Abstract
An electrode surface is presented that enables the characterisation of redox-active membrane enzymes in a native-like environment. An ubiquinol oxidase from Escherichia coli, cytochrome bo3 (cbo3), has been co-immobilised into tethered bilayer lipid membranes (tBLMs). The tBLM is formed on gold surfaces functionalised with cholesterol tethers which insert into the lower leaflet of the membrane. The planar membrane architecture is formed by self assembly of proteoliposomes and its structure is characterised by surface plasmon resonance (SPR), electrochemical impedance spectroscopy (EIS) and tapping-mode atomic force microscopy (TM-AFM). The functionality of cbo3 is investigated by cyclic voltammetry (CV) and is confirmed by the catalytic reduction of oxygen. Interfacial electron transfer to cbo3 is mediated by the membrane-localised ubiquinol-8, the physiological electron donor of cbo3. Enzyme coverages observed with TM-AFM and CV coincide (2–8.5 fmol·cm−2) indicating that most - if not all - cbo3 on the surface is catalytically active and thus retains its integrity during immobilisation.
Introduction
Since the first electrochemistry on a protein was reported almost 30 years ago (cytochrome c),1,2 much research has focused on the preparation of ‘protein-friendly’ electrode surfaces. For cytochrome c alone more than a hundred papers have appeared that compare different electrode surfaces.3 A key issue has been, and still is, to interface redox-active proteins and enzymes to an electrode surface without changing their structural integrity and functionality. A particularly powerful method has been to immobilise enzymes on the surface, removing the rate-limiting step of diffusion. Electrochemical studies of adsorbed enzymes allow an in-depth study of the fundamental parameters that typify these enzymes, like catalysis, inhibition and electron flow.4–8
Although many reports have emerged on the electrochemistry of soluble proteins, far less has been published on membrane proteins in spite of their central role in processes such as the mitochondrial electron-transport chain and photosynthesis. Indeed, many redox-active proteins are located in the mitochondrial, chloroplast or plasma membranes.9 As cytochrome c has become an important test case for adapting electrode surfaces to globular proteins, so too has cytochrome c oxidases (CcO) become an important test case for membrane proteins.10–21 Different strategies have been reported each with their own advantages and drawbacks, such as direct adsorption of detergent solubilised protein on modified surfaces,18–20 the incorporation of CcO in supported membranes21 or hybrid bilayers14–17 and attaching the membrane proteins to the surface followed by the reconstitution of the lipid bilayer.10–12
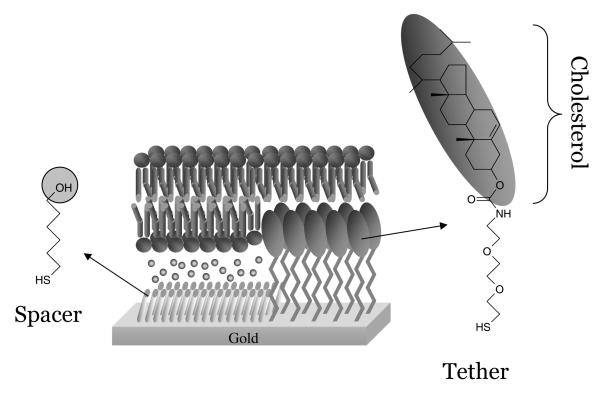
Our aim is to develop an electrode surface that allows the investigation of a range of membrane proteins in a native(-like) environment and is relatively easy to prepare using self-assembly mechanisms. To fulfil these aims we have used the methodology of tethered bilayer lipid membranes (tBLM).12,13,22–31 In tBLMs, the bilayer is attached to the surface via special chemical anchors, which on one side are bound to the surface and on the other side insert into a bilayer leaflet (Figure 1). For this purpose lipid derivatives have been synthesised, which, via a hydrophilic linker, are connected to a group that binds strongly to the surface. One such molecule we have synthesised and characterised is the cholesterol derivative shown in Figure 1. The thiol group is separated from the cholesterol lipid via an ethyleneoxy chain.28–31 Importantly, to form a phospholipid bilayer instead of a monolayer of phospholipids on top of a monolayer of cholesterol tethers, the cholesterol derivatives are mixed with small spacer molecules (6-mercaptohexanol). Self-assembled monolayers prepared from mixed thiol solutions are generally observed to form phase separated domains on the nanoscale, induced by differences in chain length32–36 or chemistry.37–39 Since the two thiol compounds used here are very distinct, both in length and chemistry, similar phase separation is expected as is schematically shown in Figure 1. This will allow phospholipids to enter both leaflets of the bilayer and provide space for transmembrane proteins.
Figure 1.
Chemical structures of 6-mercaptohexanol (spacer) and the cholesterol tether molecule used to form tBLMs. In the middle is a schematic representation of a tethered lipid bilayer formed on a mixed self-assembled monolayer of tether and spacer molecules.
Here, we describe an innovation in which the tBLM system is used to immobilise an ubiquinol oxidase, cytochrome bo3 (cbo3) from Escherichia coli, on an electrode surface. Like aa3-type cytochrome c oxidases, cbo3 is a member of the heme-copper oxidase family40–43 and is a redox-driven proton pump that couples the reduction of water to the generation of proton-motive force across the membrane. Cbo3 is a four subunit protein of 144 kDa that receives its electrons from the membrane component, ubiquinol-8. Here, we will show that tBLMs can successfully be formed from proteoliposomes and that cbo3 is co-adsorbed on the surface in a functional and integral form.
Methods
Protein purification and reconstitution
Cytochrome bo3 (cbo3) was purified as previously described from the E. coli strain, GO105/pJRhisA, in which the his-tagged protein is over-expressed,41 except that the detergent n-dodecyl-β-D-maltoside (DDM) was used throughout the purification (1% for solubilisation of the membranes and 0.05% for all subsequent steps in the purification). The purity of the sample was confirmed by SDS-PAGE. The as-isolated protein had a Soret peak at 409 nm - similar to the 408 nm found previously.42 - and the protein concentration was estimated using ∈408nm = 188 mM−1cm−1.42 Protein concentrations based on a Schaffner-Weissman assay44 were about 30% higher than those based on the Soret band. Cbo3 was reconstituted into vesicles of an E. coli ‘polar’ lipid extract (Avanti) as described by Carter et al.40 Lipid vesicles of about 100 nm diameter were prepared using a mini extruder (Avanti) to which octylglucoside (OG) and protein were added (final concentrations: ~16 mg/ml lipid and ~ 45 mM OG). Typically, 1 % protein / lipid (w/w) was used. After 5-10 min. incubation at 4 °C the lipid/protein mixture was rapidly diluted (> 100 times) with cold (4 °C) buffer and centrifuged at 100,000 g for 1 hour to spin down the proteoliposomes. The resuspended proteoliposomes were used within 2 days. Protein yields after reconstitution were determined with a Schaffner-Weissman protein assay and were between 40 and 50% of the starting material (for samples with 1 mass percent ratio). Depending on the amount of lipid lost during the preparation this results in proteoliposomes with ~0.5% mass percent of cbo3. CD spectroscopy (Jasco J715 spectropolarimeter) confirmed that cbo3 retained its conformation during the reconstitution (see supporting information). All experiments were performed in buffer (20 mM MOPS pH 7.4, 30 mM Na2SO4) at 20 °C. For the control experiments, vesicles were prepared that were treated identically to the proteoliposomes except that cbo3 was omitted.
Cytochrome c oxidase (CcO) was purified and reconstituted as previously described from the P. denitrificans strain, AO1/pPDWT.45,46 Consequent handling of the proteoliposomes was identical to that of cbo3.
Electrode materials, microscopy and spectroscopy
Gold substrates for AFM, SPR and electrochemical experiments were prepared as described previously.47 For the electrochemistry, 150 nm Gold (Advent, 99.99% ) was evaporated through a mask on a 10 nm chromium adhesion layer on cleaned glass microscope slides (Deconex, sonication, methanol) with an Edwards Auto 306 evaporator at < 2 × 10−6 bar. The mask consisted of a circular ‘electrode’ area (⊘ = 5 mm) with a 1 mm link for connection to the electrochemical equipment. The total area (circular + link) in contact with the electrolyte solutions is 0.25 ± 0.03 cm2. After evaporation the electrodes were stored until used. Before usage the slides were sonicated in DCM, rinsed with MeOH and dried under N2. Self-assembled monolayers (SAMs) were made by incubating the electrode slides for ~ 16 hours in a 2-proponal solution of 0.11 mM cholesterol tether and 0.89 mM 6-mercaptohexanol. The gold slides were thoroughly rinsed with DCM and MeOH, dried with N2 and used within a day. The surface ratio of the different thiols on the surface was checked for each electrode with impedance spectroscopy, assuming that the capacitance of a mixed SAM is equal to the summed capacitance of the pure SAMs times the area occupied. This assumption was previously checked and found to be correct with contact angle measurements.47 For the thiol mixture used, the surface area ratio was typically found to be 50% /50% cholesterol-tether / 6-mercaptohexanol.
For the electrochemical experiments a glass cell was used that housed a calomel reference electrode, Pt counter electrode and glass slide with the gold electrode. The glass cell also housed a Clark-type oxygen electrode and had an inlet for argon. The Clark-type oxygen electrode was positioned < 5 mm from the working electrodes. Two Clark electrodes were used: one from World Precision Instruments (ISO, resolution ±0.1 mg/L) and one home-built electrode, which was slower in response but had a higher resolution (±0.02 mg/L). Oxygen concentration was typically ‘regulated’ by purging the cell thoroughly with argon until anaerobic and then stopping the argon flow while continuously measuring cyclic voltammograms and monitoring the increase in oxygen concentration in time due to slow oxygen diffusion into the cell via the Argon inlet.
For the AFM and force curve experiments, template stripped gold (TSG) surfaces were prepared by evaporating 150 nm gold on silicon wafers. The gold surface was then glued with EPO-TEK 377 to glass and cured for 120 min at 120° C. After cooling, the slides were detached from the silicon wafers to expose the TSG surface, which was directly immersed in the thiol solution (0.11 mM EO3-Cholesterol and 0.89 mM 6-Mercaptohexanol) for one hour. Using electrochemistry it was confirmed that also on TSG surfaces the used thiol mixture results in a 50% /50% cholesterol-tether / 6-mercaptohexanol area ratio. AFM height images and force curves were recorded under fluid (buffer) at 25° C using a Multimode AFM on a Nanoscope IIIa or IV (Digital Instruments, Veeco Metrology Group, Inc., CA) controller and silicon nitride cantilevers (NP, Veeco Metrology Group). The cantilevers had a spring constant of approximately 0.18 or 0.52 N/m. The height images reported here are raw, unfiltered data obtained by tapping (dynamic) mode at a frequency in the range 8-9 kHz. Force curves (z-Piezo extension vs cantilever deflection) were recorder under indentical conditions and converted into true force-distance curves showing force in nN as a function of true tip-surface separation in nm.
SPR experiments were performed on prisms evaporated with 50 nm gold as described previously28 and the results were fit by a Levenberg-Marquardt procedure.48,49
Tethered bilayer formation
Proteoliposomes with cbo3 (or control vesicles) were extruded in a mini extruder (Avanti) using 100 nm nuclepore track-etched membranes (Whatman) and subsequently added to the gold surface in the presence of 10 mM Ca(II). We have shown previously that calcium ions are necessary to induce the formation of tethered lipid bilayers when lipid extracts are used that are derived from E. coli.47 (For a more thorough characterisation of the effects of calcium we refer to the work of Richter and Brisson.50) This lipid mixture contains approximately 20-35% negatively charged phospholipids (phosphatidylglycerol and cardiolipin)51,52 and the Ca(II) might be necessary to overcome the electrostatic repulsion between the vesicles. In general it was found that a tethered lipid bilayer was formed within 1 hour at 20 °C. After bilayer formation, the surface was rinsed several times with buffer solutions containing 1 mM EDTA to remove Ca(II) ions and remaining vesicles.
Results
Structural analysis
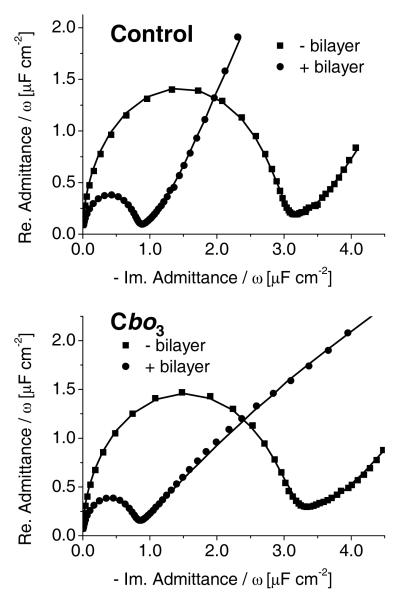
Tethered lipid bilayers were characterised with surface plasmon resonance (SPR), electrochemical impedance spectroscopy (EIS) and tapping-mode atomic force microscopy (TM-AFM). Figure 2 shows the EIS results in the form of Cole-Cole plots in which the diameter of the half circle is equivalent to the double layer capacitance. It is immediately clear that inclusion of cbo3 has almost no effect on the double layer capacitance of the tethered membrane. This indicates that cbo3 does not induce large defects in the tethered bilayer. The capacitance of the lipid bilayers (0.7–0.8 μF/cm2, see supportive information for EIS analysis) is only slightly larger than that of an ideal phospholipid bilayer (0.5 μF/cm2) and similar to that reported previously.29,47,53,54
Figure 2.
Cole-Cole plots before and after formation of a tBLM measured at 0 V vs. SCE. Top: Control (without cbo3). Bottom: With cbo3. The lines represent fits as is described in Supporting Information.
The increase in bilayer thickness after self-assembly of vesicles without cbo3 was determined with SPR to be 3.8 ± 0.1 nm (n=3). This corresponds with previous studies and is as expected when half the inner lipid layer is occupied by cholesterol tethers.28 The presence of cbo3 results in a bilayer that is not significantly thicker (4.3 ± 0.3 nm , n=9, S.D. = 0.9 nm), although we note that a variability between samples was observed. In order to study the tethered lipid bilayers further, TM-AFM images of the electrode surface were recorded.
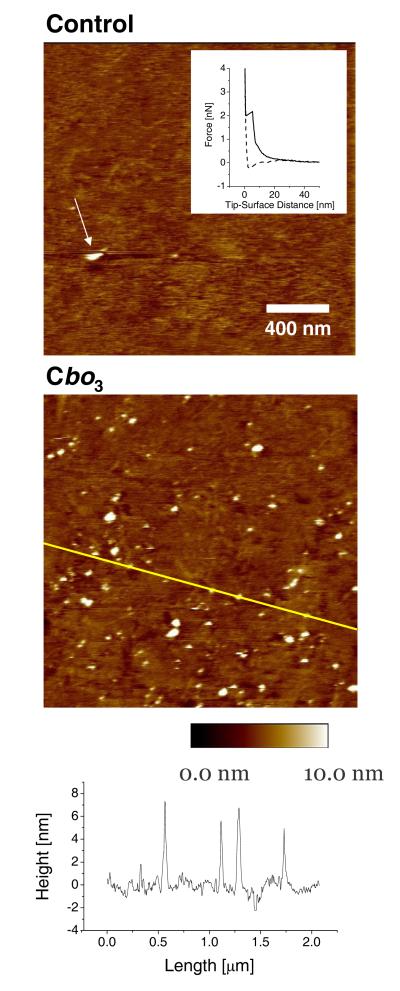
Figure 3 shows a set of TM-AFM results of tBLMs with and without cbo3. Without cbo3 a mostly flat surface (RMS < 0.5 nm) is observed with very infrequently a heightened area due to an adsorbed vesicle. The presence of a lipid bilayer is confirmed by force measurements (insert Figure 3[Top]), which shows the typical behaviour of a lipid bilayer.55 When cbo3 is present, a large number of heightened areas are observed (Figure 3[Middle]). Most of these are 5 ± 2 nm in height and roughly circular with a diameter of 30-40 nm. Some of the areas are larger with diameters up to 75 nm and a height of ~10 nm. No heightened areas were observed in contact mode AFM (CM-AFM) at high forces (> 2 nN) confirming they were not due to the underlying gold surface. Furthermore, the raised areas were damaged or dislocated by the cantilever tip in CM as shown by TM-AFM scans afterwards. On some of the prepared samples with cbo3 adsorbed vesicles were encountered. These vesicles were ~20 nm high with diameters > 150 nm. Importantly, these vesicles were often removed or dislocated by the tip during TM scanning (see Figure 3[Top] for an example), indicating that they were loosely bound. We note that adsorbed vesicles were more frequently observed for samples with cbo3 and the amount of adsorbed vesicles was variable between samples.
Figure 3.
TM-AFM images of tBLMs on template stripped gold. Top: Control experiment without cbo3. For illustration purposes we show an image with an adsorbed vesicle, although these were very infrequently encountered in control experiments. The arrow shows a single vesicle being removed during the scan (scan direction is from bottom to top). Insert: Typical force curve of tBLM layer (solid line, tip approaching surface; dashed line, tip receding from surface). Analysis of this set of force curves indicated an average penetration depth of the bilayer of 4.5 ± 0.1 nm (S.D. = 0.9 nm) and a ‘piercing force’ of 1.50 ± 0.02 nN (S.D. = 0.4 nN). Middle: tBLM with cbo3. Bottom: Height profile of the middle image indicated by the line.
Functional analysis
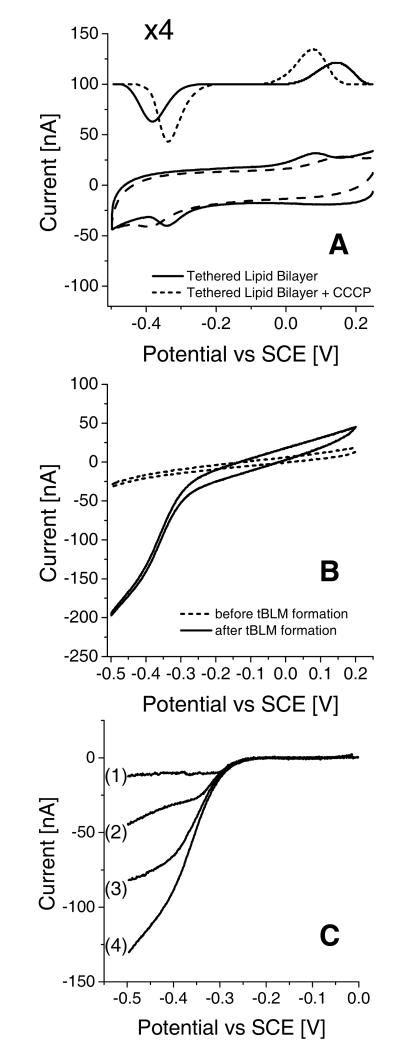
Cbo3 from E. coli is a terminal oxidase and functions as an ubiquinol-8 oxidase. Importantly, the control experiments shown in Figure 4A indicate that the E. coli lipid extract - used throughout this work - still contains ~0.3 mass percent of ubiquinol-8 (UQ-8).56 The redox signals in Figure 4A are assigned to UQ-8 based on control experiments with EggPC/UQ-10 (not shown) and values reported in literature.47,57 The peak separation increased at higher scan rates indicating that the electron transfer is kinetically controlled.58,59 This is partly due to protonation and deprotonation reactions of UQ-8 as addition of the proton ionophore, CCCP, sharpens the peaks and reduces the peak separation. Since UQ-8 is likely to be oxidised or reduced while located at the interface between the gold surface and the lipid bilayer, CCCP is thought to facilitate the proton exchange between this interface and the buffer solution on the other side of the lipid bilayer. The interfacial redox kinetics are also limited by the high co-operativity of the two-electron reaction of the quinones, which is described in more detail by Marchal et al.57 (∣ΔE0∣ ~ 0.6 V at pH 7.4). From the peak area it is determined that the average coverage of UQ-8 is ~2 pmol·cm−2.
Figure 4.
Cyclic voltammograms (CVs) of tBLM systems (electrode area = 0.25 cm2). (A) Control sample at 10 mV·s−1. tBLMs (without cbo3) before and after addition of 10 μg/ml carbonyl cyanide 3-chlorophenylhydrazone (CCCP). Baseline subtracted signals are shown with an offset and 4 times enlarged. (B) CVs at 1 mV·s−1 before and after formation of a tBLM with cbo3. O2 in both CVs is ~14 μM. (C) Baseline corrected CVs at 1 mV·s−1 of tBLM with cbo3 at different O2 concentrations, (1) ~0, (2) ~0.2, (3) ~0.9 and (4) ~14 μM. CV (4) is derived from the same data shown in (B).
When a tethered bilayer is formed with vesicles containing cbo3 a sigmoidal wave is observed starting at the same potential as the reduction of UQ-8 (Figure 4B) indicating two important points: (1) cbo3 located in the tethered membrane is at least partly active; and (2) electrons are transferred from the gold electrode to cbo3 via UQ-8. Varying the substrate (oxygen) concentration in the electrochemical cell changed the magnitude of the catalytic wave as expected (Figure 4C). The apparent Km of cbo3 for oxygen in the electrochemical setup was found to vary between experiments, which might reflect the instability of the Clark-type oxygen electrodes or variable O2 diffusion gradients. However, the
was always < 3 μM. Addition of extra UQ-10 to the membrane did not significantly increase enzyme activity, indicating that the amount of UQ-8 adsorbed on the surface is >>Km. Addition of 1 mM CN− completely inhibitted cbo3 and, consequently, UQ-8 oxidation/reduction peaks were observed that are similar to those shown in Figure 4A.
Discussion
Surface structure
The results demonstrate that it is possible to incorporate the membrane protein, cbo3, in a tBLM using self-assembly methods. Impedance spectroscopy indicates that the bilayer capacitance is identical to that of membranes without cbo3, suggesting that self-assembly of cbo3 does not induce large defects in the membrane.
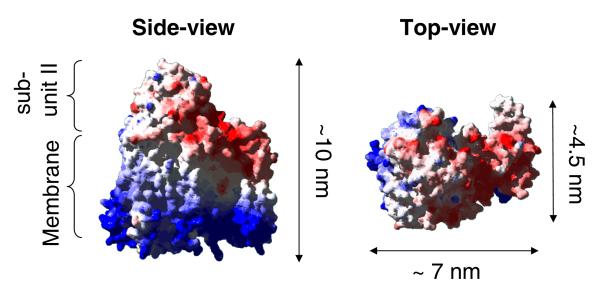
TM-AFM results show that cbo3 is randomly distributed across the surface. The height (Z direction) is consistent with that of cbo3, which is ~10 nm high (see Figure 543) and is therefore expected to extend about 5 nm above the surrounding lipid bilayer. In contrast, the size in the xy-direction is much larger (30-75 nm) than the size of cbo3. This could be due to elevation of the lipid bilayer by the protein since the space between the membrane and the surface in the tBLM system is not sufficient to accommodate the extramembranous domains of cbo3. However, it is also likely that the size is artificially enlarged by the geometry of the cantilever tip.60 We cannot exclude the possibility that multiple proteins are located within each heightened area or that some of the larger areas represent semi-fused vesicles or defects.
Figure 5.
Space-filling representation of the structure of cytochrome bo .43 A relatively large extramembranous domain of subunit II of cbo3 is indicated in the structure on left.
A reconsitution procedure very similar to the one used in this work has been shown give an unidirectional orientation of cbo3 in the vesicles.61 However, in order to acquire good quality tBLMs the reconstituted vesicles had to be extruded through a 100 nm track-etched membrane, which might randomise the protein orientation. Regretfully, due to the low resolution, the orientation of cbo3 could not be unambiguously determined with AFM.
Enzyme activity
The tethered bilayers are formed by self-assembly of vesicles with ~0.5 mass percent of cbo3 (see Methods). If it is assumed that all cbo3 assembles when the tethered bilayer is formed and that half the surface is occupied by cholesterol tethers, cbo3 coverage is expected to be 8 fmol·cm−2.62 This is confirmed by the AFM results. Particle analysis of the data shown in Figure 3 (middle) gives 150-175 molecules in a 2 × 2 μm area, corresponding to 6 - 7.5 fmol·cm−2. Analysis over a larger set of data shows that the coverage varies between 2 and 7.5 fmol·cm−2. Importantly, the same enzyme coverage is indicated by the electrochemical results. Using the published maximum activity of cbo3 (Vmax in solution is 730 e−·s−1)41 and the catalytic activity on the electrode (400 - 600 nA·cm−2, Figure 4B) the active enzyme coverage can be estimated at 5.6 and 8.5 fmol·cm−2. Analysis of more electrodes shows that the active enzyme coverage varies between 3 - 8.5 fmol·cm−2. These estimates confirm that a large part - if not all - of the cbo3 on the surface is active. In this respect it is important to note that maximum enzyme activity is achieved at O2 levels below 10 μM (< 0.3 ppm), which is well below the level at which gold-catalysed oxygen reduction (i.e., the background) becomes a significant part of the voltammogram.
Comparison with other systems
To our knowledge, five groups have reported on the immobilisation of cytochrome c oxidase (CcO) on electrode surfaces.10–21 Of these, slow, but direct electron transfer from the electrode to the immobilised protein has been described by Hawkridge et al.,14–17 Dong et al.19,20 and Tollin et al.21 However, as is also noted by Dutton et al.,18 in none of these systems did the interfacial electron transfer induce catalytic oxygen reduction by CcO. In contrast, sigmoidal reduction waves typifying catalytic activity have been observed by Naumann et al.,10 Dutton et al.18 and in the work described here. Both Naumann et al. and Dutton et al. used the physiological redox partner, cytochrome c, to mediate electron transfer from the electrode to the immobilised enzyme.10,18 Our work corresponds with these observations since the physiological electron donor, UQ-8, was needed to mediate electron transfer between the electrode and cbo3. Furthermore, no catalytic activity was observed when tBLMs were formed with CcO from P. denitrificans nor was direct interfacial electron transfer to CcO detected. To observe clearly a sigmoidal wave in the voltammograms, slow scan speeds are required (< 2 mV·s−1) so that the signals of the mediators (which depend on scan rate) will be small relative to the catalytic currents. The catalytic currents obtained after baseline subtraction are ~50,18 ~13310 and ~500 (this work) nA·cm−2. These relatively small catalytic currents (compared to most other voltammetric results of globular enzymes) either reflect low enzyme coverage (as shown for here for cbo3), partially deactivated enzyme or limiting activity of the electron mediator. Further comparison between the work described here and previous work on CcO is difficult since the electron transfer between cytochome c and CcO is very different in nature than between ubiquinol and cbo3.
Concluding remarks
We have shown that redox-active membrane proteins can conveniently be co-immobilised in tBLM systems using self-assembly methodology. For enzymes that interact with the quinone pool, electrochemical interaction is possible using quinone as the natural mediator. It is therefore expected that this technique is readily applicable to a range of enzyme systems. The tBLMs are prepared from proteoliposomes which require purified proteins and reconstitution into vesicles. Although these techniques are now used routinely, they are still far from trivial for many proteins. We are, therefore, currently investigating whether similar tBLM systems can be prepared with total membrane extracts, omitting the purification and reconstitution steps. In this respect we note that recently the group of Bourdillon has succeeded in preparing tBLM structures from inner mitochrial membranes.63
Supplementary Material
Acknowledgements
CcO was purified from a P. denitrificans strain, AO1/pPDWT, which was generously provided by Dr. A. Puustinen and Prof. M. Wikström (University of Helsinki). This research was supported by an UK BBSRC David Phillips fellowship to LJ, (24/JF/19090), and by BBSRC and EPSRC grants to PJFH, SDE and RJB.
Abbreviations
- Cbo3
Cytochrome bo3
- CcO
Cytochrome c Oxidase
- CV
Cyclic Voltammetry
- EIS
Electrochemical Impedance Spectroscopy
- TM-AFM
Tapping-Mode Atomic Force Microscopy
- SPR
Surface Plasmon Resonance
- UQ
UbiQuinol
Footnotes
Supporting information available CD spectra of vesicle-reconstituted cbo3 (proteoliposomes); further analysis of the impedance data shown in Figure 2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Yeh P, Kuwana T. Chem. Lett. 1977:1145–1148. [Google Scholar]
- 2.Eddowes MJ, Hill HAO. J. Chem. Soc.-Chem. Commun. 1977:771–772. [Google Scholar]
- 3.Fedurco M. Coord. Chem. Rev. 2000;209:263–331. [Google Scholar]
- 4.Armstrong FA. Curr. Opin. Chem. Biol. 2005;9:110–117. doi: 10.1016/j.cbpa.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Léger C, Elliott SJ, Hoke KR, Jeuken LJC, Jones AK, Armstrong FA. Biochemistry. 2003;42:8653–8662. doi: 10.1021/bi034789c. [DOI] [PubMed] [Google Scholar]
- 6.Rusling JF. Acc. Chem. Res. 1998;31:363–369. [Google Scholar]
- 7.Shleev S, Tkac J, Christenson A, Ruzgas T, Yaropolov AI, Whittaker JW, Gorton L. Biosens. Bioelectron. 2005;20:2517–2554. doi: 10.1016/j.bios.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Willner I, Katz E. Angew. Chem. Int. Ed. 2000;39:1180–1218. doi: 10.1002/(sici)1521-3773(20000403)39:7<1180::aid-anie1180>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 9.Ly JD, Lawen A. Redox Rep. 2003;8:3–21. doi: 10.1179/135100003125001198. [DOI] [PubMed] [Google Scholar]
- 10.Ataka K, Giess F, Knoll W, Naumann R, Haber-Pohlmeier S, Richter B, Heberle J. J. Am. Chem. Soc. 2004;126:16199–16209. doi: 10.1021/ja045951h. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich MG, Giess F, Naumann R, Knoll W, Ataka K, Heberle J, Hrabakova J, Murgida DH, Hildebrandt P. Chem. Commun. 2004:2376–2377. doi: 10.1039/b410998h. [DOI] [PubMed] [Google Scholar]
- 12.Giess F, Friedrich MG, Heberle J, Naumann RL, Knoll W. Biophys. J. 2004;87:3213–3220. doi: 10.1529/biophysj.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naumann R, Schmidt EK, Jonczyk A, Fendler K, Kadenbach B, Liebermann T, Offenhausser A, Knoll W. Biosens. Bioelectron. 1999;14:651–662. [Google Scholar]
- 14.Burgess BK, Rhoten MC, Hawkridge FM. Langmuir. 1998;14:2467–2475. [Google Scholar]
- 15.Burgess BK, Jones VW, Porter MD. Langmuir. 1998;14:6628–6631. [Google Scholar]
- 16.Cullison JK, Hawkridge FM, Nakashima N, Yoshikawa S. Langmuir. 1994;1994 [Google Scholar]
- 17.Rhoten MC, Burgess JD, Hawkridge FM. Electrochim. Acta. 2000;45:2855–2860. [Google Scholar]
- 18.Haas AS, Pilloud DL, Reddy KS, Babcock GT, Moser CC, Blasie JK, Dutton PL. J. Phys. Chem. B. 2001;105:11351–11362. [Google Scholar]
- 19.Dong SJ, Li JH. Bioelec. Bioener. 1997;42:7–13. [Google Scholar]
- 20.Li JH, Cheng GJ, Dong SJ. J. Electroanal. Chem. 1996;416:97–104. [Google Scholar]
- 21.Salamon Z, Hazzard JT, Tollin G. Proc. Natl. Acad. Sci. U. S. A. 1993;90:6420–6423. doi: 10.1073/pnas.90.14.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brink G, Schmitt L, Tampe R, Sackmann E. Biochim. Biophys. Acta-Biomembr. 1994;1196:227–230. doi: 10.1016/0005-2736(94)00218-5. [DOI] [PubMed] [Google Scholar]
- 23.Bunjes N, Schmidt EK, Jonczyk A, Rippmann F, Beyer D, Ringsdorf H, Graber P, Knoll W, Naumann R. Langmuir. 1997;13:6188–6194. [Google Scholar]
- 24.Krishna G, Schulte J, Cornell BA, Pace R, Wieczorek L, Osman PD. Langmuir. 2001;17:4858–4866. [Google Scholar]
- 25.Raguse B, Braach-Maksvytis V, Cornell BA, King LG, Osman PDJ, Pace RJ, Wieczorek L. Langmuir. 1998;14:648–659. [Google Scholar]
- 26.Schiller SM, Naumann R, Lovejoy K, Kunz H, Knoll W. Angew. Chem.-Int. Edit. 2003;42:208–211. doi: 10.1002/anie.200390080. [DOI] [PubMed] [Google Scholar]
- 27.Sinner EK, Knoll W. Curr. Opin. Chem. Biol. 2001;5:705–711. doi: 10.1016/s1367-5931(01)00269-1. [DOI] [PubMed] [Google Scholar]
- 28.Williams LM, Evans SD, Flynn TM, Marsh A, Knowles PF, Bushby RJ, Boden N. Langmuir. 1997;13:751–757. [Google Scholar]
- 29.Jenkins ATA, Boden N, Bushby RJ, Evans SD, Knowles PF, Miles RE, Ogier SD, Schonherr H, Vancso GJ. J. Am. Chem. Soc. 1999;121:5274–5280. [Google Scholar]
- 30.Jenkins ATA, Bushby RJ, Evans SD, Knoll W, Offenhausser A, Ogier SD. Langmuir. 2002;18:3176–3180. [Google Scholar]
- 31.Cheng YL, Boden N, Bushby RJ, Clarkson S, Evans SD, Knowles PF, Marsh A, Miles RE. Langmuir. 1998;14:839–844. [Google Scholar]
- 32.Bain CD, Whitesides GM. J. Am. Chem. Soc. 1989;111:7164–7175. [Google Scholar]
- 33.Chen S, Li L, Boozer CL, Jiang S. Langmuir. 2000;16:9287–9293. [Google Scholar]
- 34.Shon Y-S, Lee S, Perry SS, Lee TR. J. Am. Chem. Soc. 2000;122:1278–1281. [Google Scholar]
- 35.Folkers JP, Laibinis PE, Whitesides GM, Deutch J. J. Phys. Chem. 1994;98:563–571. [Google Scholar]
- 36.Tamada K, Hara M, Sasabe H, Knoll W. Langmuir. 1997;13:1558–1566. [Google Scholar]
- 37.Chambers RC, Inman CE, Hutchison JE. Langmuir. 2005;21:4615–4621. doi: 10.1021/la050104t. [DOI] [PubMed] [Google Scholar]
- 38.Smith RK, Reed SM, Lewis PA, Monnell JD, Clegg RS, Kelly KF, Bumm LA, Hutchison JE, Weiss PS. J. Phys. Chem. B. 2001;105:1119–1122. [Google Scholar]
- 39.Stranick SJ, Parikh AN, Tao YT, Allara DL, Weiss PS. J. Phys. Chem. 1994;98:7636–7646. [Google Scholar]
- 40.Carter K, Gennis RB. J. Biol. Chem. 1985;260:986–990. [PubMed] [Google Scholar]
- 41.Rumbley JN, Nickels EF, Gennis RB. Biochim. Biophys. Acta-Protein Struct. Molec. Enzym. 1997;1340:131–142. doi: 10.1016/s0167-4838(97)00036-8. [DOI] [PubMed] [Google Scholar]
- 42.Osborne JP, Cosper NJ, Stalhandske CMV, Scott RA, Alben JO, Gennis RB. Biochemistry. 1999;38:4526–4532. doi: 10.1021/bi982278y. [DOI] [PubMed] [Google Scholar]
- 43.Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek N, Laakkonen L, Puustinen A, Iwata S, Wikström M. Nat. Struct. Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 44.Schaffner W, Weissmann C. Anal. Biochem. 1973;56:502–514. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 45.Riistama S, Laakkonen L, Wikström M, Verkhovsky MI, Puustinen A. Biochemistry. 1999;38:10670–10677. doi: 10.1021/bi990885v. [DOI] [PubMed] [Google Scholar]
- 46.Jasaitis A, Verkhovsky MI, Morgan JE, Verkhovskaya ML, Wikström M. Biochemistry. 1999;38:2697–2706. doi: 10.1021/bi982275l. [DOI] [PubMed] [Google Scholar]
- 47.Jeuken LJC, Connell SD, Nurnabi M, O’Reilly J, Henderson PJF, Evans SD, Bushby RJ. Langmuir. 2005;21:1481–1488. doi: 10.1021/la047732f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes in C. Second edition ed Cambridge University Press; Cambridge: 1992. [Google Scholar]
- 49.Hansen W. J. Opt. Soc. Am. 1968;58:380–390. [Google Scholar]
- 50.Richter RP, Brisson AR. Biophys. J. 2005;88:3422–3433. doi: 10.1529/biophysj.104.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkinson SG. Gram-negative bacteria. In: Ratlegde C, Wilkinson SG, editors. Microbial Lipids. Vol. 1. Academic Press; London: 1988. [Google Scholar]
- 52. See also www.avantilipids.com.
- 53.Naumann R, Schiller SM, Giess F, Grohe B, Hartman KB, Karcher I, Koper I, Lubben J, Vasilev K, Knoll W. Langmuir. 2003;19:5435–5443. [Google Scholar]
- 54.Terrettaz S, Mayer M, Vogel H. Langmuir. 2003;19:5567–5569. [Google Scholar]
- 55.Dufrêne YF, Boland T, Schneider JW, Barger WR, Lee GU. Faraday Discuss. 1998:79–94. doi: 10.1039/a807637e. [DOI] [PubMed] [Google Scholar]
- 56.The concentration of UQ-8 in the tBLM on the surface is approximately 2 pmol·cm−2. Using the bilayer thickness of 3.8 nm (this is without the cholesterol tethers as determined with SPR) and assuming an average weight density of the lipid bilayer of 1.2 g/mL, the concentration of UQ-8 is 0.3 mass percent.
- 57.Marchal D, Boireau W, Laval JM, Bourdillon C, Moiroux J. J. Electroanal. Chem. 1998;451:139–144. [Google Scholar]
- 58.Laviron E. Voltammetric Methods for the Study of Adsorbed Species. In: Bard AJ, editor. Electroanalytical Chemistry. Vol. 12. Marcel Dekker; New York: 1982. [Google Scholar]
- 59.Hirst J, Armstrong FA. Anal. Chem. 1998;70:5062–5071. doi: 10.1021/ac980557l. [DOI] [PubMed] [Google Scholar]
- 60.The lipid are expected to modify the AFM probe when imaging bilayer (this has also been observed by other groups performing AFM on bilayers, see ref.50 and references therein). A tip with radius 10 nm would have its asperities submerged and smoothed out and broadens the tip radius to 13 nm. If 5 nm of cbo3 is sticking out of the bilayer, then an AFM tip with a radius of 13 nm would result in an image of a hemispherical blob ≈30 nm in diameter and losing the fine detail.
- 61.Verkhovskaya ML, GarciaHorsman A, Puustinen A, Rigaud JL, Morgan JE, Verkhovsky MI, Wikstrom M. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10128–10131. doi: 10.1073/pnas.94.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.To estimate the coverage in fmol·cm−2 it is assumed that the surface area/mass ratio of the lipid bilayer is 0.05 Å2·Da−1. To our knowledge, this value for the total lipid extract of E. coli has not been determined, but the mean area of most phospholipids lies between 55 and 85 Å2 while their molecular weight lies between 700 and 800 Da. The surface area/mass ratio of cbo3 can be estimated at 0.02 Å2·Da−1 (see Figure 5, MW of cbo3 is 144 kDa). Thus when 0.5% of the mass of the proteoliposomes is cbo3, 0.2% of the surface is occupied by cbo3. 2×10−4/3×1017 m2 is 7×1012 cbo3 molecules·m−2, which is 10 fmol·cm−2. Since half the surface is occupied by cholesterol tethers, a quarter of the total tBLM will be filled by the tether molecules, lowering the cbo3 coverage on the surface by a quarter.
- 63.Elie-Caille C, Fliniaux O, Pantigny J, Maziere JC, Bourdillon C. Langmuir. 2005;21:4661–4668. doi: 10.1021/la046973k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.