Abstract
Rhabdomyosarcoma (RMS) is the most common type of pediatric soft tissue sarcoma. The MET receptor has an important role in the biology of RMS, and its overexpression and hyperactivation correlate with the metastatic ability of RMS. Consequently, interfering with MET expression or functionality may constitute a sound strategy for reducing the progression and metastatic potential of RMS. Our study reveals that downregulation of the MET receptor leads to changes in the morphology of ARMS cell in vivo. Tumors acquire a spindle shape that is characteristic of muscle fibers. Inhibition of MET expression or function leads to (i) a decreased expression of the early myogenic marker MyoD, (ii) a decreased ability of ARMS cells to metastasize to bone marrow cavities, (iii) downregulation of CXCR4 receptor expression and (iv) a decreased migration of MET-depleted cells towards gradients of HGF and SDF-1. Finally, we demonstrate that in vitro differentiation of alveolar RMS cells decreases their metastatic behavior by reducing both the expression of the MET and CXCR4 receptors and their migratory response to HGF and SDF-1. These findings suggest that blockers of MET receptor function and inducers of RMS cells differentiation may be clinically useful for reducing the aggressiveness and metastatic potential of RMS and may have significant implications for its treatment.
Keywords: MET, CXCR4, differentiation, tumor growth, metastasis, rhabdomyosarcoma
Rhabdomyosarcoma (RMS) is the most common type of pediatric soft tissue sarcoma.1 On the basis of histopathological features, RMS can be divided into four subtypes, with embryonal RMS (ERMS) and alveolar RMS (ARMS) being the most prevalent.2 Cases of ARMS exhibit a significantly worse prognosis than ERMS case, as indicated by a higher risk of both relapse and metastasis in ARMS cases. This poor prognosis has been linked with the particular characteristic of ARMS cells undergoing chromosomal translocation, leading to the expression of the fusion proteins PAX3-FKHR (t(2:13)(q35:q14)) and PAX7-FKHR (t(1:13)(p36:q14)). In contrast, ERMS tumors do not carry a distinct genetic lesion and generally follow a more favorable course; however, a 11p15-loss of heterozygosity has been found in some cases.3, 4, 5, 6, 7
The origin of RMS has yet to be fully elucidated. According to one hypothesis, it develops from satellite cells,7 while a contrasting theory proposes it has a mesenchymal origin.8, 9 However, the common assumption is that the development of this tumor is closely related to defects in the proper differentiation of stem or progenitor cells.10
The MET receptor, encoded by MET proto-oncogene, belongs to a family of growth factor receptors that have intrinsic tyrosine kinase activity.11, 12 MET activation by its ligand, hepatocyte growth factor (HGF) (also known as scatter factor), leads to pleiotropic effects in various target cell types, including morphogenetic transformation, induction of cell proliferation, cell motility and invasiveness under both normal and pathological conditions.13 MET has been reported to be overexpressed in a variety of human tumors, where it has a crucial role in malignant transformation.14, 15, 16 Moreover, RMS tumors have been shown to express the MET receptor, with the ARMS subtype exhibiting the highest level of expression.17 It has been postulated that the deregulation of MET activity is a key event underlying tumor metastasis,12, 18 as its overexpression and hyperactivation have been shown to robustly correlate with the metastasis of epithelial cancer cells,13 colon cancer cells19 and head and neck squamous cancer cells.20 Intriguingly, the downregulation of MET receptors on fibrosarcoma cells has also been reported to reduce tumor growth, invasion, and motility.21 Additionally, treatment with MET receptor inhibitors has been found to reduce tumor burden and decrease the metastasis of ovarian,22, 23 breast, lung and glioma tumor models.24
Hence, decreasing MET receptor expression or functionality may constitute a sound strategy for reducing the progression and metastasis of RMS. Our group demonstrated previously that the downregulation of the MET receptor not only reduces tumor growth but also reduces the short-term engraftment potential of ARMS cells.25
The chemokine receptor CXCR4, expressed by RMS cells, has also been found to affect the metastatic potential of this tumor.26, 27 Additionally, CXCR4 overexpression and hyperactivation has been shown to correlate with the metastatic ability of breast cancer cells.28 Consistent with this finding, blocking of CXCR4 receptor expression inhibits tumor growth in mouse models of breast cancer.29
A tumor can be regarded as a disease of cell differentiation. However, the molecular mechanism that renders cells unable to differentiate during carcinogenesis is not well understood and most likely varies between tissue and cancer types.30 Several agents with the ability to induce differentiation have been studied over the past few years, with some already in clinical use.10, 31, 32
Previous research has not evaluated the influence of MET receptor expression on RMS cell differentiation in detail. Physiologically, MET is rapidly downregulated at the onset of myogenic differentiation.33 Previously, Taulli et al.34 demonstrated that the re-expression of microRNA-206 (miR-206) in human RMS cells induces myogenic differentiation and blocks tumor growth in xenografted mice by switching the global mRNA expression profile to one resembling that of mature muscle. In the study, the authors postulated that the differentiation process was connected to MET receptor expression, underscoring its importance for RMS.
Because the MET and CXCR4 receptors have a key role in the progression of RMS tumors, we evaluated the influence of MET downregulation on the metastatic potential of RMS cells, both in vivo and in vitro. Furthermore, because rapid downregulation of the MET and CXCR4 receptors takes place during the onset of myogenic differentiation, we investigated how the differentiation of RMS cells influences MET and CXCR4 receptor expression and activation.
Results
MET-dependent differentiation of RMS cells in vivo
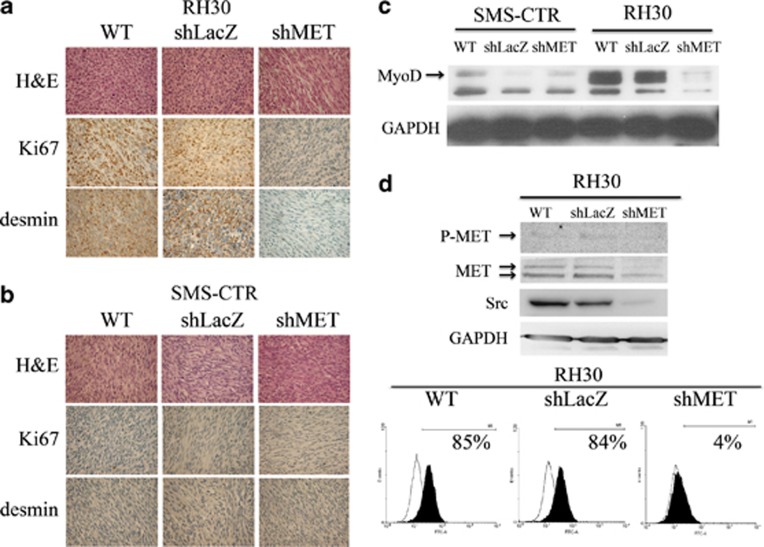
As previously reported,25 NOD-SCID mice were injected with two types of control cells (WT and shLacZ) as well as shMET RH30 and SMS-CTR cells. Hematoxylin and eosin staining revealed that the morphology of tumors derived from the control cells of both subtypes differed significantly. The tumors derived from control alveolar RH30 cells had a disorganized and immature appearance (Figure 1a), while the architecture of tumors from embryonal SMS-CTR cells showed cell linings that were close to each other, similar to muscle fibers (Figure 1b). Interestingly, the morphology of tumors derived from shMET RH30 cells also resembled the morphology of the more mature and differentiated, spindle-shaped SMS-CTR cells (Figure 1a). Immunohistochemical examination revealed that control RH30 cells had a high expression of desmin and Ki67, whereas tumors derived from shMET RH30 cells were characterized by a lack of both Ki67 and desmin expression (Figure 1a). SMS-CTR control and shMET SMS-CTR cells did not exhibit expression of these markers (Figure 1b).
Figure 1.
Histopathological, molecular and flow cytometry analysis of control and shMET ARMS and ERMS cells. The comparison of ARMS, RH30, (a) and ERMS, SMS-CTR, (b) tumors derived from WT and shMET transduced RMS cells showed differences in morphology, Ki67 and desmin expression. (a) H&E staining (upper panel) revealed that the morphology of MET-depleted ARMS tumors resembles the morphology of more differentiated ERMS tumors; Ki67-staining shows a reduced level of proliferation in MET-depleted tumors (middle panel). MET-depleted tumors did not express desmin, suggesting they have a more mature phenotype (lower panel). (b) H&E staining (upper panel) shows the spindle shaped morphology of ERMS tumors regardless of MET receptor expression; Ki67-staining shows a reduced proliferation rate (middle panel), and these tumors did not express desmin (lower panel). (c) Western blot analysis of MyoD expression shows weak staining in SMS-CTR cells and complete downregulation of MyoD expression in shMET RH30 cells. (d) Western blot analysis of MET tyrosine-phosphorylation shows a lack of MET receptor phosphorylation and a reduced level of total MET and Src expression in shMET cells (upper panel). Flow cytometry analysis of the MET receptor expression (lower panel). Representative staining of one tumor out of six is shown. Western blot and FACS analysis were performed at least three times with similar results. Representative results are shown
As the morphology of tumors derived from shMET RH30 cells indicated a more differentiated phenotype, we investigated the influence of MET receptor downregulation on the expression of MyoD, a marker of undifferentiated muscle cells. In contrast to control cells, shMET RH30 cells exhibited a strong decrease in the expression of MyoD (Figure 1c). In contrast, SMS-CTR cells showed a low expression of MyoD regardless of MET receptor status (Figure 1c). These data are the first to indicate that MET receptor expression may be linked to the undifferentiated status of ARMS tumors.
Src kinase expression and activation have been linked to MET receptor signaling. Therefore, we also investigated the levels of MET-receptor phosphorylation and the expression of Src kinase in the RH30 cell line. Phosphorylated MET protein appeared as a very faint band in control cells and was absent in shMET RH30 cells (Figure 1d). Unexpectedly, MET receptor downregulation caused a strong decrease in Src kinase expression. Using flow cytometry analysis to assess the level of MET receptor downregulation in the RH30 cell line, we found that expression was robustly decreased from ∼85% in control cells to ∼4% in shMET cells (Figure 1d).
MET-depleted cells exhibit a reduced ability to metastasize to bone marrow (BM) in vivo
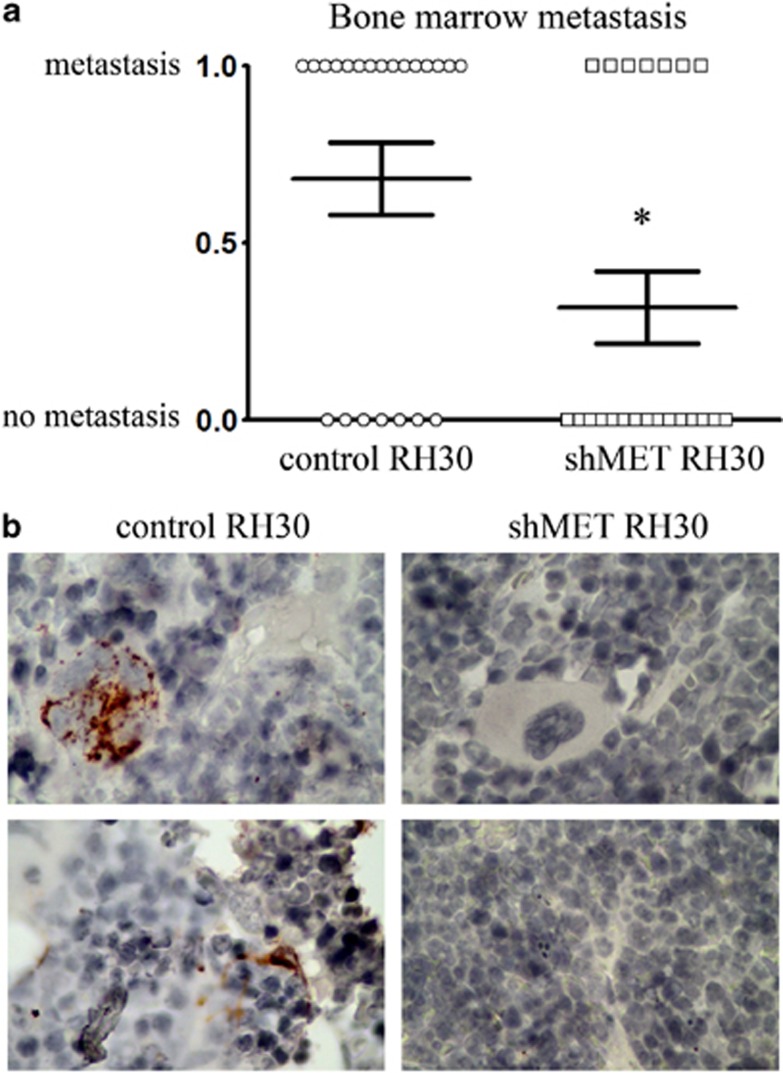
Our studies of the metastatic potential of RH30 cells show that mice injected intra subcutaneous (i.sc.) with control RH30 cells had considerably more BM metastasis than mice injected with shMET RH30 cells after 30 days of tumor growth. In mice injected with control RH30 cells, 68% of legs examined had BM micrometastasis. However, only 36% of the legs of animals injected with shMET cells contained BM micrometastasis (Figure 2a). The confirmation of the appearance of human RMS cells in murine BM is presented in Figure 2b (tumor cells were stained immunohistochemically) and in Supplementary Figure 1 (tumor cells were detected by immunofluorescence). These data suggest that MET receptor expression is important for the metastatic process of RMS tumors.
Figure 2.
BM metastatic ability of tumors derived from control and shMET cells. (a) The graph shows the percentage of legs with and without metastasis in the BM for control RH30 and shMET RH30 cells. Each point on the graph represents one leg (WT–circle, shMET-square). These experiments were performed twice with n=6 and n=5, giving the overall number of 22 legs for each group. The results were combined and submitted to χ2 analysis. χ2, df=4.464. *P<0.05. (b) Immunohistochemistry of murine BM. Human RMS cells are visualized using anti-vimentin staining. 600 × magnification
Mouse hepatocyte growth factor does not activate migration of human RMS cells
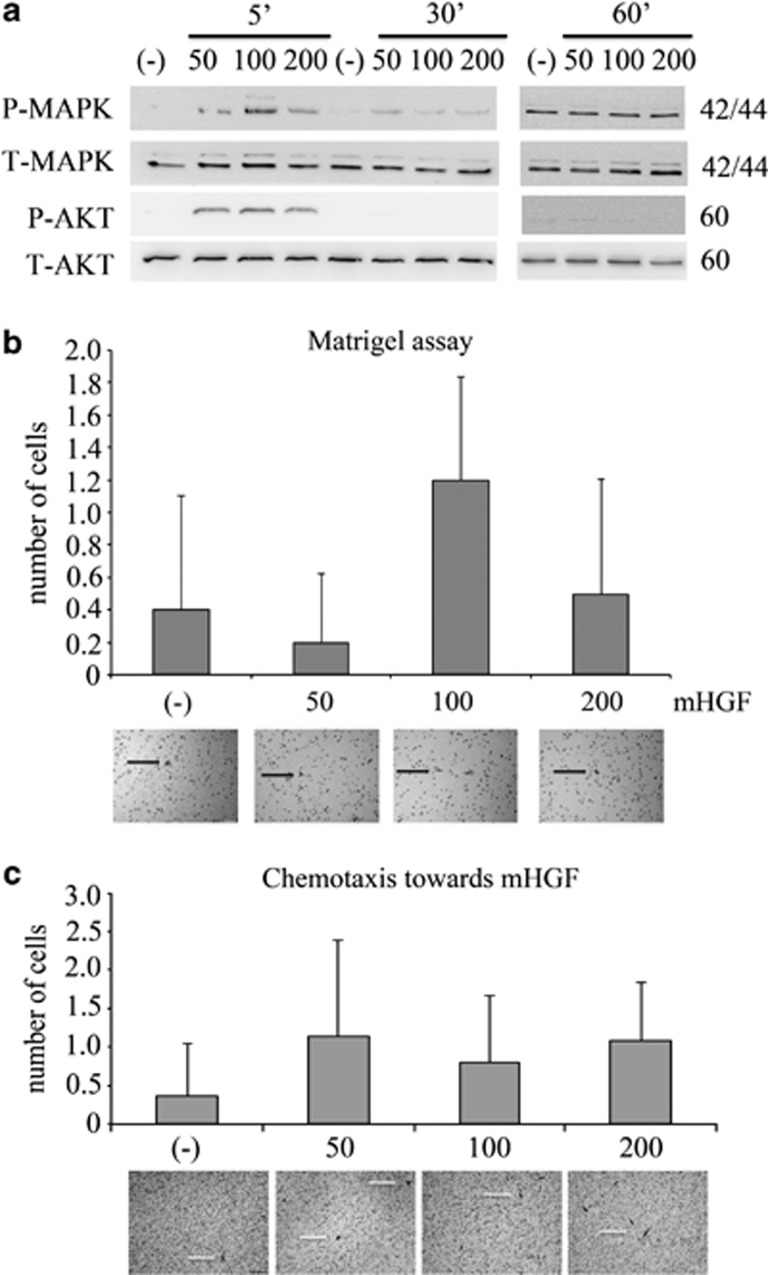
Even though recent findings have revealed that mouse HGF is able to activate human cells,35 some authors have postulated that murine HGF does not activate human MET receptor.36, 37 When we examined phosphorylation levels of the AKT and MAPK kinases, which are proteins involved in cell movement and directional chemoattractant response, we observed kinase activation after 5 min. However, after 30 and 60 min of stimulation, AKT and MAPK were not phosphorylated (Figure 3a). In addition, this short activation time of MET receptor was not sufficient to cause either an invasion or chemotaxis of RH30 cells toward a mouse HGF gradient (Figures 3b and c).
Figure 3.
Activation of RH30 cells by mouse HGF. (a) Western blot analysis of AKT and MAPK phosphorylation following stimulation with different doses of mouse HGF. Activation of AKT and MAPK kinases was observed after 5 min of stimulation with mouse HGF at concentration levels of 50, 100 and 200 ng/ml. (b) In invasion assays, RH30 cells did not exhibit any migratory responses through the Matrigel towards a mouse HGF gradient (50, 100, 200 ng/ml). (c) RH30 cells did not show any chemotactic activity towards murine HGF after stimulation with 50, 100 and 200 ng/ml of mouse HGF. Cells that migrated through pores in the membranes in invasion and chemotaxis assays are indicated by white arrows. Bar=50 μm
Decrease of CXCR4 expression and signaling in MET receptor-depleted cells
Because mouse HGF was insufficient to induce metastasis in RMS cells, we subsequently looked for other surface receptors influenced by the presence of MET receptor expression that could be responsible for the decreased ability of RMS cells to engraft to the BM of NOD-SCID mice. One receptor known to be involved in RMS BM metastasis is CXCR4.26, 27, 28
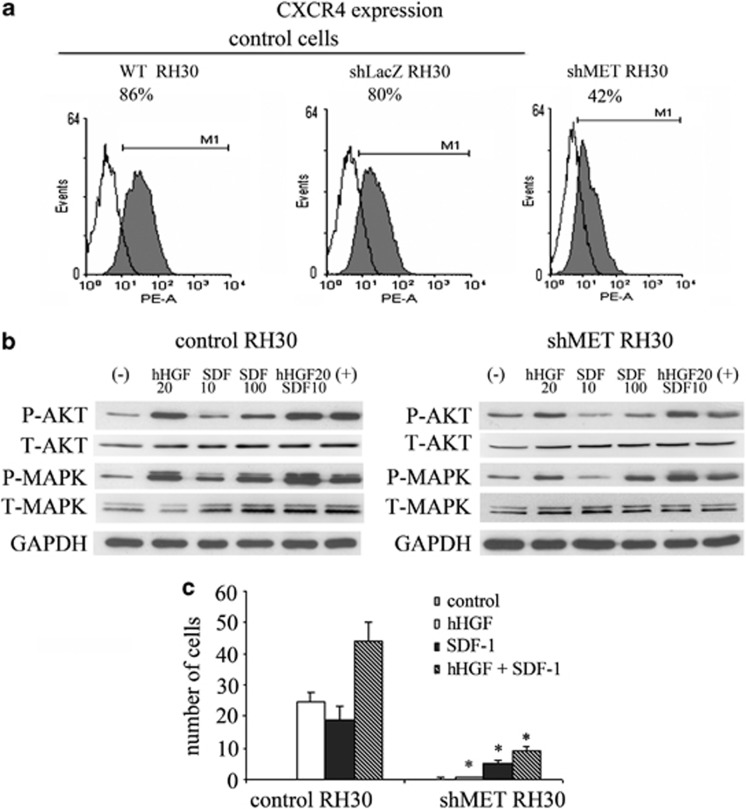
To evaluate how MET receptor downregulation mediates the expression and activation of the CXCR4 receptor, we used MET-depleted cells to examine both CXCR4 expression and activation. We found that cytofluorometric characterization of shMET RH30 cells exhibited a decreased expression of the CXCR4 receptor (Figure 4a). Downregulation of the MET receptor also led to reduced phosphorylation of MAPK and AKT following SDF-1 stimulation in ARMS cells (Figure 4b). Moreover, the reduced expression of CXCR4 after MET receptor downregulation correlated strongly with the severely reduced chemotactic ability of RMS cells in response to a SDF-1 gradient (Figure 4c). These results partially confirm our hypothesis that CXCR4 may be responsible for the BM micrometastasis of RMS cells.
Figure 4.
Expression and function of the CXCR4 receptor in MET-depleted cell. (a) FACS analysis of CXCR4 staining in RH30 cells. A considerable decrease in the expression of the receptor in shMET RH30 cells when compared with control cells was noted. (b) Western blot analysis of AKT and MAPK activation in shMET RH30 cells showed a decrease in the phosphorylation of these kinases following stimulation with human HGF (20 ng/ml) and SDF-1 (10 ng/ml and 100 ng/ml). (c) Chemotaxis assay showed a significant decrease in the cell migration of shMET RH30 cells towards both HGF and SDF-1 gradients when compared with control cells. *P<0.05
Inhibition of MET activation decreases CXCR4 expression and responsiveness to SDF-1 in RMS cells
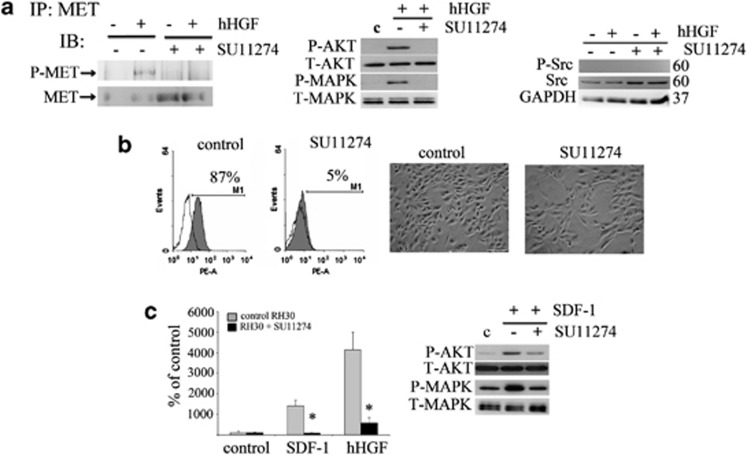
Next, we wanted to know whether blocking MET receptor activation influences the expression and functionality of the CXCR4 receptor. To answer this question, we used the specific MET receptor phosphorylation inhibitor, SU11274. To verify the ability of SU11274 to block the activity of the MET receptor, RH30 cells were stimulated with human HGF. The MET receptor was phosphorylated on tyrosine 1234/1235 following human HGF stimulation, whereas preincubation with SU11274 strongly inhibited MET receptor phosphorylation (Figure 5a left panel). As expected, downstream phosphorylation of MAPK and AKT kinases by human HGF was also decreased in inhibitor-treated cells (Figure 5a middle panel). The level of total Src seemed to increase slightly after SU11274 treatment; however, the phosphorylated form of Src was undetectable both in the control and in SU11274-treated cells (Figure 5a right panel). Interestingly, we observed a significant decrease in CXCR4 receptor expression on the surface of RH30 cells following treatment with SU11274 using flow cytometry analysis (Figure 5b). Moreover, we noticed that the morphology of the MET inhibitor-treated cells changed, becoming more elongated and spindle-shaped (Figure 5b). The phosphorylation of AKT and MAPK kinases after SDF-1 stimulation was also inhibited in the RH30 cells treated with SU11274 (Figure 5c). In addition, we found defects in the chemotactic response of treated cells to both SDF-1 and HGF gradients (Figure 5c). At the same time, their proliferation potential remained unchanged (data not shown). These findings further confirm our in vivo data and clearly show that MET and CXCR4 expression has a functional role in the migration and differentiation of RMS cells.
Figure 5.
Negative influence of the MET inhibitor (SU11274) on CXCR4 signaling. (a) The expression of phospho-MET after human HGF stimulation of control cells. Preincubation with SU 11274 abolished this phosphorylation. Phospho-MET was immunoprecipitated from 1 mg of protein extracts with an anti-MET antibody. Corresponding blots were developed after staining with anti-phospho-MET (Tyr 1234/1235) (left panel). Western blot analysis of AKT and MAPK activation in inhibitor treated cells showed a reduced activation of these signaling pathways following stimulation with 20 ng/ml human HGF (middle panel). Western blot analysis of phospho-Src showed no expression of this protein, whereas the total level of Src increased slightly after MET receptor inhibition using SU11274 (right panel). (b) Flow cytometry analysis of CXCR4 receptor expression and morphology assessment after inhibitor treatment. RH30 cells were treated with 5 μM of SU11274 for 16 h before experiments. The decrease in the level of surface expression was observed and the changes in the morphology of SU11274-treated cells were noted. Cells became more elongated and resembled differentiating cells. (c) The chemotactic analysis of RH30 cells. The ability of RH30 cells to migrate toward the human HGF (20 ng/ml) and SDF-1 (100 ng/ml) gradients was severely reduced after SU11274 treatment (left panel). Western blot analysis of AKT and MAPK phosphorylation after SDF-1 stimulation (100 ng/ml) showed a reduced activation of these signaling pathways following incubation with SU11274 (right panel). Western blot and FACS analysis was performed a minimum of three times and representative results are shown. The chemotaxis assay was performed three times in duplicates. Bar=50 μm; *P<0.05
Differentiation of RMS cells causes decreased expression and signaling of the MET and CXCR4 receptors
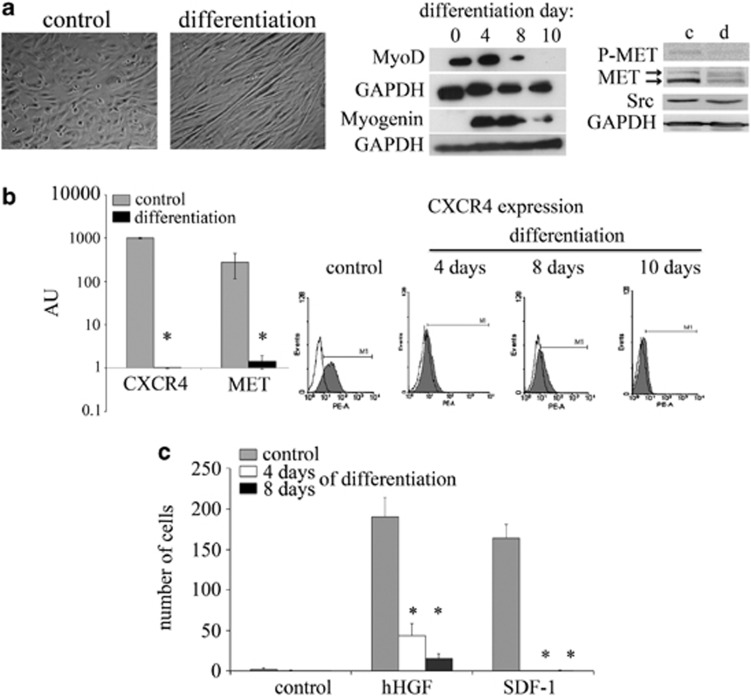
On the basis of our in vivo observation that MET-depleted tumors were more differentiated and our in vitro observation that murine C2C12 cell differentiation led to loss of chemotactic responsiveness to a SDF-1 gradient (Supplementary Figure 2), we speculated that the differentiation process influences the expression and/or functionality of the MET and CXCR4 receptors. To confirm our hypothesis, RH30 cells were subjected to differentiation. During the differentiation process, the cells altered their morphology, becoming elongated and spindle-shaped (Figure 6a left panel). An analysis of muscle differentiation markers revealed a decrease in the expression of MyoD and an increase in the expression of myogenin (Figure 6a middle panel). During the differentiation process, a significant decrease in MET expression was observed, in conjunction with a decrease in MET tyrosine phosphorylation and a slight decrease in Src expression (Figure 6a right panel). Remarkably, we also observed a decrease in CXCR4 receptor expression at both the mRNA (Figure 6b left panel) and protein levels (Figure 6b right panel). Subsequent evaluation of the migratory properties of differentiated RH30 cells revealed a strong inhibition of human HGF- and SDF-mediated migration (Figure 6c), which positively correlated with the decreased expression of these receptors. These data offer further evidence that MET receptor modulates ARMS cell differentiation and strongly influences the metastatic ability of ARMS cells.
Figure 6.
Decreased expression and signaling of the MET and CXCR4 receptors in differentiated RMS cells. (a) The morphology of RH30 cells under differentiation inducing conditions. The differentiation process of RH30 cells was induced using low serum (2% HS) and TPA (100 nM). The cells were analyzed on days 4, 8 and 10. Beginning on day 4, morphological changes became apparent. The formation of characteristic elongated cellular structures was observed in cells cultured under differentiation conditions. The morphological changes were accompanied by the changes in the expression of early (MyoD) and late (myogenin) muscles differentiation markers. The expression of MyoD decreased to an undetectable level during the differentiation process. At the same time, the level of myogenin increased during the first 8 days (middle panel). Western blot analysis of phospho-MET and total MET showed significantly reduced signals for these proteins after 10 days of differentiation, in addition to a slight decrease of total Src protein. GAPDH was used as a loading control (right panel). The expression of the MET and CXCR4 receptors was evaluated in both undifferentiated and differentiated RH30 cell lines. (b) RT-PCR analysis of MET and CXCR4 receptor expression. The significant downregulation of the MET and CXCR4 receptors was observed at the mRNA level (left panel). Flow cytometry analysis of CXCR4 receptor expression in RH30 cells. The expression of the CXCR4 receptors decreased in a time-dependent manner and was completely gone after 10 days of differentiation (right panel). (c) Chemotactic assay of RH30 cells. The differentiated cells were subjected to a chemotaxis assay towards SDF-1 and human HGF gradients. A strong inhibition in the migration properties to both chemoattractants was observed. The experiment was repeated three times with similar results. Bar=50 μm; *P<0.05
Discussion
Despite recent advances, the prognosis for cases of RMS in advanced metastatic stages is still bleak. In our study, we examined how the downregulation of MET receptor expression and the induction of RMS cell differentiation affect the metastatic potential of RMS cells. Our group25 and others34 have previously shown that MET downregulation causes a decrease in the size of tumors derived from RMS cells transplanted into NOD-SCID mice. These findings provide further explanation for this mechanism that may be responsible for the reduction in tumor size. We demonstrate, for the first time, that MET receptor downregulation, in conjunction with the inhibition of HGF- induced MET activation, results in profound differentiation of ARMS tumor cells in vivo. MET-depleted ARMS cells acquire the morphology of the more differentiated spindle-shaped ERMS cells.
Our immunohistochemical analysis also confirmed that MET-depleted tumors had a reduced proliferation potential (as evidenced by the lack of Ki67 expression) and lacked expression of the marker desmin. This lack of desmin expression was also observed in SMS-CTR cells, regardless of MET expression. Previous studies have found desmin expression in both alveolar and embryonal RMS's.38 Lack of desmin was found to be a sign of a less differentiated RMS type,39 and the later appearance of desmin has been correlated with the differentiation process.40 However, there have also been reports showing that the downregulation of desmin expression, as a result of chemotherapy, correlates with morphological changes of RMS cells into differentiated strap-like cells.41
Nevertheless, there are no published data evaluating the expression of desmin in RMS-derived cultured cancer cell lines. Our observation that the inhibition of HGF-MET interactions through MET downregulation decreases desmin expression extends recently published suggestions that HGF may upregulate desmin expression in mesenchymal cells.42 Moreover, Anastasi et al.33 found that constitutive expression of the MET receptor causes morphological changes and prevents the differentiation of mouse satellite C2C12 cells. Thus, our data suggest that the reduction in tumor size following MET receptor downregulation is caused by the induction of tumor cell differentiation. This result directly links the expression of the MET receptor with the differentiation status of RMS cells.
To examine whether downregulation of the MET receptor influences the metastatic potential of RMS cells, NOD-SCID mice were injected subcutaneously with both control cells and MET-depleted RMS cells. After 30 days, the presence of these cells in bone marrow, one of the primary sites of RMS metastasis, was evaluated (Figure 2). Analysis revealed that the downregulation of the MET receptor does significantly reduce the metastasis of RH30 cells. These findings support our previous data showing that downregulation of the MET receptor expression inhibits the ability of RMS cells to engraft into BM during short-term assays.25 Recently, the MET receptor was also shown to have an important role in the BM metastasis of breast cancer.43
Because previous studies,36, 37 as well as our own observations (Figure 3), suggest that human RMS cells do not respond to murine HGF gradients in chemotactic assays and murine HGF cannot stimulate the migration of human RMS cells in an in vivo murine model, we speculated that a different mechanism mediated the reduced metastatic ability and lowered short-term engraftment potential of shMET RH30 cells.25
Our published data have revealed that the CXCR4 receptor is an extremely potent inducer of RMS cell migration.26 Moreover, we have previously shown that this receptor is expressed on murine satellite cells44 and our current research indirectly indicates that CXCR4 receptor expression decreases during differentiation of murine satellite cells (Supplementary Figure 2). Interestingly, MET receptor activation is correlated with CXCR4 receptor expression in both RMS and breast cancer cells.45, 46
On the basis of these findings, we became interested in the fate of the CXCR4 receptor in shMET RH30 cells. In this study, we demonstrate that the downregulation of MET receptor expression in RH30 cells causes a simultaneous downregulation of CXCR4 receptor expression and reduces its activity in these cells (Figure 4). To examine the mechanism underlying the decrease in CXCR4 expression and activation in MET-depleted cells, we used a specific inhibitor of the MET receptor. We found that inhibiting MET receptor activity was sufficient for decreasing the expression and activation of the CXCR4 receptor in RH30 cells. Because the viability and proliferation rates of treated and non-treated RMS cells were similar, the defect in HGF and SDF-1 responsiveness was not due to a toxic effect of the MET inhibitor.
Thus, for the first time we have demonstrated that the functional MET receptor is necessary for the expression and activation of CXCR4 in RMS cells. This finding indicates the existence of a direct relationship between these two receptors at the level of MET receptor activation. This finding also indicates that the expression and activation of the CXCR4 receptor is tightly regulated through a signaling pathway that is in turn regulated by MET receptor activation. Therefore, our results are consistent with the previous report of Jankowski et al.27 and further elucidate the relationship between the MET and CXCR4 receptors in RMS cells.
Similar data regarding the interrelation between these two receptors have been reported for both low and high invasive breast carcinoma cells. HGF stimulation was shown to lead to the activation of c-Src (signaling downstream of HGF/MET), and depending upon the signaling pathway, resulted in the activation or suppression of NF-κB and Ets1 transcription factors, causing a subsequent increase or decrease of CXCR4 expression, respectively.45 On the basis of these observations, it seems probable that the relationship between MET and CXCR4 in RMS cells is also connected with the c-Src, NF-κB and Ets1 signaling pathways. Our results show that the downregulation of MET receptor expression causes a strong decrease in Scr expression and that the differentiation of RMS cells also leads indirectly to a slight decrease in Scr expression.
In the case of RMS, other authors have linked the expression of the MET and CXCR4 receptors to the expression of the PAX3-FKHR fusion protein.46 On the basis of our results, we cannot challenge the correlation between the PAX3-FKHR transcription factor and the expression of the MET and CXCR4 receptors. However, recent reports on the effect of the direct binding of PAX3-FKHR to the CXCR4 promoter on CXCR4 expression have been questioned26, 47 These studies reveal that binding of PAX3-FKHR to regions other than the core promoter might be responsible for the expression of PAX3-FKHR-dependent genes.47 Our data not only corroborate this recent finding but also indicate that while the presence of PAX3-FKHR is necessary, it is insufficient for inducing CXCR4 expression without the concurrent activation of the MET receptor.
We also examined the effect of differentiation on the metastatic behavior of RMS cells. We observed an increased expression of myogenin and MYHC2, as well as myostatin (late muscles differentiation markers), in RH30, CW9019 RMS and C2C12 cells following the induction of differentiation (Supplementary Figure 3). Furthermore, the induction of RMS cell differentiation decreased the expression and activation of the MET and CXCR4 receptors, both of which are important for the metastatic ability of RMS tumors, especially the alveolar subtype.26, 27 The differentiation of RMS cells also strongly reduced their metastatic ability and decreased their migration toward HGF and SDF-1 gradients in vitro. Thus, for the first time, we have shown that the differentiation of RMS cells is directly connected with their ability to disseminate to distant organs.
Because satellite cells express both the MET and CXCR4 receptors, akin to RMS cells, we induced the differentiation of satellite cells and checked their ability to migrate toward HGF and SDF-1 gradients. In both cases, a strong decrease in migratory ability toward HGF or SDF-1 was observed. This similarity suggests that ARMS development is more likely linked to the differentiation defect of the satellite cells rather than to the differentiation defect of the mesenchymal cells.
The role of MET receptor activation in the pathogenesis of RMS has been well documented; however, the influence of MET expression on the differentiation of RMS has not been studied at all. Recent work by Taulli et al.34 showed that the re-expression of microRNA-206 in human RMS cells promoted myogenic differentiation and blocked tumor growth in xenografted mice by switching the global mRNA expression profile to one resembling mature muscle. Forced induction of miR-206 expression at the onset of normal myogenesis promotes this differentiation by modulating more than 700 genes, including the MET receptor, causing MET receptor downregulation in murine satellite cells and two RMS cell lines (ERMSRH4 and ARMSRH18),34 and the authors speculated that the differentiation process might be connected to MET receptor expression. A different study showed that both miR1 and miR133a induce differentiation in embryonal RMS, but not in alveolar RMS.48 These authors speculated that the presence of the PAX3-FKHR fusion gene might be responsible for the inhibition of tumor cell differentiation. In our study, the downregulation of MET expression was sufficient to induce the differentiation of ARMS, suggesting that out of those 700 genes, the deregulated MET receptor gene may be the most important factor in blocking the differentiation RMS cells. The fact that the downregulation of the MET receptor, regardless of the mechanism, restores the differentiation ability of these cells indicates there is a connection and dependency between the downstream signaling pathways of HGF-MET and other genes associated with differentiation.
Differentiation therapy using an ATRA agent was found to be beneficial for the treatment of APL patients,32 establishing a paradigm for the use of differentiation therapy in the treatment of other tumors. Because several different studies have linked the development and progression of RMS tumors with a defect in early muscle or mesenchymal cells,49 we postulate that the induction of RMS cell differentiation (using, for example, MET receptor blockers and inhibitors, similarly to ATRA) could be a new therapeutic strategy for advanced, aggressive, and metastatic RMS tumors.
Materials and Methods
Cell lines
C2C12 cells (ATCC), RMS cell lines used in this study (SMS-CTRERMS, RH30, CW9019ARMS) (kindly provided by Dr. PJ Houghton (Center for Childhood Cancer, Columbus, OH, USA)) were maintained in DMEM (Gibco,BRL Grand Island, NY, USA) supplemented with 10% heat-inactivated FBS (Gibco, BRL Grand Island, NY, USA), 100 IU/ml penicillin, 10 mg/ml streptomycin (Gibco, BRL Grand Island, NY, USA). Cells were cultured at 37 °C, 5% CO2 and 95% humidity. They were split usually twice a week with medium change. RMS cells were transduced directly with viral supernatants and subsequently selected with blasticidin (Invitrogen, Carlsbad, CA, USA) as has been shown.25 MET shRNA sequence 5′-AGUCCGAGAUGAAUGUGAAtt-3′ has been designed with use of available algorithm from Ambion http://www.ambion.com/techlib/misc/siRNA finder.html.
Reagents
Met Inhibitor SU11274: (3Z)-N-(3-Chlorophenyl)-3-((3,5-dimethyl-4-((4-methylpiperazin-1-yl) carbonyl) -1H-pyrrol-2-yl)methylene)-N-methyl-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide (Calbiochem, Darmstadt, Germany) was dissolved in DMSO and used at a concentration of 5 μM.50, 51, 52
Differentiation protocol
RH30 and CW9019 cells were subjected to differentiation by culturing in the differentiation medium composed of DMEM (Gibco BRL, Grand Island, NY, USA), 2% horse serum (HS) (Gibco BRL, Grand Island, NY, USA) and 100 nM TPA (Sigma, St Louis, MO, USA).10, 53 The cells were cultured in this medium for 4, 8 and 10 days, and subsequently used in appropriate experiments. C2C12 cells were subjected to the differentiation and myotube formation by culturing in the differentiation medium composed of DMEM (Gibco BRL, Grand Island, NY, USA) and 10% HS (Gibco BRL, Grand Island, NY, USA), according to ATCC recommendations.
Flow cytometry
For evaluation of CXCR4 receptor expression, cells were incubated with monoclonal PE-labeled anti-human CD184 (CXCR4) antibody, clone 12G5 (BD Pharmingen, San Diego, CA, USA).26 For detection of MET receptor expression, mononclonal FITC-labeled anti-human HGFR/c-MET antibody, clone 95106 (R&D) was used. Briefly, 1 × 105 cells suspended in 100 μl of staining buffer (PBS, 2% FBS) were added to a test tube containing the appropriate amount of each antibody. The cells were incubated in the dark for 30 min at 4 °C. The stained cells were then washed and collected using a FACSCanto cytometer (Becton Dickinson, San Jose, CA, USA) and finally analyzed with FACS Diva software (Becton Dickinson).
RNA extraction and reverse transcription
The total RNA was extracted using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) followed by DNAse treatment (Promega, Madison, WI, USA). The reverse polymerase transcription was performed using MMLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol.
Quantitative real-time PCR (qRT-PCR) analysis
Gene expression was determined by qRT-PCR analysis on ABI PRISM 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) using a commercially available TaqMAN PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The primers sequences were as follows: Human: GAPDH Hs99999905_m1, MET (Hs01565589_m1), CXCR4 (Hs00237052_m1), MYOSTATIN (Hs00976237_m1), MYOGENIN (Hs01032275_m1), MYHC2 (Hs00430042_m1), mouse: GAPDH (Mm99999915_g1), MYOSTATIN (Mn01254559_m1), MYOGENIN (Mn00446195_g1), MYHC2 (Mn01332564_m1); all from (Applied Biosystems, Foster City, CA, USA). The mRNA expression level for all samples was normalized to the housekeeping gene GAPDH. 2−ΔΔCt method allowed to calculate relative expression of the genes.
Western blot and Immunoprecipitation
Western blots were done on extracts prepared from cells as previously described.25 Briefly, RMS cells were lysed (for 10 min) on ice in M-Per lysing buffer (Pierce Rockford, IL, USA) containing protease and phosphatase inhibitors (Sigma, St. Louis, MO, USA). Subsequently, the extracted proteins were separated on a 12% sodium dodecyl sulfate–PAGE gel, and the fractionated proteins were transferred into a PVDF membrane (BioRad; BioRad Laboratories, Hecules, CA, USA). Phosphorylation of MET was detected by anti-phospho-MET (Tyr1234/1235) (Cell Signaling, Danvers, MA, USA) antibody. The phosphorylation of AKT and MAPK was assessed using primary rabbit anti-phospho-AKT (Ser 473) and mouse anti-phospho-MAPK (Thr202/Tyr204) antibodies both from Cell Signaling Danvers, MA, USA. The phosphorylation of Src was detected by monoclonal mouse anti-phospho-Src (Tyr416) antibody (clone 9A6; EMD Millipore Corp., Billerica, MA, USA) and the total level of Src was determined by monoclonal mouse anti-Src (clone GD11). The total level of MET and MyoD proteins was assessed using primary rabbit polyclonal antibodies (Santa Cruz Biotech., Santa Cruz, CA, USA), the expression of myogenin was assessed using primary mouse monoclonal antibody (Santa Cruz Biotech., Santa Cruz, CA, USA), and they were subsequently detected with horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse IgG secondary antibodies (Santa Cruz Biotech., Santa Cruz, CA, USA). The membranes were developed with an enhanced chemiluminescence (ECL) reagent (Amersham Life Sciences, Little Chalfont, UK), dried and subsequently exposed to the HyperFilm (Amersham Life Sciences, Little Chalfont, UK) or were imaged by Gel Logic Imaging System 1500 (Kodak; Molecular Imaging System Corestea Health Inc., Rochester, NY, USA). An equal loading in the lanes was evaluated by probing with an rabbit anti-GAPDH, rabbit anti-AKT, and mouse anti-MAPK antibody all from Cell Signaling, Danvers, MA, USA. Phospho-MET was immunoprecipitated from 1 mg of protein extracts with anti-MET rabbit polyclonal antibody (MET C-12: sc-10; Santa Cruz, CA, USA). Corresponding blots were developed with anti-phospho-MET (Tyr1234/1235) (Cell Signaling, Danvers, MA, USA).
Chemotaxis assay
The directional movement of cells toward the HGF gradient was evaluated using modified Boyden's chamber with 8-μm pore polycarbonate membrane inserts (Transwell; Costar Corning, Cambrige, MA, USA). The cells detached with 0.25% trypsin were seeded into the upper chamber of an insert at a density of 2.5 × 104 in 100 μl. The lower chamber was filled with a prewarmed medium containing hHGF (20 ng/ml), mouse HGF (50, 100, 200 ng/ml) both from R&D System, Minneapolis, MN, USA or SDF-1 (100 ng/ml) (PeproTech EC, London, UK). 0.5% BSA DMEM medium was used as a negative control. Inserts were removed from the transwell after 24 h and the cells were fixed with methanol. The cells that did not migrate were scraped off with cotton wool from the upper membrane and cells that had transmigrated to the lower side of the membrane were stained with Wright solution (Merck, Darmstadt, Germany) and counted under high power field (HPF) with inverted microscope (Olympus IX70, Olympus Optical Co., LTD., Tokyo, Japan). Five fields were counted each time and the mean number of cells per HPF was calculated. Chemotaxis assay was performed at least three times, always in duplicates.
Invasion assay
GFR Matrigel invasion inserts (BD Pharmingen, San Diego, CA, USA) were rehydrated with DMEM for 2 h and transferred to the wells containing medium with chemoattractant (mouse HGF 50–200 ng/ml) or medium alone (DMEM with 0.5% BSA) as a control. The cells were then harvested by trypsinization, washed, resuspended in DMEM medium containing 0.5% BSA, and seeded at a density of 2.5 × 104 in 0.5 ml to the inside of the inserts. After 24 h, the cells that invaded the Matrigel were counted on the undersides of filters following fixation and staining with Wright solution. The experiments were carried out twice in duplicates.
Long- and short-term murine models
For long-term-assay 5 × 106, RMS cells were injected subcutaneously into 6–8 weeks NOD-SCID mice. After 30 days the mice were killed and their tumors and BM cells harvested. The tumors were weighed and fixed in formalin. Immunohistochemical evaluation was performed using primary mouse monoclonal antibodies from DakoCytomation, Denmark, UK, anti-Ki67 (clon MIB-1; 1 : 75), anti-desmina (clon D33; 1 : 50) and EnVision Detection Systems Peroxidase/DAB, Rabbit/Mouse (DakoCytomation, Denmark, UK).54 Each experimental group comprised 5–6 animals and all the experiments were repeated three times.
The appearance of RMS cells in the bone marrow, as a metastasis from ectopic injection, was evaluated by real-time PCR using human GAPDH specific primers-probe set (Hs99999905_m1; Applied Biosystems, Foster City, CA, USA) as a marker of human RMS cells.55 A control experiment showed no cross-reactivity with murine GAPDH (Mm99999915_g1; Applied Biosystems, Foster City, CA, USA). In order to study the potential of these cells to metastasize into BM cavities, 30 days after injection, the cells were isolated from two of the legs of each mouse and their presence was examined using qRT-PCR to distinguish between human and murine cells. Next, in order to confirm the presence of human RMS cells in the BM of injected mice, the bones were fixed in formalin and stained immunohistochemically, using monoclonal mouse anti-vimentin antibody (clone v9; 1 : 100) (DakoCytomation, Denmark, UK). In order to visualize human cells in the BM in short-term engraftment, RMS cells were labeled with 9 μM DiI (Molecular Probes) for 30 min at 37 °C in PBS supplemented with glucose (1 mg/ml) and injected intravenously (1 × 106/mouse). After 24 h cells isolated from mice legs were analyzed using fluorescent microscope (Olympus).
Statistical analysis
Statistical analysis was performed using a one-way non-paired Student's t-test with Microsoft Excel or test χ2 with post-hoc Fisher's test using Graphpad software (GraphPad Software Inc., La Jolla, CA, USA). P-values lower than 0.05 were considered as significant. The data from three separate experiments was analyzed.
Acknowledgments
This work was supported by research grants to KM from the Polish Ministry of Science and Higher Education: N N401 229734, N N401 054839, N N401 142339, NN 4010036.
Glossary
- AKT
protein kinase B
- APL
acute promyelocytic leukemia
- ARMS
alveolar rhabdomyosarcoma
- ATRA
all-trans retinoic acid
- ERMS
embryonal rhabdomyosarcoma
- Ets1
v-ets erythroblastosis virus E26 oncogene homolog 1
- HGF
hepatocyte growth factor
- MAPK
mitogen-activated protein kinases
- miR1
microRNA 1
- miR133a
microRNA 133a
- MYHC2
myosin heavy chain 2
- MyoD
myogenic differentiation antigen
- NF-B
nuclear factor-kappaB
- NOD-SCID
nonobese-diabetic/severe combined immunodeficiency
- RMS
rhabdomyosarcoma
- SDF-1
stromal cell-derived factor-1
- SU11274
a selective Met tyrosine kinase inhibitor
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschella′
Supplementary Material
References
- Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med. 1999;341:342–352. doi: 10.1056/NEJM199907293410507. [DOI] [PubMed] [Google Scholar]
- Parham DM, Barr FG.Skeletal muscle tumorsIn: Fletcher CDM, Unni KK, Mertens F (eds). Pathology and Genetics of Tumors of Soft Tissue Sarcoma IARC press: Lion; 2002141–154. [Google Scholar]
- Davis RJ, D'Cruz CM, Lovell MA, Biegel JA, Barr FG. Fusion of PAX7 to FKHR by the Variant t(1;13)(p36;q14) Translocation in Alveolar Rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- Barr FG, Nauta LE, Davis RJ, Schäfer BW, Nycum LM, Biegel JA. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- Parham DM, Ellison DA. Rhabdomyosarcomas in adults and children: an update. Arch Pathol Lab Med. 2006;130:1454–1465. doi: 10.5858/2006-130-1454-RIAACA. [DOI] [PubMed] [Google Scholar]
- Ren YX, Finckenstein FG, Abdueva DA, Shahbazian V, Chung B, Weinberg KI, et al. Mouse mesenchymal stem cells expressing PAX-FKHR form alveolar rhabdomyosarcomas by cooperating with secondary mutations. Cancer Res. 2008;68:6587–6597. doi: 10.1158/0008-5472.CAN-08-0859. [DOI] [PubMed] [Google Scholar]
- Charytonowicz E, Cordon-Cardo C, Matushansky I, Ziman M. Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell. Cancer Lett. 2009;279:126–136. doi: 10.1016/j.canlet.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Carey KA, Segal D, Klein R, Sanigorski A, Walder K, Collier GR, et al. Identification of novel genes expressed during rhabdomyosarcoma differentiation using cDNA microarrays. Pathol Int. 2006;56:246–255. doi: 10.1111/j.1440-1827.2006.01958.x. [DOI] [PubMed] [Google Scholar]
- Mazzone M, Comoglio PM. The Met pathway: master switch and drug target in cancer progression. ASEB J. 2006;20:1611–1621. doi: 10.1096/fj.06-5947rev. [DOI] [PubMed] [Google Scholar]
- Lesko E, Majka M. The biological role of HGF-MET axis in tumor growth and development of metastasis. Front Biosci. 2008;13:1271–1280. doi: 10.2741/2760. [DOI] [PubMed] [Google Scholar]
- Weidner KM, Sachs M, Birchmeier W. The Met receptor tyrosine kinase transduces motility, proliferation, and morphogenic signals of scatter factor/hepatocyte growth factor in epithelial cells. J Cell Biol. 1993;121:145–154. doi: 10.1083/jcb.121.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- Chu SH, Feng DF, Zhang H, Chen ET, Duan ZX, Li XY. C-Met targeted RNA interference inhibits growth and metastasis of glioma U251 cells in vitro. J Neurooncol. 2009;93:183–189. doi: 10.1007/s11060-008-9772-5. [DOI] [PubMed] [Google Scholar]
- Merlin S, Pietronave S, Locarno D, Valente G, Follenzi A, Prat M. Deletion of the ectodomain unleashes the transforming, invasive, and tumorigeneic potential of the MET oncogene. Cancer Sci. 2009;100:633–638. doi: 10.1111/j.1349-7006.2008.01079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG. Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res. 1998;58:3542–3546. [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Bilchik A, Saha S, Turner R, Wiese D, Tanaka M. c-MET expression level in primary colon cancer: a predictor of tumor invasion and lymph node metastases. Clin Cancer Res. 2003;9:1480–1488. [PubMed] [Google Scholar]
- Sun S, Wang Z. Head neck squamous cell carcinoma c-Met+ cells display cancer stem cell properties and are responsible for cisplatin-resistance and metastasis. Int J Cancer. 2011;15:2337–2348. doi: 10.1002/ijc.25927. [DOI] [PubMed] [Google Scholar]
- Guo Y, Xie J, Rubin E, Tang YX, Lin F, Zi X, et al. Frzb, a secreted Wnt antagonist, decreases growth and invasiveness of fibrosarcoma cells associated with inhibition of Met signaling. Cancer Res. 2008;68:3350–3356. doi: 10.1158/0008-5472.CAN-07-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillhardt M, Christensen JG, Lengyel E. An orally available small-molecule inhibitor of c-Met, PF-2341066, reduces tumor burden and metastasis in a preclinical model of ovarian cancer metastasis. Neoplasia. 2010;12:1–10. doi: 10.1593/neo.09948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillhardt M, Park SM, Romero IL, Sawada K, Montag A, Krausz T. Foretinib (GSK1363089), an orally available multikinase inhibitor of c-Met and VEGFR-2, blocks proliferation, induces anoikis, and impairs ovarian cancer metastasis. Clin Cancer Res. 2011;17:4042–4051. doi: 10.1158/1078-0432.CCR-10-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakes FM, Chen J, Tan J, Yamaguchi K, Shi Y, Yu P. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- Lukasiewicz E, Miekus K, Kijowski J, Drabik G, Wilusz M, Bobis-Wozowicz S, et al. Inhibition of rhabdomyosarcoma's metastatic behavior through downregulation of MET receptor signaling. Folia Histochem Cytobiol. 2009;47:485–489. doi: 10.2478/v10042-009-0108-x. [DOI] [PubMed] [Google Scholar]
- Libura J, Drukala J, Majka M, Tomescu O, Navenot JM, Kucia M, et al. CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood. 2002;100:2597–2606. doi: 10.1182/blood-2002-01-0031. [DOI] [PubMed] [Google Scholar]
- Jankowski K, Kucia M, Wysoczynski M, Reca R, Zhao D, Trzyna E, et al. Both hepatocyte growth factor (HGF) and stromal-derived factor-1 regulate the metastatic behavior of human rhabdomyosarcoma cells, but only HGF enhances their resistance to radiochemotherapy. Cancer Res. 2003;63:7926–7935. [PubMed] [Google Scholar]
- Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65:967–971. [PMC free article] [PubMed] [Google Scholar]
- Thomas D, Kansara M. Epigenetic modifications in osteogenic differentiation and transformation. J Cell Biochem. 2006;98:757–769. doi: 10.1002/jcb.20850. [DOI] [PubMed] [Google Scholar]
- Pálmer HG, Sánchez-Carbayo M, Ordóñez-Morán P, Larriba MJ, Cordón-Cardó C, Muñoz A. Genetic signatures of differentiation induced by 1alpha,25-dihydroxyvitamin D3 in human colon cancer cells. Cancer Res. 2003;63:7799–7806. [PubMed] [Google Scholar]
- Hansen LA, Sigman CC, Andreola F, Ross SA, Kelloff GJ, De Luca LM. Retinoids in chemoprevention and differentiation therapy. Carcinogenesis. 2000;21:1271–1279. [PubMed] [Google Scholar]
- Anastasi S, Giordano S, Sthandier O, Gambarotta G, Maione R, Comoglio P, et al. A natural hepatocyte growth factor/scatter factor autocrine loop in myoblast cells and the effect of the constitutive Met kinase activation on myogenic differentiation. J Cell Biol. 1997;137:1057–1068. doi: 10.1083/jcb.137.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, et al. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previdi S, Maroni P, Matteucci E, Broggini M, Bendinelli P, Desiderio MA. Interaction between human-breast cancer metastasis and bone microenvironment through activated hepatocyte growth factor/Met and beta-catenin/Wnt pathways. Eur J Cancer. 2010;46:1679–1691. doi: 10.1016/j.ejca.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Rong S, Bodescot M, Blair D, Dunn J, Nakamura T, Mizuno K, et al. Tumorigenicity of the met proto-oncogene and the gene for hepatocyte growth factor. Mol Cell Biol. 1992;12:5152–5158. doi: 10.1128/mcb.12.11.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Su Y, Lanning N, Gustafson M, Shinomiya N, Zhao P, et al. Enhanced growth of human met-expressing xenografts in a new strain of immunocompromised mice transgenic for human hepatocyte growth factor/scatter factor. Oncogene. 2005;24:101–106. doi: 10.1038/sj.onc.1208181. [DOI] [PubMed] [Google Scholar]
- Seidal T, Kindblom LG, Angervall L. Myoglobin, desmin and vimentin in ultrastructurally proven rhabdomyomas and rhabdomyosarcomas. An immunohistochemical study utilizing a series of monoclonal and polyclonal antibodies. Appl Pathol. 1987;5:201–219. [PubMed] [Google Scholar]
- Molenaar WM, Oosterhuis JW, Oosterhuis AM, Ramaekers FCS. Mesenchymal and muscle-specific intermediate filaments (vimentin and desmin) in relation to differentiation in childhood rhabdomyosarcomas. Hum Pathol. 1985;16:838–843. doi: 10.1016/s0046-8177(85)80256-2. [DOI] [PubMed] [Google Scholar]
- Carter RL, McCarthy KP, Machin LG, Jameson CF, Philp ER, Pinkerton CR. Expression of desmin and myoglobin in rhabdomyosarcomas and in developing skeletal muscle. Histopathology. 1989;15:585–595. doi: 10.1111/j.1365-2559.1989.tb01624.x. [DOI] [PubMed] [Google Scholar]
- Godbole P, Outram A, Wilcox DT, Duffy PG, Sebire NJ. Myogenin and desmin immunohistochemistry in the assessment of post-chemotherapy genitourinary embryonal rhabdomyosarcoma: prognostic and management implications. J Urol. 2006;176:1751–1754. doi: 10.1016/j.juro.2006.03.117. [DOI] [PubMed] [Google Scholar]
- Forte G, Minieri M, Cossa P, Antenucci D, Sala M, Gnocchi V, et al. Hepatocyte growth factor effects on mesenchymal stem cells: proliferation, migration, and differentiation. Stem Cells. 2006;24:23–33. doi: 10.1634/stemcells.2004-0176. [DOI] [PubMed] [Google Scholar]
- Previdi S, Abbadessa G, Dalò F, France DS, Broggini M. Breast cancer-derived bone metastasis can be effectively reduced through specific c-MET inhibitor tivantinib (ARQ 197) and shRNA c-MET knockdown. Mol Cancer Ther. 2012;11:214–223. doi: 10.1158/1535-7163.MCT-11-0277. [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Majka M, Kucia M, Drukala J, Pietrzkowski Z, Peiper S, et al. Expression of functional CXCR4 by muscle satellite cells and secretion of SDF-1 by muscle-derived fibroblasts is associated with the presence of both muscle progenitors in bone marrow and hematopoietic stem/progenitor cells in muscles. Stem Cells. 2003;21:363–371. doi: 10.1634/stemcells.21-3-363. [DOI] [PubMed] [Google Scholar]
- Matteucci E, Ridolfi E, Maroni P, Bendinelli P, Desiderio MA. c-Src/histone deacetylase 3 interaction is crucial for hepatocyte growth factor dependent decrease of CXCR4 expression in highly invasive breast tumor cells. Mol Cancer Res. 2007;5:833–845. doi: 10.1158/1541-7786.MCR-07-0054. [DOI] [PubMed] [Google Scholar]
- Diomedi-Camassei F, McDowell HP, De Ioris MA, Uccini S, Altavista P, Raschellà G, et al. Clinical significance of CXC chemokine receptor-4 and c-Met in childhood rhabdomyosarcoma. Clin Cancer Res. 2008;14:4119–4127. doi: 10.1158/1078-0432.CCR-07-4446. [DOI] [PubMed] [Google Scholar]
- Tarnowski M, Grymula K, Reca R, Jankowski K, Maksym R, Tarnowska J, et al. Regulation of expression of stromal-derived factor-1 receptors: CXCR4 and CXCR7 in human rhabdomyosarcomas. Mol Cancer Res. 2010;8:1–14. doi: 10.1158/1541-7786.MCR-09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PK, Missiaglia E, Shields L, Hyde G, Yuan B, Shepherd CJ, et al. Distinct roles for miR-1 and miR-133a in the proliferation and differentiation of rhabdomyosarcoma cells. FASEB J. 2010;24:3427–3437. doi: 10.1096/fj.09-150698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlino G, Helman LJ. Rhabdomyosarcoma—working out the pathways. Oncogene. 1999;18:5340–5348. doi: 10.1038/sj.onc.1203038. [DOI] [PubMed] [Google Scholar]
- Berthou S, Aebersold DM, Schmidt LS, Stroka D, Heigl C, Streit B, et al. The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants. Oncogene. 2004;23:5387–5393. doi: 10.1038/sj.onc.1207691. [DOI] [PubMed] [Google Scholar]
- Ma PC, Jagadeeswaran R, Jagadeesh S, Tretiakova MS, Nallasura V, Fox EA, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res. 2005;65:1479–1488. doi: 10.1158/0008-5472.CAN-04-2650. [DOI] [PubMed] [Google Scholar]
- Seiwert TY, Jagadeeswaran R, Faoro L, Janamanchi V, Nallasura V, El Dinali M, et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res. 2009;69:3021–3031. doi: 10.1158/0008-5472.CAN-08-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguanno S, Bouchè M, Adamo S, Molinaro M. 12-O-tetradecanoylphorbol-13-acetate-induced differentiation of a human rhabdomyosarcoma cell line. Cancer Res. 1990;50:3377–3382. [PubMed] [Google Scholar]
- Pollock L, Rampling D, Greenwald SE, Malone M. Desmin expression in rhabdomyosarcoma: influence of the desmin clone and immunohistochemical method. J Clin Pathol. 1995;48:535–538. doi: 10.1136/jcp.48.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Terashima M, Hoshino Y, Ohtani S, Kashimura S, Kanzaki N. Detection of cancer cells disseminated in bone marrow using real-time quantitative RT-PCR of CEA, CK19, and CK20 mRNA in patients with gastric cancer. Gastric Cancer. 2006;9:308–314. doi: 10.1007/s10120-006-0398-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.