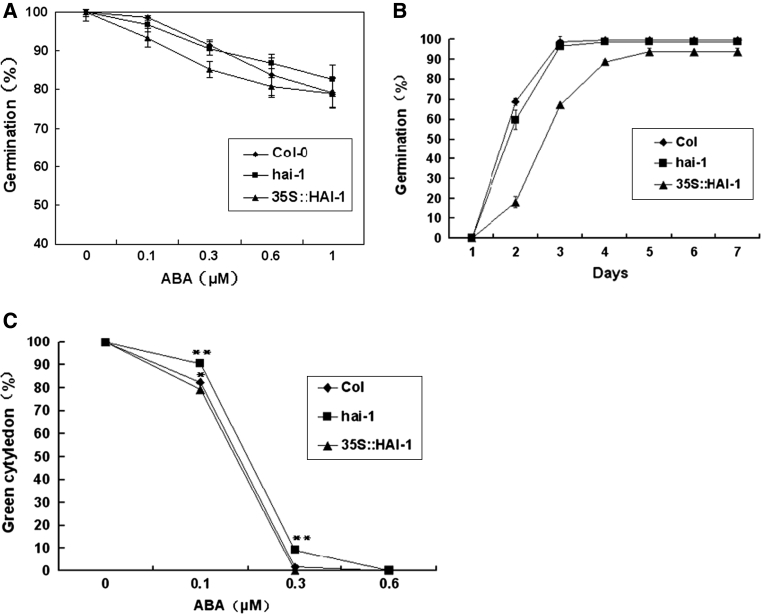
We then subjected the mutant seeds in the presence of various concentrations of ABA (0, 0.1, 0.3, 0.6 or 1 μM). The germination kinetic profile showed that more loss-of-function insertion mutant seeds germinated than wild-type seeds and over-expression mutant seeds at 3 days after stratification (Fig. 7a). The mutants of hai-1 had higher seed germination rates than the wild type and over-expression transgenic plants for 5 days in the presence of 0.1 μM ABA (Fig. 7b), the germination rates of all mutants eventually reach to 100 % at 7 days (Fig. 7b). The more obvious reduction in germination rate for the over-expression transgenic plants than wild-type and loss-of-function mutant plants. A dose–response curve showed that after 7 days in the presence of 0, 0.1, 0.3 or 0.6 μM ABA, the green cotyledon percentage of hai-1 mutants was always higher than that of over-expression transgenic plants and wild type plants (Fig. 7c).
Fig. 7.
Mutants of hai-1 were insensitive to ABA in seed germination inhibition. a Germination for ABA dose–response was scored at 3 days after cold treatment. b Over-expression transgenic mutants 35S::HAI-1 showed lower seed germination kinetic profiles with 0.1 μM ABA. c The green cotyledon percentage kinetic profiles of various mutants and wild type are shown in the presence of various ABA concentrations (0, 0.1, 0.3 or 0.6 μM) at 7 days after cold treatment. Values are averages ± SD (n = 40–50), three replicates. *P ≤ 0.05, **P ≤ 0.01
Adaptive responses to drought stress
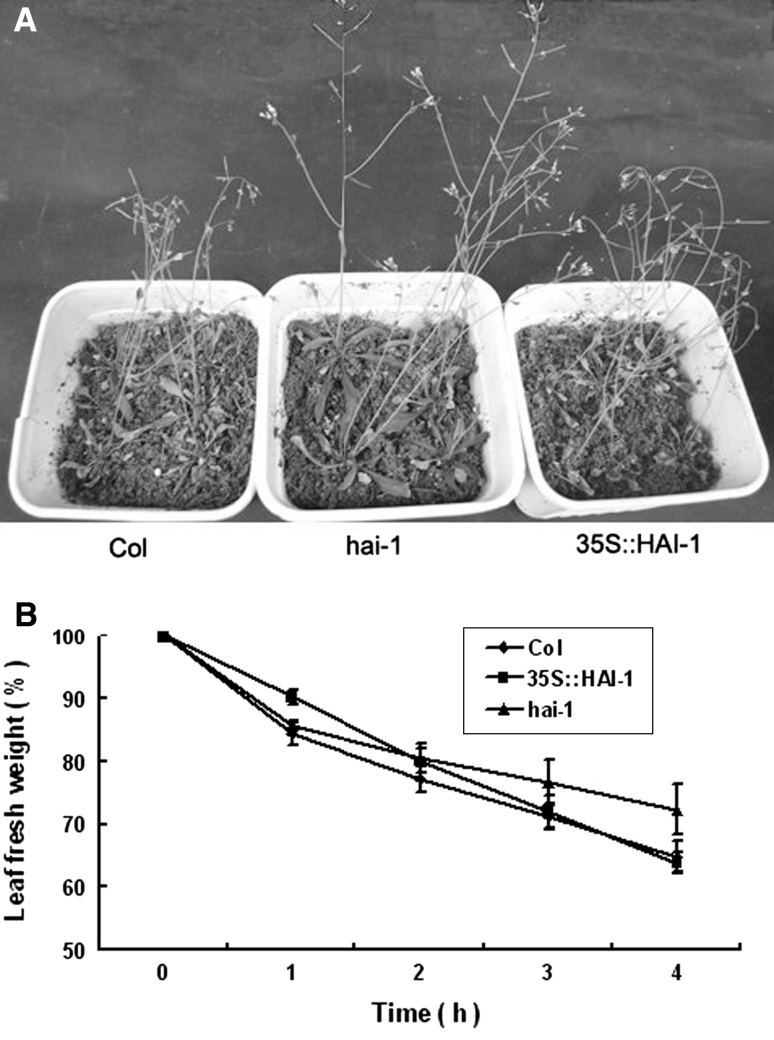
Water-loss data was obtained, under greenhouse conditions, after exposing 20-day-old plants to drought stress by completely withholding water. Figure 9a showed after 12 days without watering, wild-type plants wilted and many rosette leaves yellowed. A limited improvement was observed under these conditions, hai-1 mutants showed a reduced water loss as compared to wild type and over-expression transgenic plants.
Fig. 9.
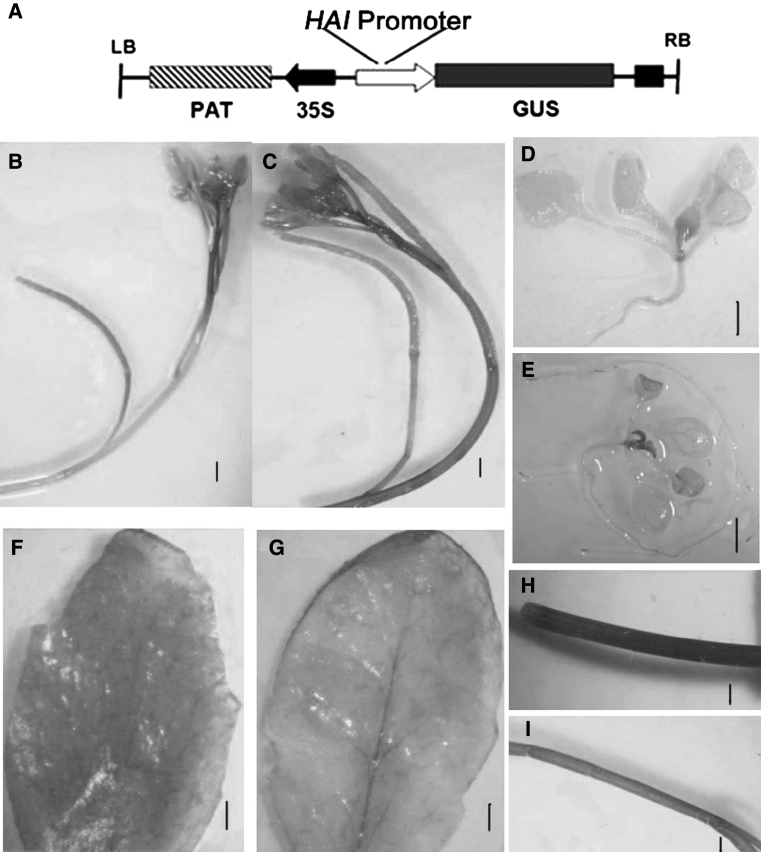
Expression patterns of HAI-1 as revealed by pHAI-1::GUS. a The schematic diagram of expression vector Promoter::Gw-GUS. b An inflorescence with flowers and stems without ABA retreatment. c An inflorescence with flowers and stems with 1 μM ABA retreatment for 2 h. d 8-day-old seedlings without ABA retreatment. e 8-day-old seedlings with 1 μM ABA retreatment. f Unwounded mature leave, g wounded mature leave. h Wounded stem cut-side. I, stem with 1 μM ABA retreatment for 2 h. b, c, f–i different type parts of a 30-day-old transgenic plant grown on soil. Bars 0.3 cm (b, c), 1 cm (d, e), 0.2 cm (f, g). 0.5 cm (h, i)
Under these experimental conditions, The HAI-1 over-expression transgenic plants did not exhibit significant differences in the transpiration rate of detached leaves compared to wild type (Fig. 8b). However, loss-of-function mutants exhibited a reduced water loss (Fig. 8b).
Fig. 8.
Water-loss assays was obtained a Enhanced drought resistance of hai-1 mutant plants with respect to wild type or over-expression transgenic plants. Photographs were taken 12 days after withholding water. b Reduced water loss measured in detached leaves of hai-1 mutants as compared to wild type or its over-expression transgenic plants. Values are averages from two independent experiments (n = 5)
Detached-leaf water-loss assays are likely not sensitive enough as to detect such variations [11], which are apparent after long periods of drought. Thus, whereas wild-type plants exhibited a marked water loss under these conditions, the ABA-hypersensitive mutants exhibited a reduced water loss, particularly in the case of over-expression transgenic plants. This result was consistent with previous studies conducted in USA [2].
The expression of HAI-1 in pHAI1: GUS transgenic plants
The 1,681 bp upstream region and 221 bp downstream region of the translational start codon ATG of HAI-1 was fused to the GUS gene, and the resulting construct was transformed into wild-type Arabidopsis plants to produce a set of 18 independent pHAI1:GUS lines. GUS activities were not detected in 8-day-old transgenic seedling’s leaf vein, root, or stem, and except only a little expressed in stem primordia. While the GUS staining can be detectable in whole seeding after induced 2 h by 1 μM ABA, including in root tip, in stem primordia, and in leaf vein (Fig. 9c, d). GUS activities were abundantly detected in the stem and leaf vein of 3–4 week-old mature transgenic plants, but barely detected in young silique after ABA implied (Fig. 9a, b). In addition, we also found that the HAI-1 promoter was induced under ABA and abiotic stresses such as wounding, GUS activities were largely observed at the wound around, such as in leaf vascular system and in around cut-side stem (Fig. 9e–h).
HAI-1 protein was localized to the nucleus
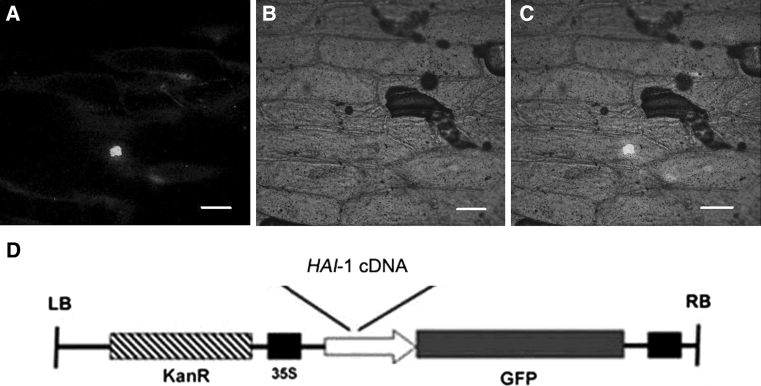
Proper elucidation of the subcellular localization of clade A PP2Cs is an important goal to better understand their role in plant physiology. To investigate the subcellular localization of HAI-1 protein, we fused the coding region of HAI1 to N-terminus of GFP under the control of cauliflower mosaic virus (CaMV) 35S promoter. The HAI-1-GFP fusion was both tested by bombardment assay into onion epidermal cells. Gene expression was revealed by the green fluorescence of the GFP marker and observed under a confocal microscope. The HAI-1-GFP fusion protein was localized to the nucleus in onion epidermal cells (Fig. 10).
Fig. 10.
Transient expression of HAI-1-GFP in the bombarded onion epidermal peels. The HAI-1-GFP fluorescence was found to be localized to the nucleus. a–c Fluorescence image, bright field image and merged fluorescence image of 35S::HAI-1-GFP. Bars 100 μm. d The schematic diagram of HAI-1:GFP fusion vector
Conclusion and discussion
According to the results of Q-PCR, the expression level of HAI-1 was very low (Fig. 2). At5g59220 is not expressed in seeds [34, 36, 57], but it is expressed in seedlings or different tissues of adult plants according to public microarray data [3].
ABA has an inhibitory effect on root growth, consequently, ABA-insensitive mutants are resistant to this ABA-mediated process [17]. In seeds, ABA has been shown to play an important role in the formation, maintenance of dormancy, and inhibition of germination [4]. The mutants of hai-1 had higher seed germination rates and green cotyledon percentages than the wild type and over-expression transgenic plants in the presence of exogenous ABA (Fig. 7). The results were amazingly consistent with the prior findings; the abi1-1 mutant displays a lower sensitivity than the wild type to the inhibition of seed germination by exogenous ABA [24].
In this study, we identify and characterize At5g59220 (HAI-1) gene disruption and over-expression phenotypes in Arabidopsis. The loss-of-function mutants hai-1 result in a strongly increased insensitivity to ABA during seed germination (Fig. 7). Moreover, constitutive expression of HAI-1 gene enhanced ABA-hypersensitive to exogenously applied ABA. ABA biosynthetic and signaling pathways can be considered as potential targets to improve plant performance under drought. Thus, it has been demonstrated that transgenic plants producing high levels of ABA display improved growth under drought stress than wild type [20, 42]. Alternatively, mutants affected in ABA signal transduction might also show an enhanced ABA response leading to stress-tolerant phenotypes.
Many examples of ABA-hypersensitive mutants have been reported [57]; however, in spite of the critical role of ABA to coordinate plant response to drought, a general correlation between enhanced response to ABA and drought tolerance has not been well established. The results showed after 12 days without watering, wild-type plants wilted and many rosette leaves yellowed (Fig. 8a). While amplified segment expression mutants showed the same phenotype as loss-of-function mutants (Fig. 8a). A limited improvement was observed under these conditions the single hai-1 mutants showed a reduced water loss as compared to wild type and over-expression mutants (Fig. 8).
Although some mutants with enhanced response to ABA have been shown to cause reduced water consumption [14, 38, 41], many examples of mutants that do not match this assertion is known. For instance, the mutants, which show ABA- hypersensitive inhibition of seed germination and super-induction of ABA-responsive genes, have compromised tolerance to drought stress [15, 53]. However, Mutants displayed ABA hypersensitivity and enhanced expression of ABA signaling genes did not correlate with stress-tolerance phenotypes [23, 38, 39]. The over-expression transgenic plants exhibited an enhanced water loss (Fig. 8b), the reason might be ABA- hypersensitive inhibition of seed germination had compromised tolerance to drought stress.
Previous leaf water loss analyses from detached leaves have shown that these assays can show phenotypic differences in mutations in which steady-state stomatal apertures already differ from wild-type controls prior to excising leaves [8, 27, 32]. The results showed mutants hai-1 display ABA hyposensitivity phenotype, however, correlate with stress-tolerance phenotypes, it needs to be further proved.
Together, these results point to an important function of HAI-1 as a negative regulator of ABA signal transduction events. The identification of a negative regulator in ABA signaling based on a cDNA over-expression screen shows that this approach can be used to isolate mutants in genes that modulate complex signaling networks in plants [16, 18, 46].
The expression of HAI-1 is very low under normal growth condition, detected ubiquitously in various organs according to microarray analysis (AtGenExpress), expressed mainly in the stem. However, the expression of HAI-1 is stronger induced by biotic and abiotic stress factors. Our results demonstrated that GUS activity could be detected in pHAI-1::GUS transgenic plants subjected to ABA and wounding. In unstressed conditions, ABI1 transcripts were the most abundant of the group-A PP2Cs, whereas the HAI-1 expression level was comparatively lower. Conversely, under water stress conditions, HAI-1 was drastically up-regulated in comparison with ABI1 [13]. In this study, stronger GUS activity could be detected in pHAI-1::GUS in the background of WT plants either treated or untreated (e.g. ABA or wounding) conditions. The GUS activity could be detected at the distal part of the roots (Fig. 9d). After induction by stresses, the GUS activity was stronger induced in roots, stem and around wounded area such as leaf vein (Fig. 9e). Basal transcript levels of At5g59220 are lower than those reported for other clade A PP2Cs; however, its expression is highly induced by ABA or osmotic stress [13, 56]. The experiment results are consistent with the previous research.
Interaction of PP2CA with the plasma membrane transporters AKT2 and SLAC1 has been reported [6, 28] and interaction of PP2CA and At5g59220 with SnRK2 s was localized to both nucleus and cytosol [13]. We found At5g59220 localized in the nucleus through bombardment assay (Fig. 10).
Therefore, At5g59220 overexpression leads to reduce seed germination, as compared with untransformed plants under inhibitory concentrations of ABA or high-osmoticum media. Finally, super-induction of ABA- and stress-inducible genes in overexpression transgenic plants do not appear to be sufficient to induce drought avoidance. Biochemical analysis of HAI-1 gene are lack, need to further studies on its activity.
Acknowledgments
This research was supported by the grants from National Natural Science Foundation of China (Nos. 30770200, 30871325 and 31071076), the Program for New Century Excellent Talents in University (NCET-10-0363) and by Hunan Provincial Natural Science Foundation of China (2010FJ4065 and 11JJ6015).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Contributor Information
Xinhong Guo, Email: gxh@hnu.edu.cn.
Xuanming Liu, Email: hnubio@126.com.
References
- 1.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 2.Angela S, Nadia R, Mohammad HM, Julian IS, Ramon Sand Pedro LR. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 2006;141:1389–1399. doi: 10.1104/pp.106.081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PL. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 2012;158(2):970–980. doi: 10.1104/pp.111.188623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady SM, Sarkar SF, Bonetta D, McCourt P. The abscisic acid insensitive 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003;34:67–75. doi: 10.1046/j.1365-313X.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- 6.Cherel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB. Physical and functional interaction of the Arabidopsis K (+) channel AKT2 and phosphatase AtPP2CA. Plant Cell. 2002;14:1133–1146. doi: 10.1105/tpc.000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacteriu-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 8.Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol. 2005;15:1196–1200. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 9.De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003;33:543–555. doi: 10.1046/j.1365-313X.2003.01652.x. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell Suppl. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelstein RR, Somerville CR. Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis defines genes that control overlapping subsets of ABA responses. Plant Physiol. 1990;94:1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii H, Verslues PE, Zhu JK. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi SK. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50(12):2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD. Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell. 2001;13:667–769. [PMC free article] [PubMed] [Google Scholar]
- 15.Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell. 1999;11:1897–1909. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hetherington AM, Woodward FI. The role of stomata in sensing and driving environmental change. Nature. 2003;424:901–908. doi: 10.1038/nature01843. [DOI] [PubMed] [Google Scholar]
- 17.Himmelbach A, Iten M, Grill E. Signaling of abscisic acid to regulate plant growth. Philos Trans R Soc Lond B. 1998;353:1439–1444. doi: 10.1098/rstb.1998.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himmelbach A, Yang Y, Grill E. Relay and control of abscisic acid signaling. Curr Opin Plant Biol. 2003;6:470–479. doi: 10.1016/S1369-5266(03)00090-6. [DOI] [PubMed] [Google Scholar]
- 19.Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 21.Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Bio Rep. 1987;5:387–450. doi: 10.1007/BF02667740. [DOI] [Google Scholar]
- 22.Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KN, Cheong YH, Grant JJ, Pandey GK, Luan S. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell. 2003;15:411–423. doi: 10.1105/tpc.006858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koornneef M, Reuling G, Karssen CM. The isolation and characterization of abscisic acid–insensitive mutants of Arabidopsis thaliana. Plant Physiol. 1984;61:377–383. doi: 10.1111/j.1399-3054.1984.tb06343.x. [DOI] [Google Scholar]
- 25.Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwak JM, Murata Y, Baizabal-Aguirre VM, Merrill J, Wang M, Kemper A, Hawke SD, Tallman G, Schroeder JI. Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol. 2001;127:473–485. doi: 10.1104/pp.010428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SC, Lan W, Buchanan BB, Luan S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA. 2009;106:21419–21424. doi: 10.1073/pnas.0910601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell. 2004;16:596–615. doi: 10.1105/tpc.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J. Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science. 1994;264:1448–1452. doi: 10.1126/science.7910981. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a relevance vector machine. Genome Res. 2006;16:414–427. doi: 10.1101/gr.4237406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang YK, Dubos C, Dodd IC, Holroyd GH, Hetherington AM, Campbell MM. AtMYB61, an R2R3-MYB transcription factor controlling stomatal aperture in Arabidopsis thaliana. Curr Biol. 2005;15:1201–1206. doi: 10.1016/j.cub.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 33.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 34.Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- 35.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007;50:935–949. doi: 10.1111/j.1365-313X.2007.03107.x. [DOI] [PubMed] [Google Scholar]
- 37.Nishimura N, Yoshida T, Murayama M, Asami T, Shinozaki K, Hirayama T. Isolation and characterization of novel mutants affecting the abscisic acid sensitivity of Arabidopsis germination and seedling growth. Plant Cell Physiol. 2004;45:1485–1499. doi: 10.1093/pcp/pch171. [DOI] [PubMed] [Google Scholar]
- 38.Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004;16:1912–1924. doi: 10.1105/tpc.021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 2005;139:1185–1193. doi: 10.1104/pp.105.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of start proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science. 1998;282:287–290. doi: 10.1126/science.282.5387.287. [DOI] [PubMed] [Google Scholar]
- 42.Qin X, Zeevaart JA. Over-expression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 2002;128:544–551. doi: 10.1104/pp.010663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quintero FJ, Ohta M, Shi H, Zhu JK, Pardo JM. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc Natl Acad Sci USA. 2002;99:9061–9066. doi: 10.1073/pnas.132092099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signaling. Plant J. 2004;37:354–369. doi: 10.1046/j.1365-313X.2003.01966.x. [DOI] [PubMed] [Google Scholar]
- 46.Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:627–658. doi: 10.1146/annurev.arplant.52.1.627. [DOI] [PubMed] [Google Scholar]
- 47.Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Spratt BG, Hedge PJ, Heesen ST, Edelman A, Broome-Smith JK. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 49.Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang QM, Tu XJ, Deng KQ, Zeng JX, Zhao XY, Tang DY, Liu XM (2009) A defect in double B-box zinc finger protein causes abnormal floral development in Arabidopsis. J Plant Biol 6(52):543–549
- 51.Wang QM, Zeng JX, Deng KQ, Tu XJ, Zhao XY, Tang DY, Liu XM. DBB1a, involved in gibberellin homeostasis, functions as a negative regulator of blue light-mediated hypocotyls elongation in Arabidopsis. Planta. 2011;233:13–23. doi: 10.1007/s00425-010-1274-y. [DOI] [PubMed] [Google Scholar]
- 52.Xie X, Wang Y, Williamson L, Holroyd GH, Tagliavia C, Murchie E, et al. The identification of genes involved in the stomatal response to reduced atmospheric relative humidity. Curr Biol. 2006;16:882–887. doi: 10.1016/j.cub.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 53.Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK. FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev. 2001;15:1971–1984. doi: 10.1101/gad.891901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, et al. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002;43:1473–1483. doi: 10.1093/pcp/pcf188. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis PP2Cs. Plant Physiol. 2006;140:115–126. doi: 10.1104/pp.105.070128. [DOI] [PMC free article] [PubMed] [Google Scholar]