Abstract
Transcriptome alterations of leukocytes from soldiers who underwent 8 weeks of Army Ranger training (RASP, Ranger Assessment and Selection Program) were analyzed to evaluate impacts of battlefield-like stress on the immune response. About 1400 transcripts were differentially expressed between pre- and post-RASP leukocytes. Upon functional analysis, immune response was the most enriched biological process, and most of the transcripts associated with the immune response were downregulated. Microbial pattern recognition, chemotaxis, antigen presentation and T-cell activation were among the most downregulated immune processes. Transcription factors predicted to be stress-inhibited (IRF7, RELA, NFκB1, CREB1, IRF1 and HMGB) regulated genes involved in inflammation, maturation of dendritic cells and glucocorticoid receptor signaling. Many altered transcripts were predicted to be targets of stress-regulated microRNAs. Post-RASP leukocytes exposed ex vivo to Staphylococcal enterotoxin B showed a markedly impaired immune response to this superantigen compared with pre-RASP leukocytes, consistent with the suppression of the immune response revealed by transcriptome analyses. Our results suggest that suppression of antigen presentation and lymphocyte activation pathways, in the setting of normal blood cell counts, most likely contribute to the poor vaccine response, impaired wound healing and infection susceptibility associated with chronic intense stress.
Keywords: microarray, immune response, battlefield-like stress, Army Rangers, RASP
Introduction
A thorough understanding of stress reactions is likely to produce better strategies to manage stress and improve health.1 Stress modulates gene expression, behavior, metabolism and immune function.2, 3, 4, 5 Chronic stress has been shown to impair lymphocyte proliferation, vaccination efficacy,6, 7, 8, 9 wound healing10, 11 and natural killer (NK) cell activity, and to increase the risk for bacterial and viral infection12 and cancer.13 Comprehensive descriptions of stress-affected molecular networks and pathways are needed to reveal strategies to reduce and prevent the pathophysiologic effects of prolonged intense stress.
Here, we report gene expression changes occurring in leukocytes collected from soldiers before and after undergoing the 8-week Ranger Assessment and Selection Program (RASP). The RASP is designed to emulate extreme battlefield scenarios: sleep deprivation, calorie restriction, strenuous physical activity and survival emotional stresses that push the soldiers to their physical and psychological limits. These grueling conditions produce a number of short-term physiologic functional alterations, including impaired cognitive function, reduced 3,5,3′-triiodothyroxine and testosterone, and increased amount of cortisol and cholesterol.14, 15 The RASP provides a rare opportunity for systems level studies of the effects of extreme battlefield-like stress, and intense chronic stress in general.
Using cDNA microarrays, we have analyzed the effect of battlefield-like conditions on leukocyte transcriptome, corroborated the results with oligonucleotide, microRNA (miR) and real-time quantitative PCR (RT–QPCR) arrays, and confirmed them using individual RT–QPCR and enzyme-linked immunosorbent assays (ELISA). This study is the first use of genome-wide transcript analysis to assess the immune response to severe chronic battlefield-like stress. About 1400 transcripts were differentially regulated due to the battlefield-like stress. Ontological enrichment and biological network analyses of the gene expression data identified changes in functional and regulatory pathways indicative of extensive immune suppression. In addition, some stress-induced miRs and a number of stress-inhibited transcription factors (TFs) were predicted to regulate or be regulated by many of the differentially expressed immune response genes. Suppressed immune response genes stayed suppressed even after post-RASP leukocytes were exposed ex vivo to the mitogenic toxin Staphylococcal enterotoxin B (SEB). The anergic state of post-RASP leukocytes to SEB and the suppressed transcripts of immune-response processes are both indicative of compromised protective immunity as a result of extreme battlefield-like stress.
Results
Physical and cellular examination of study subjects
Soldiers, who undergo the RASP, experience an average daily calorie deficit of 1000–1200 kcal, random sleep for less than 4 h per day, strenuous and exhaustive physical toiling, and emotional survival stressors. Five of the initial fifteen soldiers enrolled in our study were replaced with five others due to attrition. This was done to maintain 15 study subjects at each time point. All study subjects had complete and differential blood counts performed, and were observed for infections and injuries. At the end of the RASP, the group showed reductions in body weight (178.6–173.2 lb, P<0.05), body mass index (25.7–24.8, P<0.02) and diastolic blood pressure (85.6–61.2 mm Hg, P<5.5E-5). There was an increase in systolic blood pressure (120.4–127.3 mm Hg, P<0.04) and body temperature (96.9–98.2 °F, P<0.02).
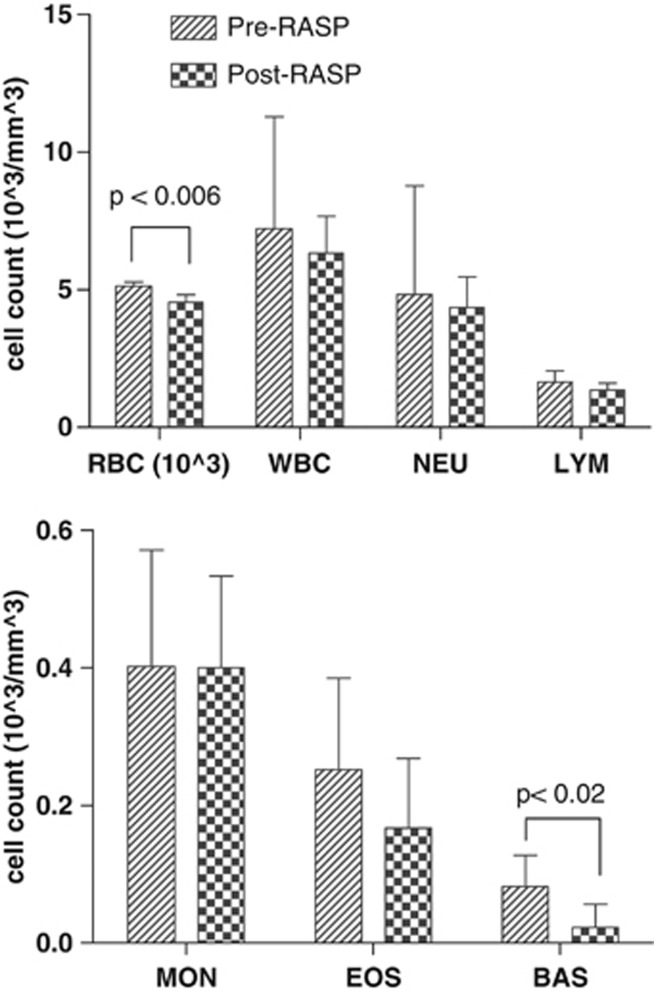
Differential and complete blood counts between pre- and post-RASP samples showed that only red blood cells (P<0.006) and basophils (P<0.02) were significantly reduced after RASP, but they remained within their normal ranges (Figure 1). Equal numbers of pre- and post-RASP leukocytes were used for RNA isolation, and equal amounts of isolated RNAs were used for microarrays and RT–QPCR assays.
Figure 1.
Differential and complete leukocyte counts of soldiers before and after RASP. Differential and complete blood counts for pre- and post-RASP subjects included red blood cells (RBC), white blood cells (WBC), neutrophils (NEU), lymphocytes (LYM), monocytes (MON), eosinophils (EOS) and basophils (BAS). Using comparative t-test, only RBC (P<0.006) and BAS (P<0.02) were significantly changed (reduced) after RASP. The ranges of cell counts including RBC and BAS (shown by the vertical lines) were within normal ranges. Normal ranges are WBC 5–12 × 103 mm−3; NEU 2–8 × 103 mm−3; LYM 1–5 × 103 mm−3; MON 0.1–1 × 103 mm−3; EOS 0.0–0.4 × 103 mm−3; BAS 0.0–0.2 × 103 mm−3.
Transcriptome profiling of pre- and post-RASP leukocytes
We performed gene expression profiles on total RNAs isolated using two different methods: Trizol (Invitrogen Inc., Grand Island, NY, USA) and PAXgene (Qiagen Inc., Valencia, CA, USA). To cross-validate our findings, we used three transcriptome profiling techniques: cDNA and oligonucleotide microarrays, and RT–QPCR arrays.
cDNA microarray analyses
For gene expression profiling, we used custom cDNA microarrays containing ∼10 000 well-characterized cDNA probes of 500–700 bp representing ∼9000 unique human gene targets. Welch's (unpaired unequal variance) t-test along with false discovery rate (FDR) correction applied to the normalized expression data, and we identified 1983 significantly changed transcripts (q⩽0.05), with 1396 of 1983 transcripts showing ⩾1.5-fold change between pre- and post-RASP samples (Supplementary Figure S1A). Among the 1396 differentially regulated genes, 288 genes (Figure 2, Supplementary Figure S1A) were significantly changed at q⩽0.001, and 87 of these showed differential regulation >threefold (Supplementary Table S1). Of these 87 genes, 72 were downregulated, and 68 of the 72 downregulated genes have direct roles in the immune response. These results strongly suggest that the RASP stressors suppress the immune response, and this finding was corroborated by functional and pathway enrichment analyses.
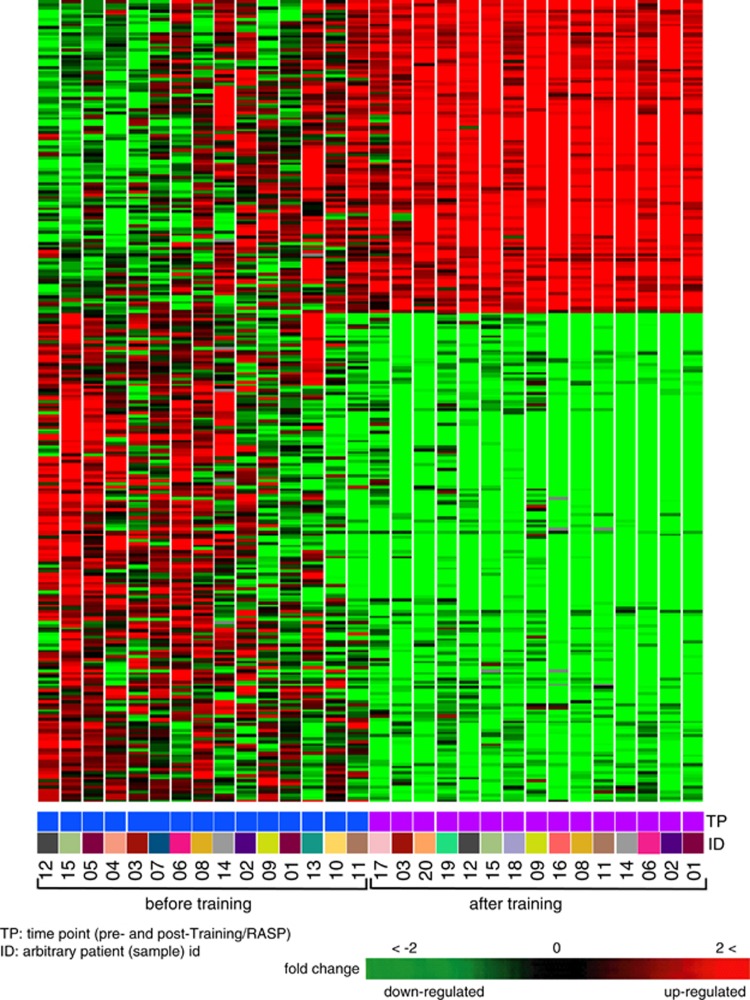
Figure 2.
Heat map of differentially expressed genes in leukocytes of soldiers before and after RASP. Heat map shows hierarchical clustering of 288 genes that passed Welch's t-test with FDR correction (q<0.001) and had expression alteration of ⩾1.5-fold. Each lane shows the 288 genes and their leukocyte expression level for each subject before (left panel) or after (right panel) RASP in comparison to human universal reference RNA.
Functional enrichments of significantly regulated genes using both hypergeometric test (FDR correction, q⩽0.05), and Fisher's exact test identified the immune system as the most affected biological process. Apoptosis, stress response, response to wounding, metabolism, hormone receptor signaling (peptide and steroid), cell cycle and unfolded protein response signaling were also significantly associated with differentially regulated transcripts. However, the immune system process was the most significantly over-represented (q<1.7E-16), and was associated with 177 differentially regulated genes. Of the 177 genes, 151 were downregulated and 26 were upregulated (Supplementary Figure S1B, Supplementary Table S2). The 151 downregulated genes were significantly associated with microbial pattern recognition, inflammation, chemotaxis, antigen presentation and activations of lymphocytes, mast cells and macrophages (Table 1). The 26 upregulated immune response genes were associated with response to steroid hormone stimulus, regulation of leukocyte activation, complement activation, negative regulation of gene expression and negative regulation of phosphorylation (Table 1).
Table 1. Functions significantly associated with differentially regulated immune response genes that passed Welch's t-test and FDR correction (q<0.05) and showed >1.3-fold change in post-RASP leukocytes compared with pre-RASP leukocytes.
| GO-ID | Function | Gene symbol | ||
|---|---|---|---|---|
| Functions of downregulated immune response genes | ||||
| 45321 | Leukocyte activation | MICA, CD8A, CD8B, ELF4, TLR4, ADA, CD74, CD93, CD2, FCER1G, CD4, SYK, IL4, KLF6, PTPRC, CD3D, IL8, CD3E, RELB, SLAMF7, CD40, LAT, LCK, CD79A, LCP2 | ||
| 6954 | Inflammatory response | CXCL1, ITGAL, TNF, TLR2, NFKB1, ITGB2, TLR4, CCL5, CD97, CCL20, KRT1, IL1B, IL1A, CEBPB, IL8, IL1RN, GRO3, CD40, CCL18, CD180, C8G, SCYA7, CCL13, CCR7, CYBB, CCR5, CRH, CD14 | ||
| 19882 | Antigen processing and presentation | HLA-DQB1, MICA, CD8A, HLA-DRB1, RELB, HLA-C, FCGRT, HLA-B, HLA-G, CD74, B2M, FCER1G, HLA-DPA1, HLA-DPB1, HLA-DOB, AP3B1, HLA-DRA | ||
| 46649 | Lymphocyte activation | IL4, PTPRC, KLF6, MICA, CD3D, CD8A, ELF4, CD3E, CD8B, RELB, CD40, SLAMF7, CD74, ADA, LCK, CD2, CD4, CD79A, SYK | ||
| 30097 | Hemopoiesis | IL4, PTPRC, KLF6, CD3D, LYN, HCLS1, RELB, IFI16, MYH9, CD164, CD74, LCK, CD4, SPIB, CD79A, MYST1, SYK, MYST3 | ||
| 52033 | Pathogen-associated molecular pattern recognition | PF4, CHIT1, TLR2, TLR4, SCYA7, CD14, PF4V1, CLP1, TICAM1, FPRL1, FPR1 | ||
| 6935 | Chemotaxis | IL4, CXCL1, C5AR1, IL8, GRO3, ITGB2, PF4, CCL5, CCL18, SCYB5, SCYA7, CCL13, CCR7, CCR5, PPBP, CCL20, IL1B, FCER1G, SYK | ||
| 42110 | T-cell activation | PTPRC, MICA, CD3D, CD8A, CD3E, CD8B, ELF4, RELB, CD74, ADA, LCK, CD2, CD4, SYK | ||
| 2274 | Myeloid leukocyte activation | LAT, IL8, CD93, RELB, FCER1G, TLR4, LCP2 | ||
| 50778 | Positive regulation of immune response | PTPRC, MICA, SLK, FYN, KRT1, TLR2, FCER1G, CD79A, C8G, SYK | ||
| 6959 | Humoral immune response | PSMB10, CD83, ST6GAL1, TNF, HLXB9, POU2F2, KRT1, AIRE, C8G | ||
| 1934 | Positive regulation of phosphorylation | TNF, CCND3, LYN, HCLS1, IL1B, CD4, SYK | ||
| 45087 | Innate immune response | CYBB, IL1R1, SARM1, CLP1, KRT1, TLR2, TLR4, SLAMF7, CD180, C8G | ||
| 2252 | Immune effector process | PTPRC, LAT, MICA, FCN2, KRT1, FCER1G, SLAMF7, CD74, C8G | ||
| 30593 | Neutrophil chemotaxis | IL8, FCER1G, IL1B, ITGB2, SYK | ||
| 7229 | Integrin signaling | LAT, ITGAL, ITGAX, ITGB2, MYH9, ITGAM, SYK | ||
| 45058 | T-cell selection | CD3D, CD4, CD74, SYK | ||
| 1816 | Cytokine production | IL4, CD4, ISGF3G, CD226, LCP2 | ||
| 6909 | Phagocytosis | CD93, FCN2, CLP1, FCER1G, CD14 | ||
| 2460 | Somatic recombination for adaptive response | IL4, RELB, FCER1G, TLR4, CD74, C8G | ||
| Functions associated with upregulated immune response genes | ||||
| 48545 | Response to steroid hormones | CEBPA, CAV1, HMGB2, PRKACA, CD24 | ||
| 42326 | Negative regulation of phosphorylation | CAV1, PRKACA, INHA | ||
| 6956 | Complement activation | C4B, C3, C2 | ||
| 10817 | Regulation of hormone levels | DHRS2, ACE, FKBP1B | ||
| 43434 | Response to peptide hormones | HHEX, PRKDC, PRKACA | ||
| 2762 | Negative regulation of myeloid leukocyte differentiation | FSTL3, INHA | ||
| 32088 | Negative regulation of NFκB activity | POP1, SIVA | ||
| 51384 | Response to glucocorticoids | CEBPA, CAV1, PRKACA | ||
| 16481 | Negative regulation of transcription | CEBPA, HHEX, CAV1, HMGB2, FST, HELLS | ||
Oligonucleotide microarrays
Gene expression alterations in leukocytes of soldiers before and after RASP were also analyzed using PAXgene RNA isolation and oligonucleotide microarrays representing 24 650 human gene probes. This combination of a different RNA isolation procedure and a different oligonucleotide microarray assay showed the immune system again to be the most significantly affected process. Welch's t-test (P<0.05, without multiple correction) and fold change filter (⩾1.5-fold) were used to analyze normalized expression levels. Among 1570 genes that passed these filters, 104 genes were associated with immune response processes, including microbial recognition, chemotaxis, inflammation, antigen presentation and activations of T cells, B cells and NK cells (Supplementary Figure S2).
Real-time quantitative PCR array
We used RT–QPCR arrays to confirm differential expression of genes identified using cDNA and oligonucleotide microarrays, and to survey additional immune-related genes. The PCR arrays contained more than 160 genes in antigen presentation and NFκB signaling pathways (RT2 Profiler PCR Arrays, SABiosciences, Frederick, MD, USA). The results verified downregulation of 116 immune response genes (Supplementary Figure S3, Supplementary Table S3) consistent with the microarrays data.
Transcripts important for microbial pattern recognition, inflammation, antigen presentation, T-cell activation and TFs related to immune response were suppressed across cDNA, oligonucleotide and PCR arrays (Supplementary Figure S4).
Real-time quantitative PCR
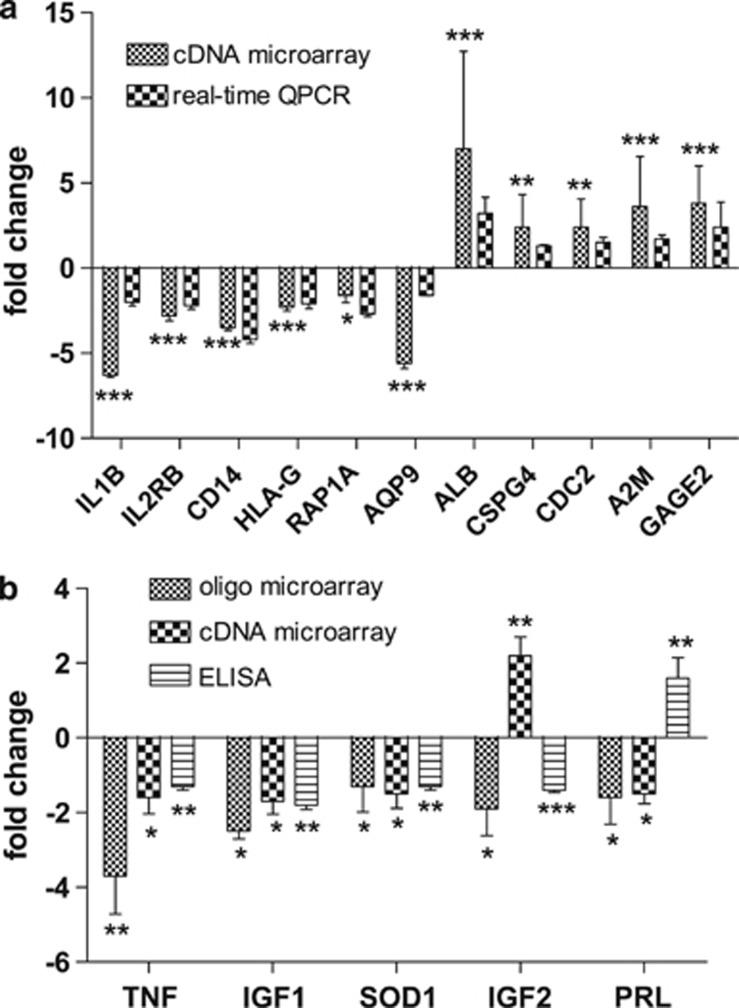
Additional RT–QPCR assays with specific primer pairs (Supplementary Table S4) were used to confirm 11 representative genes among 1396 significantly altered genes (up and downregulated). Real-time QPCR confirmed and validated the differential expression of IL1B, IL2RB, CD14, HLA-G, RAP1A, AQP9, ALB, CSPG4, CDC2, A2M and GAGE2 at the transcript level (Figure 3a).
Figure 3.
Comparison of transcript levels determined by two different assays and comparison of transcript levels with plasma protein levels. (a) Correlation of real-time QPCR and cDNA microarray data: real-time PCR reactions for each gene were carried out with three or more replicates. Trizol RNA isolation and cDNA microarrays were used. Significance levels: *0.001⩽q<0.05, **1.0E-5⩽q<0.001, ***q<1.0E-5—q stands for P-values after FDR correction. (b) Plasma protein concentrations (ELISA) compared with transcript levels (oligo and cDNA microarray): plasma concentrations of PRL, IGF1 and IGF2, TNFα and enzymatic activity of superoxide dismutase 1 (SOD1) assays were performed in triplicate on the plasma of 9 of the 10 soldiers who completed RASP. The IGF-I depletion is consistent with the finding of other investigators who measured its plasma concentration on similar subjects15 (*0.01⩽P<0.05, **0.001⩽P<0.01, ***P<0.001).
ELISA assays of plasma proteins
To evaluate protein levels for comparison with transcript alterations, we used ELISA to determine plasma levels of insulin-like growth hormones 1 and 2 (IGF1 and IGF2), prolactin (PRL), tumor necrosis factor alpha (TNFα) and enzymatic activity of superoxide dismutase 1. Relative quantities of these proteins and the levels of transcripts profiled by cDNA and oligonucleotide microarrays were compared (Figure 3b). Reduced IGF1 has been shown to be a biomarker of negative energy balance under conditions of multiple RASP stressors,14 and IGF1 transcript in leukocytes and the IGF1 protein in plasma were reduced after RASP. But IGF2 transcript levels in post-RASP leukocytes were increased when measured by cDNA arrays and decreased in oligonucleotide microarrays. Plasma concentration of PRL was upregulated, whereas transcriptome profiling showed downregulation, suggesting differential regulation of PRL at transcription and translation levels.
Genes associated with microbial recognition
Genes associated with microbial pattern recognition (which were significantly suppressed in post-RASP leukocytes) include toll-like receptors (TLR-2, 3 and 4), CD14, CD93, chitinase 1 (CHIT1), formyl peptide receptor 1 (FPR1, and FPR-like 1, FPRL1), dicer1 (DICER1), cleavage and polyadenylation factor I subunit (CLP1), platelet factor 4 (PF4, and its variant 1, PF4V1), TLR adapter molecule 1 (TICAM1) and myeloid differentiation primary response gene 88 (MYD88) (Supplementary Figure S2A, Table 1 and Supplementary Table S5). TLR6 was downregulated but it did not pass the FDR correction filter.
CD14, along with TLR4/TLR4 and TLR2/TLR6, recognizes lipopolysaccharides and peptidoglycans, respectively. TLR3, CLP1 and DICER1 bind to double stranded viral RNAs. FPR1 and FPRL1 bind bacterial N-terminal formyl-methionine peptides. CHIT1 recognizes chitin patterns of pathogens. PF4 and PF4V1 recognize patterns of plasmodium and tumor cells. TICAM1 and MYD88 are important cytosolic adapter molecules in microbial pattern recognition. Transcripts of genes important for recognition and binding of pathogen-associated molecular patterns were downregulated, suggesting a compromised innate immune response to microbial infection.
Genes associated with chemotaxis and inflammation
Stress suppressed transcripts associated with chemotaxis and inflammation included interleukins (IL1A, IL1B, IL4, IL8), interleukin receptors (IL1R1, IL1RN, IL2RB, IL10RA), chemokine (C-X-C motif) ligands (CXCL1), chemokine (C-C motif) ligands (CCL13, CCL18, CCL20), TNFα, TNF receptor super-family members 1B, 10B and 10C (TNFRSF1B, TNFRSF10B and TNFRSF10C), TNF superfamily members 3, 8, (LTB, TNFSF8), complement component 8 gamma (C8G), cytochrome b-245 beta (CYBB), CD97 and interferon gamma receptor (IFNGR2) (Supplementary Figure S2A, Table 1 and Supplementary Table S5).
Genes associated with activation of myeloid leukocytes
Supplementary Figure S2A, Table 1 and Supplementary Table S5 show suppressed transcripts associated with activation of mast cells and macrophages. These included TLRs, TNF, linker for activation of T cells (LAT), lymphocyte cytosolic protein 2 (LCP2), spleen tyrosine kinase (SYK), CD93 and IL4. Suppressed genes associated with inflammatory responses (IL1, CD14, INFGR1) were also significantly associated with activation of myeloid cells. Differentiation of myeloid leukocytes was significantly associated with interferon gamma-inducible proteins 16 and 30 (IFI16, IFI30), myosin heavy chain 9 (MYH9), IL4, Spi-B TFs (SPIB, NFκB3, RELB), MYST histone acetyltransferases (MYST1 and 3), TNF, PF4, hematopoietic cell-specific lyn substrate 1 (HCLS1), V-yes-1 Yamaguchi sarcoma viral-related oncogene homolog (LYN) and V-maf (musculoaponeurotic fibrosarcoma) oncogene homolog b (MAFB). Downregulation of hematopoietic TFs (MAFB and HCLS1) and CSF1R may indicate less viability of myeloid cells to expand or to replenish. Suppression of mRNAs of the genes listed above suggests poor activation, differentiation and proliferation of myeloid leukocytes in response to infection, and hence poor innate immune response under conditions of intense chronic stress.
Genes associated with antigen presentation
Suppressed transcripts associated with antigen preparation and presentation encompass MHC classes (I and II), CD1s, B-cell co-receptors and integrins (Supplementary Figure S2B, Table 1 and Supplementary Table S5). Transcripts of MHC class I (HLA-B, HLA-C, HLA-G, beta-2-microglobulin (B2M)), MHC class II (HLA-DRB1, HLA-DRA, HLA-DPA1, HLA-DPB1, HLA-DQA1, HLA-DQB1, CD74, HLA-DOB), B-cell co-receptors (CD79A, CD79B), Ig heavy constant gamma 1 (IGHG1), Ig heavy constant alpha 1 (IGHA1), MHC class I polypeptide-related sequence A (MICA), adapter-related protein complex 3 beta1 (AP3B1), intercellular adhesion molecules 1, 2 and 3 (ICAM1, ICAM2, ICAM3) were downregulated, implying poor antigen preparation and presentation, and hence impaired priming of the adaptive immune response.
Genes associated with activation of lymphocytes
Suppressed transcripts associated with T-cell activation, differentiation and proliferation included TCR co-receptors (CD4, CD8α, CD8β, CD3ɛ, CD3δ, CD247), LAT, TCR signaling molecules (protein kinase c theta (PRKCQ), protein tyrosine phosphatase receptor type C (PTPRC), C-SRC tyrosine kinase (CSK), SYK, lymphocyte-specific protein tyrosine kinase (LCK)), integrins CD2, CD44, integrin alpha L, M and X (ITGAL, ITGAM, ITGAX) and cyclin D3 (CCND3) (Supplementary Figure S2B, Table 1 and Supplementary Table S5).
Other downregulated transcripts, such as interleukin 4, SYK, PRKCD, CD40, PTPRC, cyclin-dependent kinase inhibitor 1A (CDKN1A), Kruppel-like factor 6 (KLF6), SLAM family member 7 (SLAMF7) and killer cell Ig-like receptor three domains long cytoplasmic tail 1 (KIR3DL1), were significantly associated with activation, differentiation and proliferation of B cells and NK cells (Supplementary Figure S2B, Table 1).
TFs associated with immune responses
TFs that are important regulators of immune response genes such as nuclear factor kappa B family (NFκB1, NFκB2, RELA, RELB), interferon-regulatory factors 1, 5, 7, 8 (IRF1, IRF5, IRF7 and IRF8), signal transducer and activator of transcription (STAT2, STAT6) and SP TFs (SP1, SP140) were suppressed (Supplementary Figure S2C, Supplementary Table S5). In addition, stress response factors, such as GA-binding protein alpha (GABPA), POU class 2 homeobox 2 (POU2F2), splicing factor 1 (SF1) and hypoxia-inducible factor 3 and alpha subunit (HIF3A), were downregulated. Steroid hormone biosynthetic factor, sterol regulatory element binding TF 2 (SREBF2) and NFκB inhibitor-like 2 (NFKBIL2) were upregulated (Supplementary Figure S2C, Supplementary Table S5).
Response of leukocytes to ex vivo treatment of SEB
SEB is a superantigen, and a potent T-cell activator known to induce proinflammatory cytokine release in vitro.16 Leukocytes of soldiers collected before and after RASP were challenged ex vivo with SEB, and immune response transcripts were analyzed. In pre-RASP leukocytes, SEB toxin induced a majority of immune response genes (Figure 4). However, in post-RASP leukocytes, the RASP-suppressed immune response genes showed no sign of re-activation even after ex vivo exposure to SEB (Figure 4 and Supplementary Figure S5). Rather, SEB exposure seemed to further suppress the expression of many of these transcripts. The impaired response of post-RASP leukocytes to SEB is consistent with the suppression of the immune response pathways and networks revealed by our transcriptome analyses.
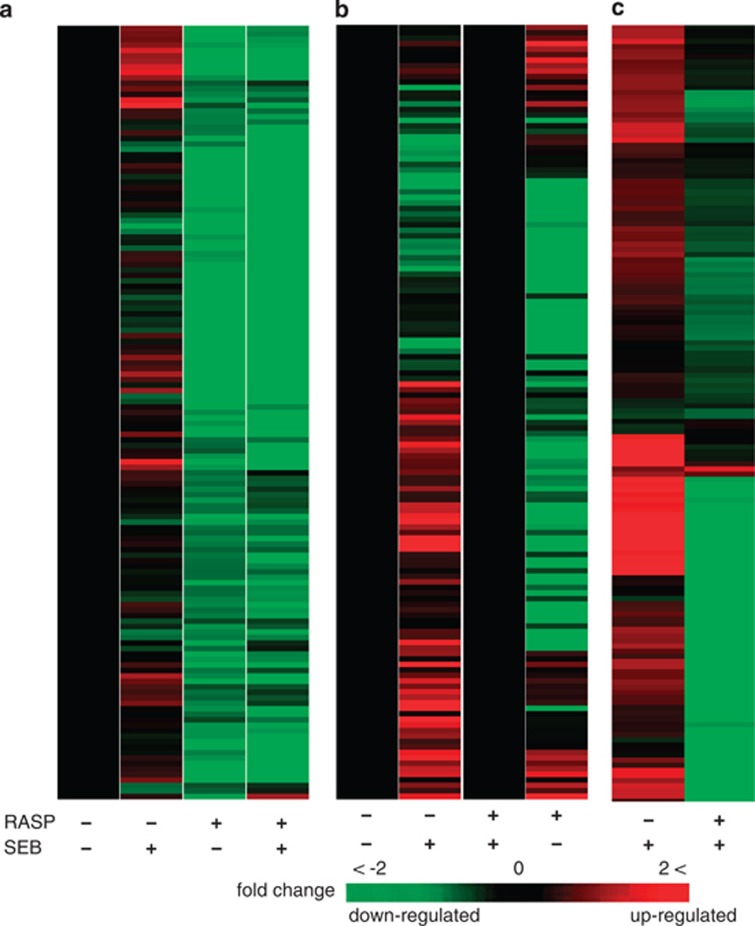
Figure 4.
Expression of immune response genes in leukocytes exposed ex vivo to SEB. Leukocytes isolated from whole blood were treated with SEB (∼106 cells ml−1 in RPMI 1640 and 10% human AB serum at a final concentration of 100 ng ml−1 SEB). Total RNA was isolated using Trizol and expression levels were profiled using cDNA microarrays. Shown here are the 151 RASP-suppressed immune response genes that passed Welch's test and FDR correction (q<0.05). (a) Lanes left to right: pre-RASP samples not exposed to SEB (control), pre-RASP samples exposed to SEB, post-RASP samples not treated with SEB, post-RASP samples exposed to SEB. For comparative visualization purpose, expression values of the other groups were transformed against the pre-RASP control samples (black lane). Heat map of the same data without transformation is given in the supplement (Supplementary Figure S5). (b) Expression values in SEB exposed leukocytes (in both the pre- and post-RASP conditions) were compared with the corresponding SEB untreated groups (pre-RASP control and post-RASP stressed groups). (c) Heat map of the 151 immune response genes in SEB treated groups (in both pre- and post-RASP leukocytes) clustered after subtraction of the corresponding baseline responses (cluster after subtraction of their expressions in the corresponding untreated groups shown in Figure 4b). Figure 4c clearly shows poor response of post-RASP leukocytes towards SEB exposure compared with pre-RASP leukocytes.
MicroRNA array analyses
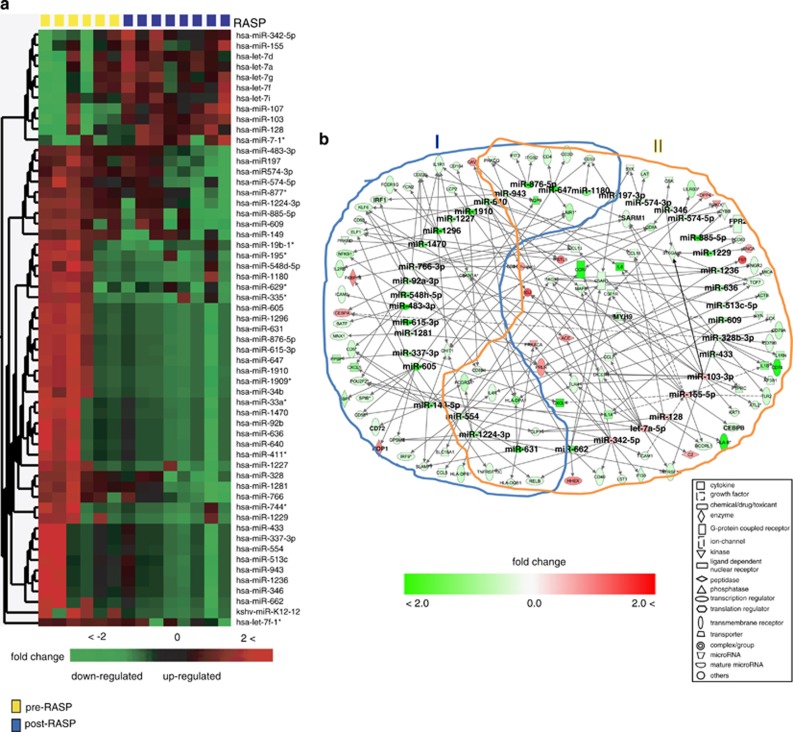
Differentially regulated miRs in pre- and post-RASP samples were assayed using Agilent's human microRNA chip (Agilent, Palo Alto, CA, USA) containing ∼15 000 probes representing 961 unique miRs. Comparison of 535 miRs (that passed normalization and flag filters) using Welch's t-test at P<0.1 with a 1.3-fold change cutoff yielded 57 miRs (Figure 5a). Welch's significance cutoff of 0.1, though relaxed, was used to initially explore 535 miRs. Expressions of 46 of the 57 miRs were suppressed and 11 miRs were induced in post-RASP leukocytes.
Figure 5.
Predicted and experimentally observed targets of RASP-regulated microRNAs. (a) Heat map of differentially regulated 57 microRNAs that passed Welch's T-test (P<0.1) and 1.3-fold change. Most (46 of 57) miRs were downregulated, and 11 miRs were upregulated in post-RASP leukocytes. (b) Predicted or experimentally observed targets of 39 of the 57 RASP-regulated miRs. Shown here are 112 RASP regulated immune response transcripts as targets of the 39 of 57 miRs. (Note: let-7a-5p is synonym for mature let-7f). Many of the miR-155 and let-7f targets were excluded for simplification and are shown next. The top functions and pathways associated with 112 target transcripts are: (I) transcription of immune response genes, communication between innate and adaptive immune cells, activation and proliferations of leukocytes, glucocorticoid receptor signaling; and (II) inflammatory response, chemotaxis, maturation of DC, communication between DC and NK cells, hemopoisis, regulation of activated leukocytes, pattern-recognition receptors and acute phase response signaling. Overall, most RASP-suppressed miRs were significantly associated with inflammatory response, and activation and proliferation of leukocytes.
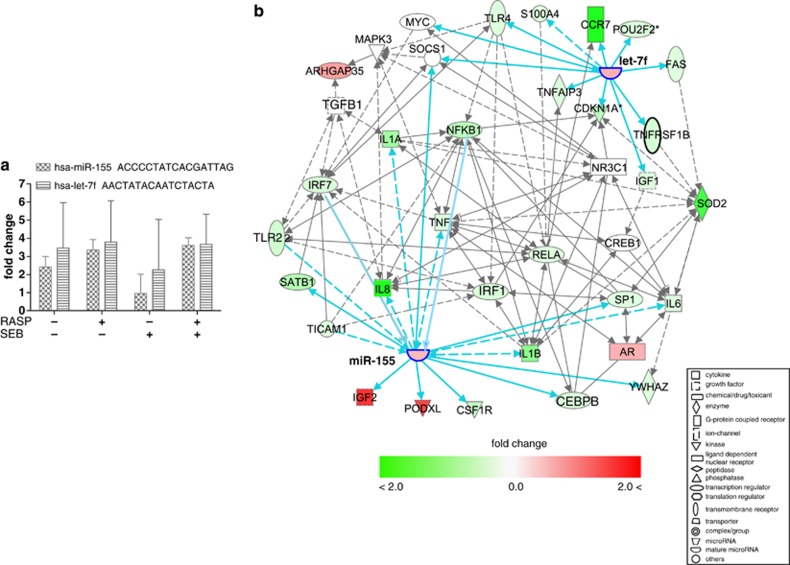
A miR target scan analyses were used to identify both high-prediction and experimentally proven targets of these 57 differentially regulated miRs (Figure 5b). Among the upregulated miRs, hsa-miR-155 and hsa-let-7f (Figure 6a) were predicted to target many RASP-suppressed transcripts, including transcription regulators of genes important for dendritic cell maturation and glucocorticoid receptor signaling (Figure 6b). Expression of miR-155 was suppressed in pre-RASP samples exposed to SEB, but it was induced in post-RASP samples treated with SEB (Figure 6a). Other stress-induced miRs are predicted to have regulatory connection with stress-affected inflammatory cytokines, antigen presenting molecules and transcription regulators of genes involved in the immune response (Figure 5b, Supplementary Figure S6A). The RASP-suppressed miRs (miR-662, miR-647, miR-876-5P, miR-631, miR-1296, miR-615-3P and miR-605) have a number of predicted targets of regulation among stress-regulated genes involved in NFκB and IRF activation pathways (Figure 5b, Supplementary Figure S6B). No targets were identified for two highly suppressed miRs, miR-1910 and 1909*.
Figure 6.
Predicted targets of miR-155 and let-7f families. (a) Expression levels of hsa-miR-155 and hsa-let-7f in pre-RASP (control), post-RASP (stressed) and pre-RASP exposed to SEB, and post-RASP exposed to SEB groups. Sequences of mature miR-155 and let-7f are also shown. (b) Regulatory target genes and modulators of miR-155 and let-7f families were identified from 1396 differentially expressed transcripts that passed Welch's t-test, FDR correction (q<0.05) and 1.5-fold filters. MiR-155 is connected to inflammatory cytokines (IL-1A,-1B, -8, TNF) and transcription factors (IRF7 and NFKB1). Solid lines represent direct and broken lines indirect regulatory connections. Enriched pathways included glucocorticoid receptor signaling, dendritic cell (DC) maturation, bacterial and viral pattern recognition, cross talk between DC and NK cells, TREM1 signaling, acute phase response signaling, and communication between innate and adaptive immunity. Others showed that in response to lipopolysaccharides (LPS), miR-155 is highly upregulated in human monocyte-derived DCs, and suppresses inflammatory cytokines of the TLR/IL-1 pathway acting as a negative feedback loop control.25 In another study, it regulates the production of inflammatory cytokines by targeting C/EBPbeta (CEBPB) in tumor-associated macrophages.26, 27 Monk et al. and others observed that the family of NFκBs and IRFs are necessary for the transcription of pri-miR-155, and its expression is modulated by the TLRs and MAPK signaling molecules28, 29 Upregulation of miR-155 in spite of suppression of the up-stream inducers of miR-155 indicate the presence of other regulators that induce expression of miR-155 under battlefield-like stress.
Expression data-based prediction of TFs and target genes
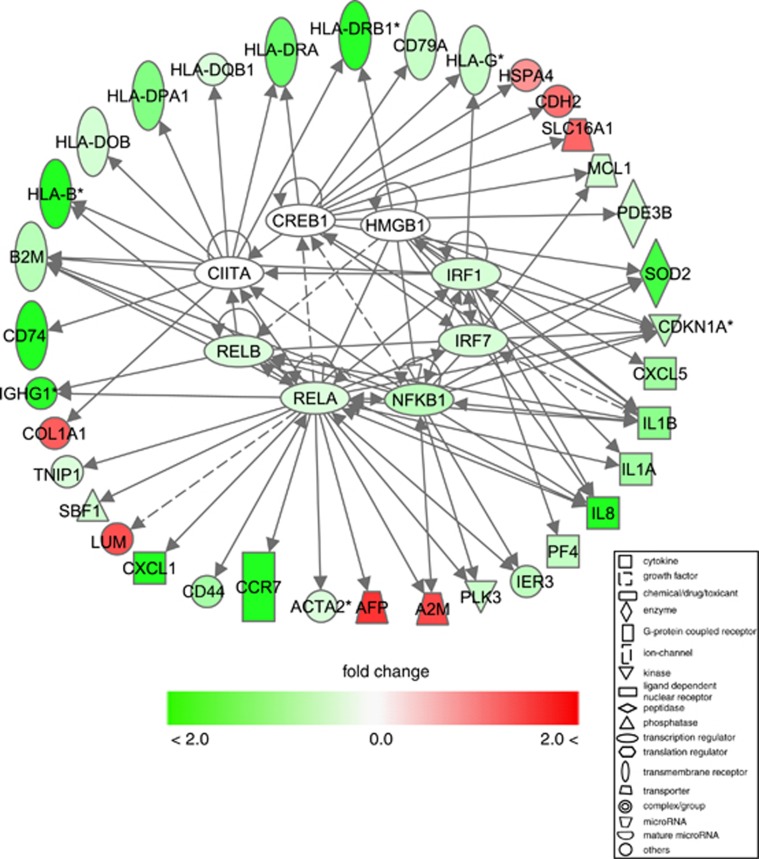
Computational data analyses tools and databases (see Materials and methods) were used for empirical and predictive association of TFs with their regulatory targets among RASP-altered genes. Activated or inhibited TFs, common regulatory sites of target genes and prediction z-scores of identified TFs were computed based on the 1369 differentially regulated genes obtained from cDNA array data (Table 2). TFs at the top of stress-inhibited list (IRF7, RELA, NFκB1, RELB, CREB1, IRF1, HMGB1 and CIITA) and their differentially expressed targets (Table 2) are involved in inflammation, priming of adaptive immune response and glucocorticoid receptor signaling (Figure 7, Supplementary Figures S7, S8). Regulatory sites for a number of TFs, including SP1, CREB1, ATF6 and cEBP, and binding sites for the defense critical NFκB TFs complex and stress response sites (STRE predicted to be regulated by MAZ and MZF1) were among common regulatory motifs identified for some of stress-suppressed genes (Supplementary Table S6). Stress-activated factors included GFI1, MYC, FOXM1, GLI2, MAX and HNF1A (Table 2), and these factors induced genes important for steroid hormone biosynthesis (for example, cortisol) and suppressed immune-related genes.
Table 2. Predicted transcription factors and targets identified among 1396 genes that passed Welch's t-test, FDR correction (q⩽0.05) and 1.5-fold change cutoff.
| TF | z-score | P-value | Target molecules in data set |
|---|---|---|---|
| Activated transcription factors and targets | |||
| GFI1 | 3.1 | 4.1E-04 | CASP1, CDKN1A, CEBPA, GUSB, ICAM1, IL1A, IL1B, IL8, IRF1, MMP7, NFKB1, NFKB2, RELA, RELB, TRAF3 |
| MYC | 3 | 1.6E-17 | ACAT1, ACTB, ACTN1, AFP, AHCY, ALB, BCAT1, BCL6, BIN1, BIRC2, BIRC5, CAPN2, CASP1 CASP10, CAV1, CCND1, CCND3, CD44, CD48, CDC20, CDH2, CDK1, CDK11A/CDK11B, CDKN1A, CEBPA, COL14A1, COL1A1, CSPG4, CYFIP2, DDX11/DDX12, DDX3X, DDX5, DUSP6, EDN1, EGR2, EIF2S2, F2, F3, FBN1 |
| FOXM1 | 2.8 | 4.8E-05 | BIRC5, CCND1, CDC20, CDK1, CDKN1A, CENPA, CENPF, FOXM1, KDR, KIF20A, MMP2, PLK4, TGFBR2 |
| GLI2 | 2.7 | 3.2E-02 | CCL5, CCND1, CDK1, CDKN1A, IL1B, ITGB1, KRT1, KRT17, PTCH1, SFRP1 |
| MAX | 2.4 | 1.4E-03 | BCL6, CDKN1A, EDN1, FTH1, ID1, KLF6, LAMP2, MTHFD1, PDGFRB, SERINC3, TSC2, UBE2C |
| HNF1A | 2.1 | 3.6E-02 | ABCC2, AFP, AKR1C4, ALB, ANPEP, APOB, AQP9, BCL6, C2, CCND1, DPP4, DUSP6, FAM107B, FBXO8, FGA, FGB, G0S2, GNB2L1, HNF4A, IGFBP1, KIF20A, KIR3DL1, LCAT, MTHFD1, NAPA, PDK1, PFKP, PIH1D1, PRLR, PZP, SERPINA7, SLC26A1, SLCO1A2, SSTR4, TRA@, UQCRC2, UROD |
| Inhibited transcription factors and targets | |||
| CEBPB | −2.2 | 1.3E-11 | ACTG2, ALB, C3, CCL5, CCND1, CD14, CDKN1A, CEBPA, CEBPB, COL1A1, CP, CSF1R, CTSC, CXCL5, CYP19A1, DDX5, DEGS1, FTL, HLA-C, HP, HSPD1, ICAM1, ID1, IGFBP1, IL1B, IL1RN, IL8, INMT, IRF9, LAMC1, LCP2, LYN, MGP, MIA, PCTP, PDGFRA, PEA15, PLAUR, PPARD, PRKCD, PR |
| JUNB | −2.3 | 2.8E-03 | ACLY, CAV1, CCND1, CD68, CDC20, COL1A1, CYP19A1, FTH1, MMP2, MVD, NCF2, PTBP2, RELB, SCD |
| CIITA | −2.4 | 1.4E-07 | B2M, CCND1, CD74, COL1A1, HLA-B, HLA-DOB, HLA-DPA1, HLA-DQA1, HLA-DQB1, HLA-DRA, HLA-DRB1 |
| POU2AF1 | −2.6 | 3.4E-03 | BCL6, CCND3, CD79A, CD79B, IGHA1, IGHG1, LCK, TRAF3 |
| STAT1 | −2.8 | 8.2E-12 | A2M, B2M, BIRC5, BTG1, C3, CASP1, CASP2, CASP4, CCL5, CCND1, CCND3, CCR7, CD14, CDKN1A, DPP4, FCER1G, GATA3, GBP1, GZMB, HLADRB1, ICAM1, IFIT3, IL1B, IL8, IRF1, IR5, IRF7, IRF9, LY96, NFE2, PDGFRB, PF4, PRL, PSMB10, PTGS2, SMAD7, SOCS3, STAT2, TLR4, TN |
| FOXO3 | −2.8 | 1.8E-04 | BIRC5, CCND1, CDKN1A, CTGF, CYR61, FOXM1, FOXO1, GPX1, IER3, IGFBP1, IL8, NAMPT, NOS3, SATB1, SOD2, TNFRSF1B, TXNIP, UBC, UBE2C |
| SPI1 | −2.9 | 1.5E-10 | ACTB, CCR7, CD14, CD68, CD79A, CD79B, CEBPA, CSF1R, CYBB, DUSP6, FCER1G, FLI1, FTH1, GNB2L1, GPX1, IGL@, IL1B, IL1RN, IRF9, ITGA5, ITGAM, ITGB2, MCL1, MMP2, NCF2, P2RY1, PIK3CG, PTGS2, PTPRC, RELA, TK1, TLR2, TLR4 |
| IFI16 | −3 | 1.8E-04 | CCL5, CCND1, CDKN1A, EDN1, GPX1, ICAM1, IFI16, IL1B, IL1RN, IL2RB, IL8, RPA3, STAT2 |
| HMGB1 | −3.1 | 1.6E-06 | CD83, CDKN1A, CXCL5, HLADRB1, ICAM1, IL1A, IL1B, IL8, MIA, PTGS2, RELB, SIRT1, TLR2, TLR4 |
| IRF1 | −3.2 | 1.0E-06 | B2M, CASP1, CASP2, CCL5, CCND1, CDKN1A, CYBB, EIF4A3, HLA-G, IFIT3, IL1B, IL8, IRF1, IRF5, IRF7, IRF9, LTB, NFE2, PF4, PSMB10, PTGS2, SOCS7, STAT2, TRIM22 |
| CREB1 | −3.4 | 1.5E-08 | ARPC3, ATP6V0B, BTG2, CCND1, CD3D, CD4, CD68, CD79A, CDH2, CEBPB, CYP19A1, CYP51A1, CYR61, DIO2, EDN1, EGR2, FN1, FOSB, GALNT1, HERPUD1, HLA-DRA, HLA-G, HMGCS1, HSPA4, IL1B, INHA, IRF7, MCL1, PDE3B, PDGFRA, PER1, PRL, PTGS2, SCD, SLC16A1, SLC2A4, SOD2, TF, TFAP2A, UPP1 |
| NFKB1 | −3.4 | 1.9E-08 | A2M, ADORA1, AKR1B1, B2M, BTG2, CCL5, CCND1, CDKN1A, COL2A1, CYBB, FANCD2, GATA3, GNB2L1, ICAM1, IER3, IFNGR2, IGHG1, IL1B, IL1RN, IL8, IRF1, LTB, MICA, NFKB1, NFKB2, PLK3, POU2F2, PRKACA, PTGS2, RELA, RELB, SOD2, TK1, TLR2, TNFAIP3 |
| RELA | −3.7 | 3.1E-17 | A2M, ABCG2, ACTA2, AFP, B2M, BIRC2, BTG2, CAV1, CCL5, CCND1, CCR7, CD44, CDKN1A, COL2A1, CXCL1, CYBB, CYP19A1, DIO2, EDN1, EWSR1, F3, GDF15, HLA-B, ICAM1, IER2, IER3, IFNGR2, IGHG1, IL1A, IL1B, IL1RN, IL8, INPP5D, IRF1, IRF7, L |
| IRF7 | −3.9 | 3.0E-03 | CASP4, CCL5, GBP1, IFI16, IFIT3, IRF1, IRF9, ISG20, ITGAM, MCL1, NAMPT, PSMB10, STAT2, TLR4, TMPO, TRIM21, TRIM22 |
Abbreviation: TF, transcription factor/regulator.
Regulation z-score; P-value of overlap.
Figure 7.
Transcription factors predicted to be inhibited by battlefield-like stressors and their targets among stress-modulated genes. Shown here transcription factors predicted to be inhibited by battlefield stressors (Table 2) and their targets among 288 stress-affected transcripts (filtered using Welch's t-test and FDR, q<0.001, and >1.5-fold change). Enriched function and pathways of these transcripts include activation and proliferation of leukocytes, maturation of DCs, communication between innate and adaptive immunity, glucocorticoid receptor signaling and antigen presentation pathway.
Summary
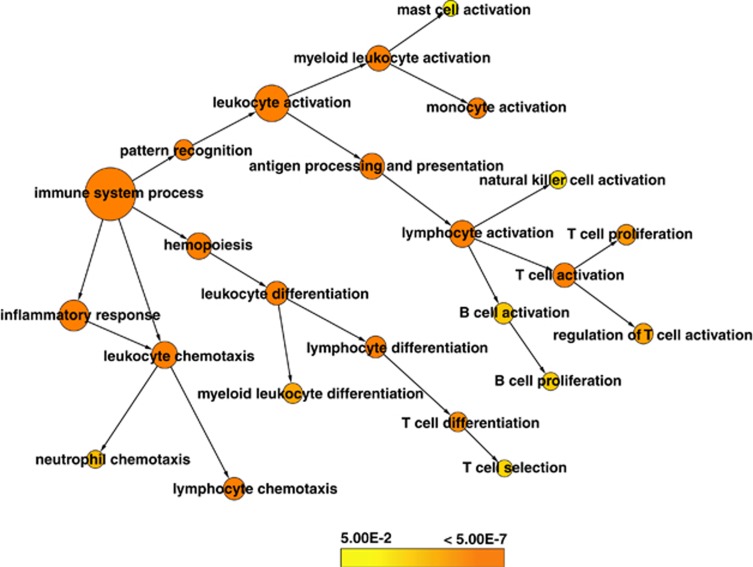
Most immune response genes were downregulated in post-RASP leukocytes compared with pre-RASP leukocytes. Functional enrichment analysis of these downregulated genes revealed their involvement in microbial pattern recognition, cytokine production and reception, chemotaxis, intercellular adhesion, immunological synapse formation, regulation of immune response, and activation and proliferation of immune cells (Figure 8).
Figure 8.
Functional network of differentially expressed genes connected by their sub-functions in the immune system. Network showing enriched functions of genes involved in the immune responses: activation of immune cells, differentiation, proliferation, antigen presentation and infection-directed migrations. Genes involved in all these functions were downregulated by the RASP stressors. Each node represents a category of gene ontology of the pathways of the immune system. Node sizes are proportional to the number of genes belonging to each category according to gene ontology, and the intensity of the node indicates significance of hypergeometric test after Bonferroni correction (q⩽0.05).
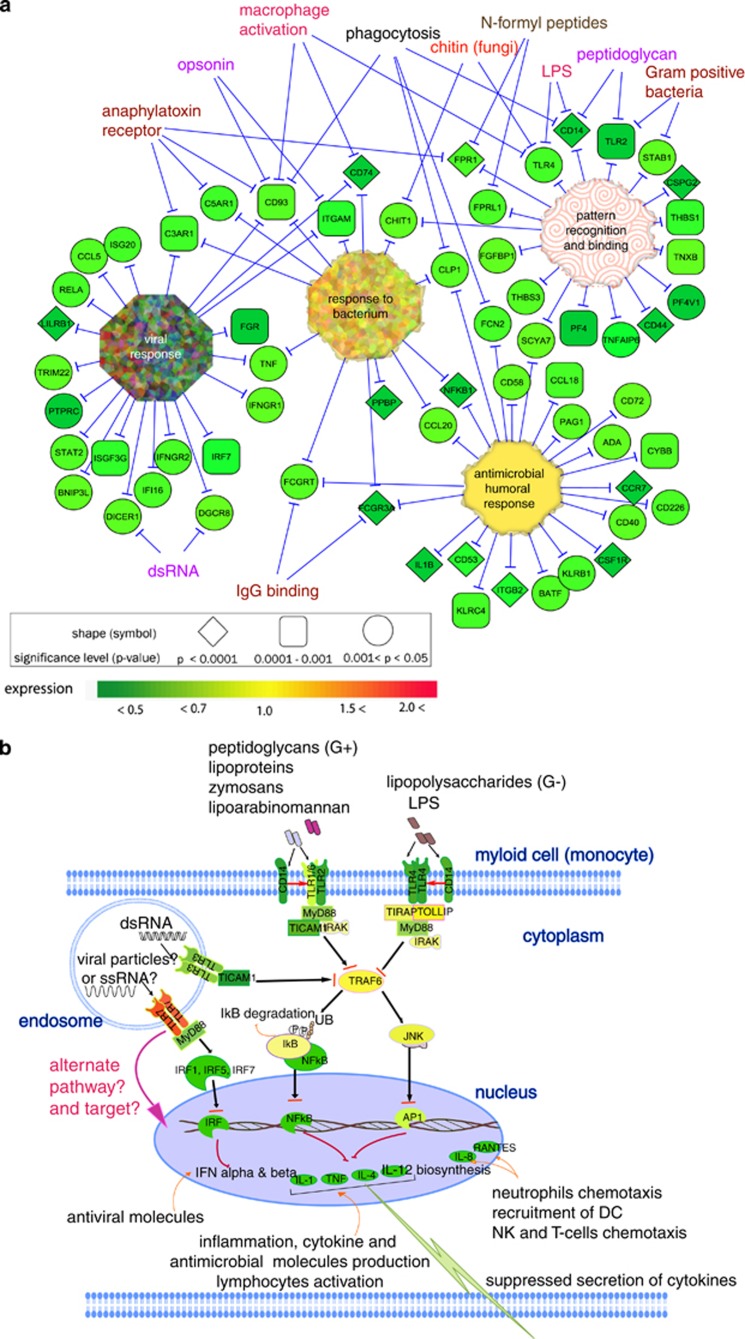
Our data suggest that the prolonged intense stress of RASP induced suppression of microbial pattern receptors of innate immunity (Figure 9a) and impairments in infection-directed maturation, activation, inflammatory response, motility and proliferation of myeloid cells (Figures 9b and c). These impaired innate cells may also fail to prime the adaptive arm of the immune response (Figure 10a).
Figure 9.
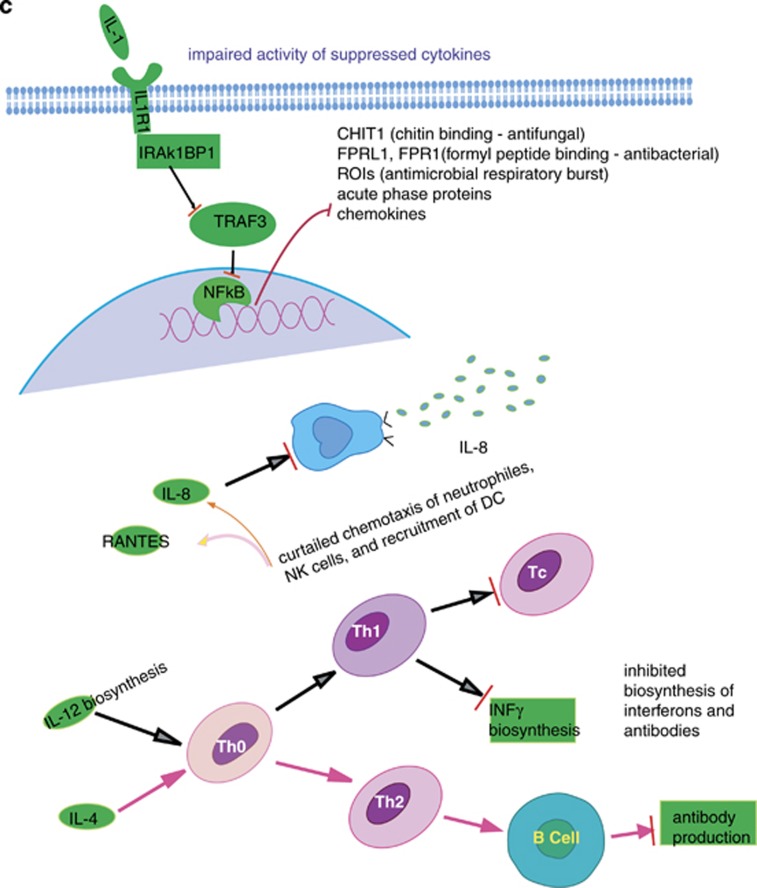
Representations of role of suppressed transcripts in the immune responses. (a) Altered immune response genes involved in pattern recognition, viral, antibacterial and effector (humoral) responses. (b) Roles of stress downregulated genes in the cellular pathways of immune response: flat ended red arrows represent suppression of the corresponding pathway (biological process). Microbial recognition receptors, inflammatory cytokines (IL1, IL1R, TNFα, CD40), chemotaxis (IL8, IL8R, RANTES, CCR5, CCR7), lymphocyte recruitment (IL4, IL12) and production of effector molecules (INFγ, IL2, IL2RB) were downregulated in post-RASP leukocytes. (c) Actions of secreted cytokines on other leukocytes: impaired activity of suppressed IL-1 on other myeloid cells to secret antimicrobial effector molecules; depleted concentration gradient of IL-8 providing curtailed guidance to neutrophils and NK cells to sites of infection, and suppressed IL-8 and RANTES unable to recruit and induce maturation of dendritic cells (for antigen presentation). Suppressed transcripts important for T-cell polarization (cellular or humoral) may mean deprivation of the host under stress from having protective immunity.
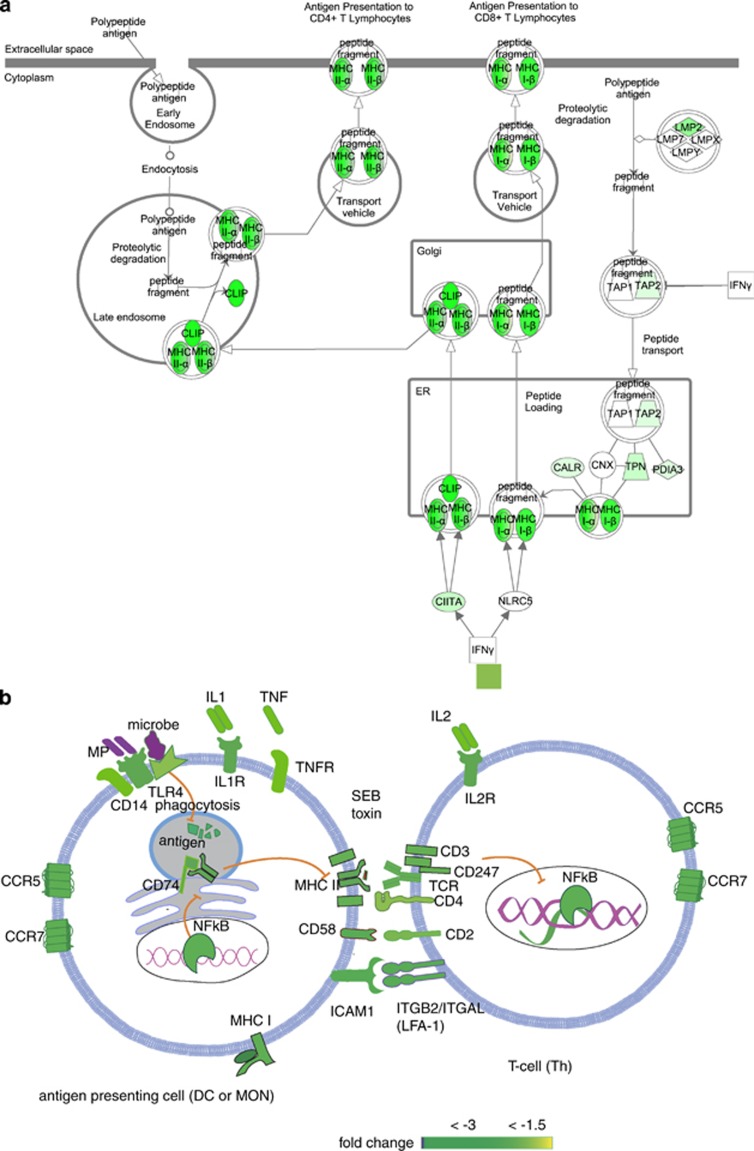
Figure 10.
Stress-suppressed genes involved in antigen presentation and synapse formation. (a) Antigen presentation pathways: this KEEG pathway taken via IPA was colored for the 288 stress-regulated genes that passed Welch's t-test, FDR correction (q⩽0.001) and changed by ⩾1.5-fold (between pre- and post-RASP groups). (b) Expression of genes important for immunological synapse formation: suppression of transcripts important in antigen preparation, presentation, chemotaxis, intercellular binding, antigen reception and downstream signaling (green nodes) may impair formation of productive immunological synapse, which may explain the poor response of post-RASP leukocytes to SEB exposure; although SEB is presented without intracellular presentation, antigen presenting and receptor molecules of the synapse were suppressed.
Adaptive cells' antigen receptors, co-receptors, signal transducers, intercellular adhesion molecules and chemokine receptors were highly suppressed in post-RASP leukocytes (Figure 10b). It is likely that these stress-debilitated lymphocytes have deficits in the activation, proliferation, differentiation and clonal expansion for mounting a defensive response against infections, which is consistent with the impaired response of post-RASP leukocytes to SEB exposure.
Discussion
The suppression of transcripts of critical immune response pathways and regulatory networks after battlefield-like stress is consistent with impaired innate and adaptive immune responses of cellular and humoral immunity. Downregulation of transcripts involved in TLR and chemokine and chemokine receptor signaling pathways indicate suppressed inflammatory response, impaired maturation of antigen presenting cells, impaired affinity maturation of integrins and impaired migration, extravasation and homing of antigen presenting cells and T cells to nearby draining lymph nodes or infection sites.
Antigen preparation and presentation was the most suppressed pathway among immune response processes (Supplementary Figure S9). Suppression of antigen presentation, including T-cell receptor and integrin pathways, indicate impaired immunological synapse formation (poor MHC-restricted antigen recognition and T-cell activation), leading to impaired adaptive and effector immune responses. Particularly, suppression of transcripts involved in cytoskeleton-dependent processes (chemokine-guided migration, integrin-mediated adhesion, immunological–synapse formation, cellular polarization, and actin–microtubule-aided receptor sequestration and signaling) likely curtails the dynamic cellular framework of T-cell activation (Figure 10b).
Unlike reports of differential regulation of Th1 and Th2 type responses observed in college students on the day of a stressful examination17 and in caregivers of chronically sick relatives,18 our data suggest that battlefield-like stressors impair not only Th1 but also Th2 type responses as shown by suppressed transcripts of TLR2 and 4, and the cytokines IL4, IL4R and IL10RA in post-RASP leukocytes. Suppression of inflammatory molecules (for example, IL1A and 1B, and IL1R1, TNF members and TNF receptors, and NFκB class of factors) and Th2 classes of cytokines are features of battlefield-like stress that are distinct from acute, and chronic psychological stresses.
Previously, miR-155 has been reported to be proinflammatory. MiR-155(−/−) mice are highly resistant to experimental autoimmune encephalomyelitis,19 and show suppression of antigen-specific T helper cell function and markedly reduced articular inflammation.20 Here miR-155 transcripts were elevated in post-RASP leukocytes (with or without SEB exposure), but were suppressed in pre-RASP leukocytes exposed to SEB (Figure 6a). This suggests that miR-155 may mediate an anti-inflammatory response to SEB toxin exposure. Also the regulatory connection of miR-155 to many stress-suppressed inflammatory cytokines may indicate a role in regulating these cytokines and glucocorticoid receptor elements, and in modulating maturation of antigen presenting cells under battlefield-like stress (Figure 6b).
The poor response of post-RASP leukocytes to SEB ex vivo challenge (Figure 4) is consistent with suppressed expression of MHCs, T-cell receptors, co-receptors and integrins that are important for activation of antigen presenting cells and T cells. Similarly, RASP-suppressed immune response genes stayed suppressed in post-RASP leukocytes exposed ex vivo to Yersinia pestis (plague) and dengue virus serotype IV as compared with pre-RASP leukocytes exposed to the same pathogens (unpublished observations).
Overall, our results clearly demonstrated that battlefield-like stressors suppress broad categories of immune response pathways, which may explain why chronically stressed individuals show poor vaccine responses, impaired wound healing and susceptibility to infections.
Conclusion
Suppressed expression of genes critical to innate, humoral and cellular immunity in post-RASP leukocytes indicate compromised protective immunity, which was confirmed by the impaired response of post-RASP leukocytes to SEB challenge. After 2 months of chronic intense stress of the RASP, the numbers and ratios of different subpopulations of leukocytes (of soldier) were within normal ranges, despite gene expression changes and impaired responses to a SEB toxin, which is indicative of anergy of post-RASP leukocytes. These observations put a caveat to the current diagnostic practice of counting immune cells to ascertain the integrity and protective ability of the immune system.
Study limitations and future directions
We have studied male cadets, and hence our results do not address gender differences in immunosuppressive responses to battlefield-like stress. Follow-up studies may be useful to check the timing and extent of recovery of the immune system to the pre-stress state, and to identify pathways or routes followed back to immune system homeostasis, which may mirror the path to immune suppression.
Molecular signatures of stress-induced immunosuppression can potentially reveal diagnostic and pharmacologic strategies to diagnose and treat it. We used a number of prediction tools to identify a set of 59 transcripts from the data reported here that can distinguish post-RASP, pre-RASP and pathogen-treated leukocytes. Such set of transcripts can be used potentially to diagnose the immune system state resulting from severe stress as distinguished from infection or malignancy.
Further transcriptomic and proteomic analyses may yield novel preventative, prognostic and therapeutic opportunities to intervene the negative consequences of severe and chronic stresses on health.
Materials and methods
Study subjects
The course of our US Army Ranger Training Subjects, now called Ranger Assessment and Selection Program (RASP), was conducted from 21 October 2004 to 15 December 2004. A total of 15 informed and consented RASP participants enrolled in our study. These trainees underwent a very stressful, three-phase training at three separate locations. The RASP program at Ft. Benning included 3 weeks of intense physical training in conjunction with calorie and sleep limitations. This was followed by about 2 weeks of training in mountainous areas in Georgia and then by 3 more weeks of training in the swamps of Florida. The attrition rate for our study subjects (out of the 15 informed and consented volunteers) was one-third during the first 3 weeks, but the 10 other volunteers remained to the end of the training. Five trainees were reinserted for ‘post training' testing. The ‘inserts' were from among the same batch of trainees and increased the n value to the constant of 15 individuals before and after the RASP program.
Blood sample collection
Whole blood (from each subject) was drawn in Leucopack tubes (BRT Laboratories Inc., Baltimore, MD, USA) before and after the 8-week RASP, and immediately spun at 200 g for 10 min. The concentrated leukocyte layer (buffy coats) was collected and treated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) for RNA isolation and then stored at –80 °C. Differential and complete blood counts were obtained immediately after blood collection using a hemocytometer, and subsequently using an ABX PENTRA C+ 60 Flow Cytometer (Horiba ABX, Irvine, CA, USA). Blood samples were also collected in PAXgene Blood RNA Tubes (VWR Scientific, Buffalo Grove, IL, USA) for direct RNA isolation.
Treatment of leukocytes with SEB toxin
Leukocytes (∼ 106 cells ml−1 in RPMI 1640 and 10% human AB serum media) were treated with SEB toxin to a final concentration of 100 ng ml−1 and SEB treated cells were incubated for 6 h at 37 °C and 5% CO2.
Treated leukocytes were centrifuged at 350 g for 15 min. The pellets were treated with 2 ml TRIzol (Invitrogen) reagent, and kept at –80 °C for RNA isolation.
RNA isolation
For cDNA microarray analysis, total RNA was isolated using the Trizol reagent according to the manufacturer's instructions. The RNA samples were treated with DNase-1 (Invitrogen) to remove genomic DNA and were re-precipitated by isopropanol. The Trizol isolated RNA was used in cDNA microarrays analysis.21 For oligonucleotide microarrays, total RNA was isolated using PAXgene tubes following the manufacturer's protocol. Isolated RNA samples were stored at –80 °C until they were used for microarray and real-time PCR analyses. The concentration and integrity of RNA were determined using an Agilent 2000 BioAnalyzer according to manufacturer's instructions. ArrayControl RNA Spikes (Ambion, Austin, TX, USA) were used to monitor RNA integrity in hybridization, reverse transcription and RNA labeling.
cDNA synthesis, labeling, hybridization and image processing
RNA was reverse transcribed and labeled using Micromax Tyramide Signal Amplification (TSA) Labeling and Detection Kit (Perkin Elmer, Inc., Waltham, MA, USA) following the manufacturer's protocol. The slides were hybridized at 60 °C for 16 h (for cDNA microarrays and Trizol isolated RNA) and at 55 °C for 16 h (for oligonucleotide microarrays and PAXgene isolated RNA). Hybridized slides were scanned and recorded using a GenePix Pro 4000B (Axon Instruments Inc., Union City, CA, USA) optical scanner, and the images were digitized using GenePix 6.0 software (Axon Instruments Inc.).
Preparation of cDNA microarrays
Human cDNA microarrays were prepared using sequence-verified PCR elements produced from ∼10 000 well-characterized human genes of The Easy to Spot Human UniGEM V2.0 cDNA Library (Incyte Genomics Inc., Wilmington, DE, USA). The PCR products, ranging between 500 and 700 bp, were deposited in 3 × saline sodium citrate at an average concentration of 165 μg ml−1 on CMT-GAPS II (γ-aminopropylsilane) coated slides (Corning Inc., Corning, NY, USA), using VersArray MicroArrayer (Bio-Rad, Hercules, CA, USA). The cDNAs were UV-cross-linked at 120 mJ cm−2 using UV Stratalinker 2400 (Stratagene, La Jolla, CA, USA). The microarrays were baked at 80 °C for 4 h. The slides were treated with succinic anhydride and N-methyl-2-pyrrolidinone to remove excess amines.
Oligonucleotide microarrays
For oligonucleotide microarray, we used the Human Genome Array-Ready Oligo Set Version 3.0 (Operon Biotechnologies, Inc., Huntsville, AL, USA), which includes 34 580 oligonucleotide probes representing 24 650 genes and 37 123 RNA transcripts from the human genome. The oligonucleotide targets were deposited in 3 × saline sodium citrate at an average concentration of 165 μg ml−1 onto CMT-GAPS II Aminopropylsilane-coated slides (Corning Inc.) using a VersArray Microarrayer. Microarrays were UV-crosslinked at 120 mJ cm−2 using UV Stratalinker 2400. Then slides were baked at 80 °C for 4 h, and were treated with succinic anhydride and N-methyl-2-pyrrolidinone to remove excess amines on the slide surface. Slides were stored in boxes with slide racks and the boxes were kept in desiccators.
Real-time QPCR
RT–QPCR arrays of 100 genes associated with inflammation, TFs, and antigen preparation and presentation pathways were carried out using Dendritic & Antigen Presenting Cell Pathway (PAHS 406) and NFκB Pathway (PAHS 25) RT2 Profiler PCR Arrays (SABiosciences) according to manufacturer's instructions. Four replicates of RNA samples isolated using PAXgene from trainees before and after RASP were assayed. The data were analyzed using SABiosciences' web-based software, RT2 Profiler PCR Array Data (SABiosciences).
Reverse transcriptase reagent (iScript) and real-time PCR master mix (QuantiTect SYBR Green PCR Kit) were obtained from Bio-Rad Inc. (Hercules, CA, USA), and Qiagen Inc., respectively. Real-time PCR were carried out in i-Cycler Real-time PCR apparatus (Bio-Rad Inc., Milpitas, CA, USA), using three to five biological replicates for each primer pair (based on sample availability). The custom oligonucleotide primers (Supplementary Table S4) were designed using Primer3 software (http://www.basic.northwestern.edu/biotools/primer3.html), or based on those from UniSTS (http://www.ncbi.nlm.nih.gov/unists) and Universal Probe Library for Human (Roche Applied Science, www.roche-applied-science.com). Their specificities were verified in the BLAST domain at NCBI. Parallel amplification reaction using 18S rRNA primers was carried out as a control. Threshold cycle (Ct) for every run was recorded and then converted to fold change using the equation: [(1+E)ΔCt]GOI/[(1+E)ΔCt]HKG, where ΔCt stands for the difference between Ct of control and treated samples of a given gene, which are either genes of interest (GOI) or housekeeping genes (HKG) and E stands for primer efficiency, calculated from slope of best fitting standard curve of each primer pair.
ELISA
Plasma concentrations of PRL, IGF-I and II, TNFα and enzymatic activity of superoxide dismutase were determined using ELISA kits from Calbiotech, Inc. (Spring Valley, CA, USA, Catalog no. PR063F), Diagnostic Systems Laboratories, Inc. (Webster, TX, USA, Catalog nos DSL-10-2800 and DSL-10-2600), Quantikine of R&D Systems, Inc. (Minneapolis, MN, USA, Catalog no. DTA00C) and Dojindo Molecular Technologies, Inc. (Gaithersburg, MD, USA, Catalog no. S311), respectively, following manufacturers' protocols.
Microarray data analyses
Background and foreground pixels of the fluorescence intensity of each spot on the microarrays were segmented using ImaGene 8.0 (BioDiscovery Inc., El Segundo, CA, USA), and the spots with the highest 20% of the background and the lowest 20% of the signal were discarded. Local background correction was applied. Genes that passed this filter in all experiments were selected for further study. Then, subgrid-based lowess normalization was performed for each chip independently. Additional per spot (dividing by control channel) and per gene (to specific samples) normalization were also performed under the Genespring GX 11.5 platform (Agilent Technologies Inc., Santa Clara, CA, USA). Statistical analysis was computed using Welch's t-test (P<0.05) with Benjamini and Hochberg FDR multiple correction for cDNA microarray data (oligonucleotide microarray data were analyzed without FDR Correction). Two-dimensional clustering was carried out based on samples and genes for visualization and assessment of reproducibility in the profiles of the significant genes across biological replicates.
Interaction networks and gene ontology enrichment
We used Bingo 2.3 (www.cytoscape.org) for gene ontology enrichment: hypergeometric distribution test with FDR or Bonferroni corrections (P<0.05). Biological processes, molecular functions and cellular components of each cluster of genes were compared with the global annotations and over-represented categories after corrections were analyzed and visualized. Functional analysis and pathways associated with stress and pathogen-regulated genes were analyzed using ingenuity pathway analysis (IPA) (Ingenuity Systems Inc., Redwood City, CA, USA). We used Cytoscape Version 2.8.2 (www.cytoscape.org) for visualizing and analyzing enriched gene ontologies, and for molecular interaction network constructions.
MicroRNA analysis
Expression profiles of miRs were assayed using Agilent's human miRNA v3 microarrays (Agilent Technologies Inc.) consisting of 15k targets representing 961 miRs. Differentially expressed miRs were analyzed using Qlucore Omices Explorer 2.2 (Qlucore AB, Lund, Sweden) and GeneSpring GX 11.5 (Agilent Technologies Inc.). Target transcripts of profiled miRs were identified using target scan of Genespring GX 11.5 (Agilent Technologies Inc.) and IPA (Ingenuity Systems Inc.). Interaction networks of differentially expressed miRs and their target mRNAs were constructed using IPA (Ingenuity Systems Inc.).
Treatment of leukocytes with SEB
Leukocytes isolated from leucopack blood samples were plated in six well tissue culture plates (∼106 cells ml−1 in GIBCORPMI 1640 media and 10% human AB serum) and treated with SEB (Toxin Technology Inc., Sarasota, FL, USA) at a final concentration of 100 ng ml−1 SEB. Cells were incubated for 6 h at 37 °C and 5% CO2. At the end of the incubation period, treated leukocytes were collected by centrifugation at 350 g for 15 min. Cell pellets were treated with 2 ml Trizol and kept at –80 °C for RNA isolation.
cDNA microarray (expression) data-based prediction of TFs, regulatory binding sites and downstream target identification
Potential regulatory sites of differentially regulated genes were identified using HumanGenome9999 (Agilent Technologies Inc.) containing partial human genome sequences (9999 bp upstream regions for 21787 genes). Statistically significant (P<0.05) common regulatory motifs of 5 to 12 nucleotides long were identified. The searching region was set to range 1 to 500 nucleotides upstream of transcription start sites. Other tools used for this purpose include MATCH and TFSEARCH. Cognate TFs regulating identified (common regulatory) sites were searched from different prediction and repository databases: DBD (www.transcriptionfactor.org), JASPAR (http://jaspar.cgb.ki.se), TRANSFAC 7.0 - Public (http://www.gene-regulation.com/pub/databases.html) using ChipMAPPER,22 ConTra v2,23 Pscan24 and IPA (Ingenuity Systems Inc.). Expression data-based prediction Z-scores and regulatory targets were analyzed using IPA. Regulator–target interaction networks and pathways were generated using Cytoscape (www.cytoscape.org) and IPA (Ingenuity Systems Inc.).
Array Data Archive
Array data are stored in the Gene Expression Omnibus (GEO) with accession number; GSE39148 at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39148.
Acknowledgments
We are grateful to the Defense Threat Reduction Agency for funding.
The views, opinions and/or findings contained in this report are those of the authors and should not be construed as official Department of the Army position, policy or decision, unless so designated by other official documentation. Citations of commercial organizations or trade names in this report do not constitute an official Department of the Army endorsement or approval of the products or services of these organizations. Research was conducted in compliance with IRB approved human subjects protocol no. 1014 for initial collection of samples, and A-16815 for continuation of data evaluation. The human use approval was obtained from the local Protection of Human subjects Office and further approved by the Human Research Protection Office, Office of Research Protections, US Army Medical Research and Materiel Command, Fort Detrick, MD, USA.
Footnotes
Supplementary Information accompanies the paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary Material
References
- Cohen S, Janicki-Deverts D, Miller GE. Psychological stress and disease. JAMA. 2007;298:1685–1687. doi: 10.1001/jama.298.14.1685. [DOI] [PubMed] [Google Scholar]
- Rokutan K, Morita K, Masuda K, Tominaga K, Shikishima M, Teshima-Kondo S, et al. Gene expression profiling in peripheral blood leukocytes as a new approach for assessment of human stress response. J Med Invest. 2005;52:137–144. doi: 10.2152/jmi.52.137. [DOI] [PubMed] [Google Scholar]
- Motoyama K, Nakai Y, Miyashita T, Fukui Y, Morita M, Sanmiya K, et al. Isolation stress for 30 days alters hepatic gene expression profiles, especially with reference to lipid metabolism in mice. Physiol Genomics. 2009;37:79–87. doi: 10.1152/physiolgenomics.90358.2008. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Miao J, Hanley G, Stuart C, Sun X, Chen T, et al. Chronic restraint stress promotes immune suppression through toll-like receptor 4-mediated phosphoinositide 3-kinase signaling. J Neuroimmunol. 2008;204:13–19. doi: 10.1016/j.jneuroim.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Padgett DA, Glaser R. How stress influences the immune response. Trends Immunol. 2003;24:444–448. doi: 10.1016/s1471-4906(03)00173-x. [DOI] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Glaser R, Gravenstein S, Malarkey WB, Sheridan J. Chronic stress alters the immune response to influenza virus vaccine in older adults. Proc Natl Acad Sci USA. 1996;93:3043–3047. doi: 10.1073/pnas.93.7.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier JN, Mathieu J, Mailfert Y, Multon E, Drouet C, Jouan A, et al. Chronic restraint stress induces severe disruption of the T-cell specific response to tetanus toxin vaccine. Immunology. 2001;102:87–93. doi: 10.1046/j.1365-2567.2001.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cowden LG, King JD, Briles DA, Schroeder HW, Stevens AB, et al. Effects of chronic stress and interleukin-10 gene polymorphisms on antibody response to tetanus vaccine in family caregivers of patients with Alzheimer's disease. Psychosom Med. 2007;69:551–559. doi: 10.1097/PSY.0b013e3180cc2c61. [DOI] [PubMed] [Google Scholar]
- Glaser R, Sheridan J, Malarkey WB, MacCallum RC, Kiecolt-Glaser JK. Chronic stress modulates the immune response to a pneumococcal pneumonia vaccine. Psychosom Med. 2000;62:804–807. doi: 10.1097/00006842-200011000-00010. [DOI] [PubMed] [Google Scholar]
- Gouin JP, Kiecolt-Glaser JK. The impact of psychological stress on wound healing: methods and mechanisms. Immunol Allergy Clin North Am. 2011;31:81–93. doi: 10.1016/j.iac.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroussi HY, Williams RL, Zhang QL, Villines D, Marucha PT. Ala42S100A8 ameliorates psychological-stress impaired cutaneous wound healing. Brain Behav Immun. 2009;23:755–759. doi: 10.1016/j.bbi.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiank C, Holtfreter B, Starke A, Mundt A, Wilke C, Schutt C. Stress susceptibility predicts the severity of immune depression and the failure to combat bacterial infections in chronically stressed mice. Brain Behav Immun. 2006;20:359–368. doi: 10.1016/j.bbi.2005.10.151. [DOI] [PubMed] [Google Scholar]
- Reiche EM, Nunes SO, Morimoto HK. Stress, depression, the immune system, and cancer. Lancet Oncol. 2004;5:617–625. doi: 10.1016/S1470-2045(04)01597-9. [DOI] [PubMed] [Google Scholar]
- Friedl KE, Moore RJ, Hoyt RW, Marchitelli LJ, Martinez-Lopez LE, Askew EW. Endocrine markers of semistarvation in healthy lean men in a multistressor environment. J Appl Physiol. 2000;88:1820–1830. doi: 10.1152/jappl.2000.88.5.1820. [DOI] [PubMed] [Google Scholar]
- Nindl BC, Barnes BR, Alemany JA, Frykman PN, Shippee RL, Friedl KE. Physiological consequences of U.S. Army Ranger training. Med Sci Sports Exerc. 2007;39:1380–1387. doi: 10.1249/MSS.0b013e318067e2f7. [DOI] [PubMed] [Google Scholar]
- Mendis C, Das R, Hammamieh R, Royaee A, Yang D, Peel S, et al. Transcriptional response signature of human lymphoid cells to Staphylococcal enterotoxin B. Genes Immun. 2005;6:84–94. doi: 10.1038/sj.gene.6364160. [DOI] [PubMed] [Google Scholar]
- Paik IH, Toh KY, Lee C, Kim JJ, Lee SJ. Psychological stress may induce increased humoral and decreased cellular immunity. Behav Med. 2000;26:139–141. doi: 10.1080/08964280009595761. [DOI] [PubMed] [Google Scholar]
- Glaser R, MacCallum RC, Laskowski BF, Malarkey WB, Sheridan JF, Kiecolt-Glaser JK. Evidence for a shift in the Th-1 to Th-2 cytokine response associated with chronic stress and aging. J Gerontol Ser A Biol Sci Med Sci. 2001;56:M477–M482. doi: 10.1093/gerona/56.8.m477. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowska-Stolarska M, Alivernini S, Ballantine LE, Asquith DL, Millar NL, Gilchrist DS, et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc Natl Acad Sci USA. 2011;108:11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Hammamieh R, Neill R, Ludwig GV, Eker S, Lincoln P, et al. Early indicators of exposure to biological threat agents using host gene profiles in peripheral blood mononuclear cells. BMC Infect Dis. 2008;8:104. doi: 10.1186/1471-2334-8-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu VD, Kohane IS, Riva A. The MAPPER database: a multi-genome catalog of putative transcription factor binding sites. Nucleic Acids Res. 2005;33 (Database issue:D91–D97. doi: 10.1093/nar/gki103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broos S, Hulpiau P, Galle J, Hooghe B, Van Roy F, De Bleser P. ConTra v2: a tool to identify transcription factor binding sites across species, update 2011. Nucleic Acids Res. 2011;39:W74–W78. doi: 10.1093/nar/gkr355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Xu Z, Ding T, Kuang DM, Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta. Cell Mol Immunol. 2009;6:343–352. doi: 10.1038/cmi.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Monk CE, Hutvagner G, Arthur JS. Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS One. 5:e13669. doi: 10.1371/journal.pone.0013669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Toomey NL, Diaz LA, Walker G, Ramos JC, Barber GN, et al. Oncogenic IRFs provide a survival advantage for Epstein-Barr virus- or human T-cell leukemia virus type 1-transformed cells through induction of BIC expression. J Virol. 85:8328–8337. doi: 10.1128/JVI.00570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.