Abstract
We obtained detailed kinetic characteristics–stoichiometry, reaction rates, substrate affinities and equilibrium conditions–of human PPIP5K2 (diphosphoinositol pentakisphosphate kinase 2). This enzyme synthesizes ‘high-energy’ PP-InsPs (diphosphoinositol polyphosphates) by metabolizing InsP6 (inositol hexakisphosphate) and 5-InsP7 (5-diphosphoinositol 1,2,3,4,6-pentakisphosphate) to 1-InsP7 (1-diphosphoinositol 2,3,4,5,6-pentakisphosphate) and InsP8 (1,5-bis-diphosphoinositol 2,3,4,6-tetrakisphosphate), respectively. These data increase our insight into the PPIP5K2 reaction mechanism and clarify the interface between PPIP5K catalytic activities and cellular bioenergetic status. For example, stochiometric analysis uncovered non-productive, substrate-stimulated ATPase activity (thus, approximately 2 and 1.2 ATP molecules are utilized to synthesize each molecule of 1-InsP7 and InsP8, respectively). Impaired ATPase activity of a PPIP5K2-K248A mutant increased atomic-level insight into the enzyme's reaction mechanism. We found PPIP5K2 to be fully reversible as an ATP-synthase in vitro, but our new data contradict previous perceptions that significant ‘reversibility’ occurs in vivo. PPIP5K2 was insensitive to physiological changes in either [AMP] or [ATP]/[ADP] ratios. Those data, together with adenine nucleotide kinetics (ATP Km=20–40 μM), reveal how insulated PPIP5K2 is from cellular bioenergetic challenges. Finally, the specificity constants for PPIP5K2 revise upwards by one-to-two orders of magnitude the inherent catalytic activities of this enzyme, and we show its equilibrium point favours 80–90% depletion of InsP6/5-InsP7.
Keywords: bis-diphosphoinositol tetrakisphosphate; cellular energy homoeostasis; diphosphoinositol pentakisphosphate; diphosphoinositol polyphosphate; inositol pyrophosphate; inositol 1,3,4-trisphosphate 5/6-kinase (ITPK1)
Abbreviations: DIPP, diphosphoinositol-polyphosphate phosphohydrolase; DTT, dithiothreitol; GST, glutathione transferase; InsP, inositol phosphate; InsP3, inositol 1,3,4trisphosphate; InsP4, inositol 1,3,4,5-tetrakisphosphate; InsP5, inositol 1,3,4,5,6-pentakisphosphate; InsP6, inositol hexakisphosphate; 1-InsP7, 1-diphosphoinositol 2,3,4,5,6-pentakisphosphate; 5-InsP7, 1-diphosphoinositol 2,3,4,5,6-pentakisphosphate; InsP8, 1,5-bis-diphosphoinositol 2,3,4,6-tetrakisphosphate; InsS6, inositol hexasulphate; IP6K, inositol hexakisphosphate kinase; ITPK1, inositol 1,3,4-trisphosphate 5/6-kinase; NCBI, National Center for Biotechnology Information; PP-InsPs, diphosphoinositol polyphosphates; PPIP5K, diphosphoinositol pentakisphosphate kinase; PPIP5K2KD, human diphosphoinositol pentakisphosphate kinase 2 kinase domain
INTRODUCTION
PP-InsPs [diphosphoinositol polyphosphates, such as 1-InsP7 (1-diphosphoinositol 2,3,4,5,6-pentakisphosphate), 5-InsP7 (5diphosphoinositol 1,2,3,4,6-pentakisphosphate) and InsP8 (1,5-bis-diphosphoinositol 2,3,4,6-tetrakisphosphate); see Figure 1] have been shown to regulate diverse cellular processes such as vesicle trafficking, exocytosis, cytoskeletal dynamics, apoptosis, environmental stress response and insulin secretion and signalling (reviewed in [1–6]). Research into the mechanisms of action of the PP-InsPs has identified a cyclin/cyclin kinase ‘receptor’ in yeast that is allosterically regulated by 1-InsP7 [7]. There is also evidence that 5-InsP7 can regulate cell-signalling by competing with PtdIns(3,4,5)P3 for binding to pleckstrin homology domains [8]. Finally, PP-InsPs have been reported to promote the diphosphorylation of certain proteins thereby regulating inherent protein activities and protein/protein interactions [9–11]. Each of these proposed mechanisms of action of the PP-InsPs are based upon a fundamental principle in signal transduction: their cellular levels are altered in a predictable manner in response to a specific intracellular or extracellular stimulus. For example, levels of 1-InsP7 are elevated during nutrient stress in yeast [7]. Total InsP7 levels also increase substantially following the addition of growth factors to cells that have been serum-starved overnight [8]. As for InsP8, its levels are elevated when cells are subjected to either hyperosmotic stress or a thermal challenge [12,13]. There are other circumstances that cause InsP7 and InsP8 levels to decrease, such as following oxidative stress [14]. Finally, InsP8 concentration decreases during bioenergetic stress [15].
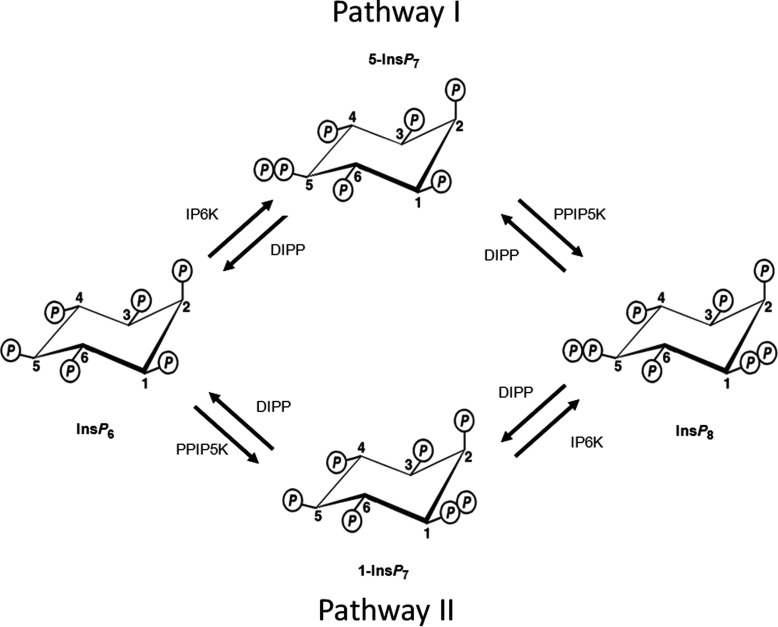
Figure 1. Pathways of enzymatic synthesis of PP-InsPs.
Figure modified from Shears 2009 [5]. The IP6Ks (myoinositol hexakisphosphate kinase; Kcs1 in yeast [48]), of which there are three isoforms in mammals, IP6K1, IP6K2 and IP6K3 [26,48,52], phosphorylate InsP6 and 1-InsP7 at the 5 position [41,53]. The PPIP5Ks (Vip1 in yeast [21]), of which there are two isoforms in mammals, PPIP5K1 and PPIP5K2 [18,19], phosphorylate InsP6 and 5-InsP7 at the 1 position [20,41]. Thus, the concerted actions of the IP6Ks and the PPIP5Ks leads to two routes of synthesis to InsP8, which are designated pathway I and pathway II after Padmanabhan et al. [17]. The diphosphate groups that are added by these kinases are hydrolysed by a family of PP-InsP phosphohydrolases (DIPPs) [44,45].
It is because of all of these signalling activities that there is a need to characterize how the turnover of PP-InsPs is regulated in intact cells. As with any signal transduction cascade, in order to understand its dynamic behaviour in vivo, it is critical to determine the kinetic parameters of its constituent signalling enzymes [16,17]. Such information–enzyme stoichiometry, reaction rates, substrate affinities and equilibrium conditions–helps us to understand the regulation of product generation as well as substrate depletion. In the current study, we provide new information concerning these properties of a PPIP5K (diphosphoinositol pentakisphosphate kinase), an enzyme that phosphorylates InsP6 (inositol hexakisphosphate) and 5-InsP7 to 1-InsP7 and InsP8, respectively (Figure 1).
Mammalian cells express two PPIP5Ks: types 1 and 2 that are 160 and 138 kDa, respectively [18,19]. The expression and purification of recombinant enzymes of such a large size are problematic. Fortunately, these are modular proteins; the N-terminal one-third of each of these proteins contains their kinase domain [18,20,21], following which is a centrally located PtdIns(3,4,5)P3-binding module [22]. The active sites of these two kinase domains appear to be structurally and catalytically indistinguishable [18,20]; the relatively small number of residues elsewhere in these domains that are dissimilar are confined to the protein surface that is distal to the active site [20]. Thus, using the kinase domain of human PPIP5K2KD (PPIP5K2 kinase domain) as a model, we have now characterized the kinetic properties of this class of enzyme.
For an ATP-dependent kinase such as IP6K (inositol hexakisphosphate kinase) or PPIP5K, its activity may not solely depend upon the concentrations of inositol phosphate substrates. There is growing interest in the possibility that changes in the levels of adenine nucleotides may also be of importance in regulating IP6K and PPIP5K activity in vivo [1,2,5,6]. This idea has taken support from the evidence that bioenergetic homoeostasis interfaces with PP-InsP turnover and cell signalling [15,23–26]. For example, some experiments indicate that bioenergetic health sets the cellular levels of PP-InsPs [15,24–26]. Other work [23] indicates that it is bioenergetic status that is regulated by the PP-InsPs. One of the goals of the current study was to increase our understanding of the role of PPIP5K in this complex inter-relationship between signalling and metabolism.
Interest in the possibility of a link between cellular bioenergetics and PP-InsP turnover began with the recognition that these molecules contain high-energy phosphoanhydride bonds that undergo rapid turnover in vivo [27,28]. Subsequently, in what was a startling exception for kinases in general [29], and inositol phosphate kinases in particular [30], a high Km (>1 mM) of the IP6K for ATP was determined [26]. IP6Ks were further shown to have a similarly high Km for ADP, facilitating an ability to act ‘in reverse’ as ATP-synthases [26]. This ATP-synthase activity was proposed to occur in vivo [26]. Thus, akin to InsP5 (inositol 1,3,4,5,6-pentakisphosphate) 2-kinase in plants [31], the IP6K reaction direction and rate of activity appear to be controlled by changes in ATP/ADP ratios [24]. These data have consolidated the idea that PP-InsP turnover interfaces with cellular bioenergetic homoeostasis.
Experiments with partially purified native preparations of PPIP5K [25] have led to the conclusion that, just like IP6K, PPIP5K is reversible under in vivo conditions, such that cellular InsP8 levels can be controlled by physiological changes in ATP/ADP ratios [25]. This hypothesis has been well received [32–34]. However, it is a natural concern that the properties of partially purified native enzyme preparations may be influenced by contaminating proteins and other small-molecule modulators. The intrinsic lability of native PPIP5K [19] is also an issue. Thus, one of the goals of the current study was to use recombinant kinase to investigate through a kinetic study the manner in which PPIP5K activity–in both the ‘forward’ and ‘reverse’ directions–may be influenced by physiological changes in levels of adenine nucleotides as well as by the concentrations of inositol phosphate substrates.
Additionally, we should note that reliable data on reaction rates, reversibility and steady-state conditions are not the only parameters that systems biologists utilize when transferring kinetic data with isolated enzymes into mathematical models of metabolic pathways in vivo; information on reaction stoichiometry is also important [16]. In the current study we determined that PPIP5K's substrate-independent ATPase activity [20] is considerably more significant than was previously appreciated, and is stimulated up to 4-fold upon the binding of InsP (inositol phosphate) substrate to the active site. We discuss the significance of this bioenergetic cost to the cell in terms of the roles of PPIPKs in vivo. We also discuss how this discovery of substrate-dependent ATPase activity of PPIP5K provided an opportunity to gain new insight into the reaction mechanism of this particular kinase.
One of the reasons that such extensive kinetic studies have not previously been performed with recombinant PPIP5Ks is the difficulties in obtaining samples of the PP-InsPs with the appropriate degree of purity. PP-InsPs are not commercially available and, at the time of writing, we are unaware of any laboratories that are currently chemically synthesizing these materials. PP-InsPs can be prepared enzymatically but these highly charged molecules are inherently difficult to isolate in a salt-free form to a high degree of purity. Recently, a new method was introduced, which accomplishes that goal [35]. We have used these procedures in our study.
MATERIALS AND METHODS
Materials
InsP6 was purchased from Calbiochem and [3H]InsP6 (~13.0 Ci/mmol) from PerkinElmer. Non-radiolabelled and [3H]-radiolabelled PP-InsPs (1-InsP7, 5-InsP7 and InsP8) were all synthesized enzymatically with the appropriate purified enzymes (see below). Deionized water was used throughout.
Protein expression and purification
The human PPIP5K2 kinase domain (residues 1–366) [hPPIP5K2KD, NCBI (National Center for Biotechnology Information) accession number NP_056031.2] and its single-site mutants were expressed in Escherichia coli and purified as previously described [20]. The protein was stored at −80°C.
The human IP6K1 (NCBI accession number NP_695005.1) was expressed and purified as previously described for human PPIP5K1 and PPIP5K2 with the following modifications [18]: the protein was expressed with an N-terminal GST (glutathione transferase) tag in BL21 (DE3) E. coli (Stratagene) by induction overnight. Cells were sonicated in lysis buffer containing an EDTA-free complete protease inhibitor (Roche). The clarified lysate was loaded on to a 5 ml GSTrap HP (GE Healthcare) column, washed and eluted with a linear gradient from 0 to 30 mM of reduced glutathione in 25 mM Tris pH 8.0, 50 mM NaCl and 1 mM DTT (dithiothreitol). The protein was stored with 0.5 mg/ml BSA and 50% glycerol at −80°C. The purity of our enzyme preparations were >95% (see Supplementary Figure S1 at http://www.bioscirep.org/bsr/033/bsr033e022add.htm).
Preparation of PP-InsPs
Enzymatic reactions were carried out in siliconized tubes (PGC Scientific) at 37°C in 20 mM Hepes, pH 6.8, 50 mM NaCl, 6 mM MgSO4, 1 mM DTT, 6 mM phosphocreatine, 24 unit/ml creatine kinase and 5 mM ATP disodium salt. Reactions (15 min) for the synthesis of [3H]-radiolabelled 1-InsP7, 5-InsP7 and InsP8 contained 9.7 μCi/ml [3H]InsP6 (~13.0 Ci/mmol) and either, respectively, 0.076 mg/ml PPIP5K2KD, 0.21 mg/ml GST–IP6K1 or 0.35 mg/ml GST–IP6K1 and 0.076 mg/ml PPIP5K2KD. Reactions for the synthesis of non-radiolabelled 1-InsP7, 5-InsP7 and InsP8 contained 1.2 mM InsP6 and were conducted as follows, respectively: 0.12 mg/ml PPIP5K2KD for 22.5 h, 0.17 mg/ml GST–IP6K1 for 3 h or 0.17 mg/ml GST–IP6K1 and 0.11 mg/ml PPIP5K2KD for 3 h. Reactions were quenched and neutralized with, respectively, 0.2 volumes 2 M HClO4 and 0.34 volumes 1 M K2CO3, 40 mM EDTA or placed at 100°C for 3–5 min, then concentrated in a SpeedVac (Savant) at 43°C to 300 μl.
The PP-InsPs were then purified via a PAGE-based method [35] with the following modifications in order to scale up the procedure. Gels (31 cm×38.5 cm×1.5 mm) were run in a model S2 sequencing gel electrophoresis system (Life Technologies Gibco BRL) at 4°C. Gels were prerun at 1000 V for 1 h prior to sample loading, then run at 1000 V for 48–60 h. The elution positions of the PP-InsPs were determined in a 1 cm wide column at the edge of the gel. The non-radiolabelled PP-InsPs were detected by staining with Toluidine Blue [35]. For the detection of PP-[3H]InsPs, the excised region of the gel was cut into 0.5 cm bands which were each mixed with 1 ml of Solusol (National Diagnostics) on a rocker (25 rev./min) for at least 5 h at room temperature, after which 4 ml of Soluscint XR (National Diagnostics) was added and samples were counted by liquid scintillation spectroscopy. To correct for any unevenness in the chromatography, the Orange-G dye front was used as a guide for determining the elution positions of the products (see Supplementary Figure S2 at http://www.bioscirep.org/bsr/033/bsr033e022add.htm). The appropriate sections of the gels were excised and washed with 5–10 ml 20% (v/v) methanol, followed by a brief rinse with 5 ml water. The gel fragments were homogenized (Tekmar Tissuemizer Mark II) in 5 ml water at 20500 rev./min at 0–4°C prior to extraction of the PP-InsP. Samples were concentrated to 1 ml (non-radiolabelled) or 4000 DPM/μl ([3H]-radiolabelled) in a SpeedVac, aliquoted and stored at −80°C. No sample degradation was detected through a dozen freeze–thaw cycles over a period of several months.
Purity of PP-[3H]InsPs was assessed by HPLC (see the Results section). Non-radiolabelled PP-InsPs were quantified by mass assay of the released orthophosphate upon complete hydrolysis by wet-ashing at 120°C for 48 h [36,37].
Enzyme assays
Kinetic parameters (Km, Vmax, kcat and kcat/Km) for adenine nucleotides and inositol phosphates were determined for PPIP5K2KD under pseudo first-order conditions [38]. Reactions (200 μl, 37°C) simulating intracellular conditions contained 20 mM Hepes-NaOH pH 7.2, 50 mM KCl, 1 mg/ml bovine serum albumen, 1 mM EDTA disodium salt, 1 mM DTT, ATP or ADP disodium salt (1.1 μM to 10 mM), inositol phosphate (10 nM–10 μM), appropriate [3H]-radiolabelled inositol phosphate tracer (4000 DPM) and PPIP5K2. Total MgSO4 varied from 2 to 12 mM depending on ATP and ADP concentrations. Free Mg2+ was 1.2±0.1 mM (estimates performed with WebMaxc Extended, http://www.stanford.edu/~cpatton/webmaxc/webmaxcE.htm, last updated 7/3/09, accessed 8/24/11 [39]). Reactions were quenched and neutralized with, respectively, 0.2 volumes 2 M HClO4 and 0.34 volumes 1 M K2CO3, 40 mM EDTA, then analysed by HPLC.
The mobile phase was initially run through a 4.6×250 mm silica presaturation column (Grace), before samples were loaded onto a 4.6×125 mm Partisphere 5 SAX column (Whatman). Buffer A was 1 mM EDTA disodium salt, Buffer B was 1.3 M (NH4)2HPO4 (Sigma) plus 1 mM EDTA disodium salt, pH 3.85 with phosphoric acid (Aldrich). The gradient (1 ml/min) was as follows: 0–5 min, 0% B; 5–10 min, B increased linearly from 0 to 45%; 10–60 min, B increased linearly from 45 to 100%; 60–75 min, B was 100%. Each fraction (~30/assay, 1 ml each) was mixed with 4 ml MonoFlow 4 scintillant (National Diagnostics) and counted on a liquid scintillation counter for 10 min. For the kinetic determinations, assay linearity was confirmed under all conditions. The Michaelis–Menten equation was fit to plots of initial velocity against substrate concentration using non-linear regression to determine Km and Vmax values (GraphPad Prism v5.03). Results are presented as the means±S.E.M. (n≥2). Similar assay conditions were used to determine equilibrium points.
ATPase activity
ATPase activity (non-productive hydrolysis of ATP to ADP that did not drive phosphorylation of an InsP) was determined by measurement of free orthophosphate in the reaction solution by the Malachite Green detection method used for quantification of PP-InsP preparations (see above) 27–270 μg/ml PPIP5K2KD was incubated at 37°C for 120 min in 50 μl reaction mixtures containing 20 mM Tris/HCl, pH 7.5, 10 mM ATP (unless otherwise indicated), 100 mM KCl, 0.1 mM EDTA and 5 mM MgCl2. Results are presented as the means±S.D. (n=3).
ATP synthesis by PPIP5K2KD
The ATP synthase activity of PPIP5K2KD was determined in a 96-well microplate format by incubating 0.3 μg/ml enzyme at 37°C for 15 min in 20 μl assays containing 1.4× reaction buffer (ATP Determination Kit, Molecular Probes), 650 μM ADP (ATP contamination<1 ppm, Apollo Scientific Limited), 250 nM InsP8 and the indicated concentrations of either InsP6 or InsS6 (inositol hexasulphate). Then 16 μl of either an ATP standard solution or reaction solution was added into 100 μl Standard Reaction Solution (ATP Determination Kit, Molecular Probes) and luminescence was recorded after 15 s using a BioTek Synergy 2 microplate reader. The IC50 values were determined using GraphPad Prism v5.03.
Crystal structure determination
Crystals of PPIP5K2KD in complex with ADP were soaked with InsP8 using previously published procedures [20].
RESULTS AND DISCUSSION
Enzymatic synthesis and purity of PP-InsPs
PP-InsPs possess energetic phosphoanhydride bonds and undergo rapid turnover [27,28], so their cellular synthesis requires a considerable bioenergetic investment. Indeed, variations in cellular bioenergetic status have been proposed to influence PP-InsP synthesis (see the Introduction section). We have investigated the role of PPIP5Ks in this process by determining the kinetic properties of the kinase domain of recombinant human PPIP5K2 (PPIP5K2KD).
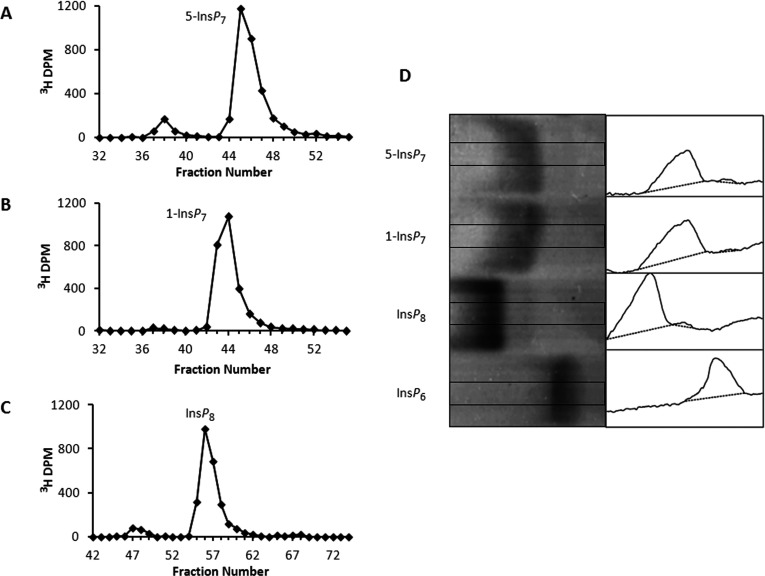
One of our goals was to analyse the ‘forward’ (ATP-consuming) and ‘reverse’ (ATP-producing) reactions; that information has not previously been derived from recombinant enzyme. We therefore required three PP-InsPs for this study, both radiolabelled and non-radiolabelled (1-InsP7, 5-InsP7 and InsP8; Figure 1). These PP-InsPs were not commercially available, and to our knowledge there was not a laboratory that was chemically synthesizing these molecules. We therefore prepared them enzymatically, and purified them by a PAGE-based technique [35]. We determined that the purities of 5-[3H]InsP7, 1-[3H]InsP7 and [3H]InsP8 were 91, 98 and 92%, respectively (Figures 2A–2C). The purities of 5-InsP7, 1-InsP7 and InsP8 were estimated to be 96, 98 and 97%, respectively (Figure 2D). This is an improvement over previous studies in which PP-InsPs were either prepared enzymatically or chemically synthesized, whereupon the final products were 75–90% pure [18,19,40–42].
Figure 2. Purity of PP-InsP preparations.
HPLC (see the Materials and methods section) of 5-[3H]InsP7 (A), 1-[3H]InsP7 (B) and [3H]InsP8 (C). The purities of 5-[3H]InsP7, 1-[3H]InsP7 and [3H]InsP8 were 91, 98 and 92%, respectively. (D) Toluidine Blue-stained PAGE analysis (16 cm gel) of purity of non-radiolabelled PP-InsP preparations (20 nmol/lane). The image was enhanced in ImageJ [54] by increasing contrast and smoothing. Purity was quantified by densitometric analysis (right-hand panels) of the central portion of each lane as indicated by the rectangles in each of the left-hand panels using ImageJ [54]. Commercial InsP6 (not further purified) served as a control. Purity was as follows: 5-InsP7, 96%; 1-InsP7, 98%; and InsP8, 97%.
Kinetic analysis of PPIP5K2KD
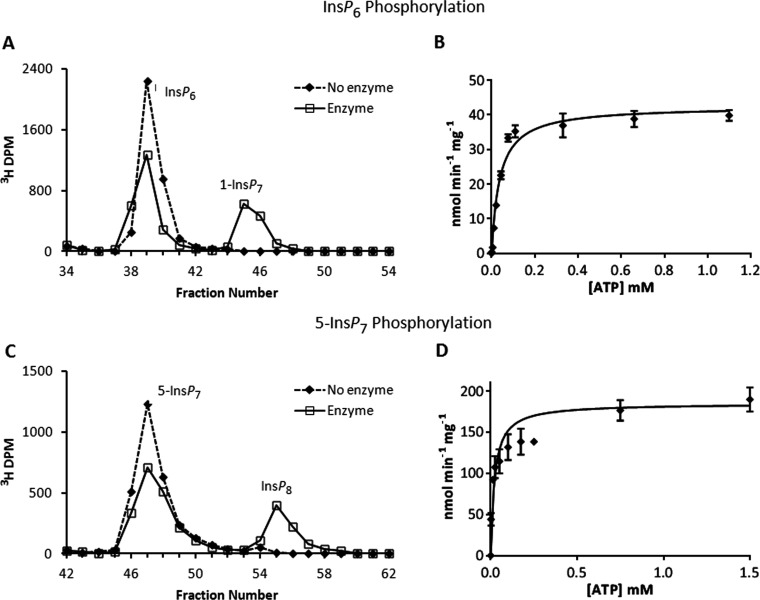
We first used HPLC to analyse the PPIP5K-mediated phosphorylation of [3H]-radiolabelled inositol phosphate substrates using saturating concentrations of ATP (see below) (Figures 3 and 4). Our data (Table 1) indicate that 5-InsP7 is the higher-affinity substrate; we found the Km value of PPIP5K2KD for InsP6 (0.4 μM; Table 1) to be 6.5-fold higher than that for 5-InsP7 (0.06 μM; Table 1). Our results are similar to the previously determined Km values for recombinant PPIP5Ks, 0.1 μM for InsP6 and 0.1–0.3 μM for 5-InsP7 [1,18,19]. These kinetic data demonstrate that the active site will be near-saturated by the levels of inositol phosphate substrates that occur in vivo [1,43]. However, one significant difference with earlier data is that we found that the Vmax values for these reactions (Table 1) were about 20–130-fold higher than those previously reported [18,19] for recombinant PPIP5K. This in turn is largely responsible for our values for kcat/Km–the specificity constants–being up to 100-fold greater (Table 1) than that of previous determinations [18,19]. This brings the specificity constant for the PPIP5Ks much closer to that of the DIPPs (diphosphoinositol-polyphosphate phosphohydrolases) [44,45], and as such is more consistent with the long-standing observation that there is high ongoing turnover of PP-InsPs in vivo [2,28]. Our much higher estimates for the Vmax, which are likely due our obtaining correctly folded protein to a greater degree of purity, counter the argument that PPIP5K activity is too low to participate in ‘phasic’ signalling events [1].
Figure 3. Phosphorylation reactions of PPIP5K2KD and substrate saturation plots for ATP and ADP.
(A) Either no enzyme (broken line) or 27 μg/ml PPIP5K2KD (solid line) was incubated with 10 μM [3H]InsP6, and 11 μM ATP for 20 min as indicated in the Materials and methods section. The reactions were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section. Representative HPLC data are shown. (B) The initial velocity of InsP6 phosphorylation was determined for forward reactions under pseudo first-order rate conditions [38] in which the concentration of the designated inositol phosphate was fixed at saturating levels (10 μM; Table 1), and the concentration of nucleotide was varied as indicated. Each individual data point was analysed by HPLC as described under the Materials and methods section. The Michaelis–Menten equation was fitted to the data using non-linear regression (GraphPad Prism v5.03). Results are presented as the means±S.E.M. (C) Either no enzyme (broken line) or 0.011 μg/ml PPIP5K2KD (solid line) was incubated with 100 nM 5-[3H]InsP7 and 10 mM ATP for 7 min as indicated in the Materials and methods section. The reactions were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section. Representative HPLC data are shown. (D) The initial velocity of 5-InsP7 phosphorylation was determined as described in the legend to panel (B) for InsP6.
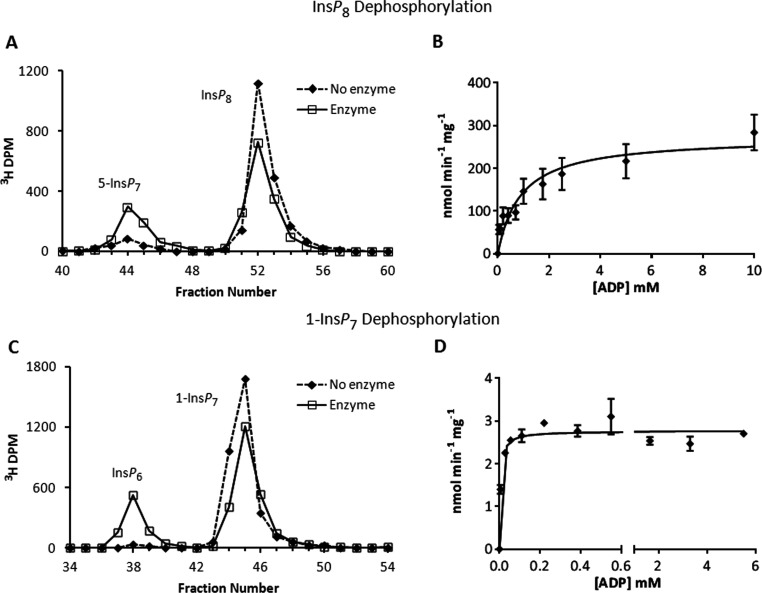
Figure 4. Dephosphorylation reactions of PPIP5K2KD and substrate saturation plots for ATP and ADP.
(A) Either no enzyme (broken line) or 0.56 μg/ml PPIP5K2KD (solid line) was incubated with 10 μM [3H]InsP8 and 5 mM ADP for 10 min as indicated in the Materials and methods section. The reactions were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section. Representative HPLC data are shown. (B) The initial velocity of InsP8 dephosphorylation was determined for reverse reactions under pseudo first-order rate conditions [38] in which the concentration of the designated inositol phosphate was fixed at saturating levels (10 μM; Table 1), and the concentration of nucleotide was varied as indicated. Each individual data point was analysed by HPLC as described under the Materials and methods section. The Michaelis–Menten equation was fitted to the data using non-linear regression (GraphPad Prism v5.03). Results are presented as the means±S.E.M. (C) Either no enzyme (broken line) or 27 μg/ml PPIP5K2KD (solid line) was incubated with 10 μM 1-[3H]InsP7 and 28 μM ADP for 45 min as indicated in the Materials and methods section. The reactions were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section. Representative HPLC data are shown. (D) The initial velocity of 1-InsP7 dephosphorylation was determined as described in the legend to panel (B) for InsP8.
Table 1. Kinetic parameters of PPIP5K2KD:PP-InsP synthesis.
Kinetic parameters were determined under pseudo first-order conditions [38] for phosphorylation of InsP6 or 5-InsP7 for both ATP and InsP substrates (see the Materials and methods section). When fixed, ATP was 10 mM and InsP 10 μM. Initial velocities were measured by conversion of an appropriate [3H]-radiolabelled inositol phosphate tracer (4000 DPM) as analysed by Partisphere 5 SAX HPLC (e.g., see Figures 3A and 3C) for each data point. The Michaelis–Menten equation was fit to plots of initial velocity against substrate concentration using non-linear regression (GraphPad Prism v5.03). No substrate inhibition was observed (results not shown). Results are presented as the means±S.E.M. (n≥2 independent experiments).
| Km (μM) | kcat/Km (M−1·s−1) | ||||||
|---|---|---|---|---|---|---|---|
| Substrates | Nucleotide | InsP | Vmax (nmol·min−1·mg−1) | kcat (s−1) | Nucleotide | InsP | |
| ATP | InsP6 | 37±5 | 0.39±0.09 | 43±2 | 0.030±0.001 | 7.9×102 | 7.6×104 |
| ATP | 5-InsP7 | 22±7 | 0.060±0.009 | 190±10 | 0.13±0.01 | 5.9×103 | 2.2×106 |
Using saturating concentrations of either InsP6 or 5-InsP7, we next derived a substrate saturation plot for ATP; this showed Michaelis–Menten kinetics (Figures 3B and 3D). The value of the Km for ATP (22–37 μM; Table 1) was similar for both InsP6 and 5-InsP7. At an [ATP] of 1 mM, the Vmax values were already attained (Figures 3B and 3D); there was also no significant increase in reaction velocity when [ATP] was further increased to 10 mM (results not shown). These data lead us to conclude that PP-InsP synthesis by PPIP5Ks is protected from physiologically relevant changes in [ATP] (1–5 mM [46]), contrary to the conclusion of a previous report [25]. As well as providing this new information about the characteristics of PPIP5Ks, our kinetic data (Table 1) can also be viewed as a successful quality control test of the recently developed PAGE-based procedures for purification of enzymatically prepared PP-InsPs [35].
Reversibility of PPIP5K and the consequences of substrate competition
Several inositol phosphate kinases have been shown to be physiologically reversible, that is, their relative rates of phosphorylation and dephosphorylation can be influenced by changes in cellular [ATP]/[ADP] ratios: IP6K [26], InsP5 2-kinase [31] and ITPK1 (inositol 1,3,4-trisphosphate 5/6-kinase) [47]. A previous report [25] has included the PPIP5Ks in this category. The degree of the ATP-synthase activities of IP6Ks and PPIP5Ks were further proposed [25,26] to be regulated by physiological changes in adenine nucleotide levels. This idea, which dovetails with the concept that PP-InsPs are highly energetic phosphate donors [26,27], has become well accepted in the field [32–34,48].
We found that PPIP5K2KD was reversible in vitro when it was incubated with ADP plus either 1-InsP7 or InsP8 (Figure 4). Substrate saturation curves showed the reactions to follow Michaelis–Menten kinetics (Figures 4B and 4D). The dephosphorylation of InsP8 proceeded at a slightly greater Vmax (Tables 1 and 2) than did the corresponding forward reaction (5-InsP7 phosphorylation). For the dephosphorylation of InsP8 the Km for ADP was 0.9 mM (Table 2), which is within the range of physiological total ADP concentrations (0.7–1.7 mM; [46]). Interestingly, for the dephosphorylation of 1-InsP7, the value for the Km of ADP was 175-fold lower, and thus much closer to the Km values for ATP in the phosphorylation reactions (Table 2).
Table 2. Kinetic parameters of PPIP5K2KD:ATP synthesis.
Kinetic parameters were determined under pseudo first-order conditions [38] for dephosphorylation (ATP synthesis) of InsP8 or 1-InsP7 for both ADP and InsP substrates (see the Materials and methods section). When fixed, ADP was 10 mM and InsP 10 μM. Initial velocities were measured by conversion of an appropriate [3H]-radiolabelled inositol phosphate tracer (4000 DPM) as analysed via Partisphere 5 SAX HPLC (e.g., see, Figures 4A and 4C) for each data point. The Michaelis–Menten equation was fit to plots of initial velocity against substrate concentration using non-linear regression (GraphPad Prism v5.03). No substrate inhibition was observed (results not shown). Results are presented as the means±S.E.M. (n≥2 independent experiments).
| Km (μM) | kcat/Km (M−1·s−1) | ||||||
|---|---|---|---|---|---|---|---|
| Substrates | Nucleotide | InsP | Vmax (nmol·min−1·mg−1) | kcat (s−1) | Nucleotide | InsP | |
| ADP | InsP8 | 900±280 | 0.022±0.011 | 270±30 | 0.19±0.02 | 2.1×102 | 8.5×106 |
| ADP | 1-InsP7 | 5.2±1.5 | 0.11±0.01 | 2.8±0.1 | 0.002±0.000 | 3.7×102 | 1.7×104 |
The high rate of dephosphorylation of InsP8 by PPIP5K2KD was somewhat surprising in view of our earlier structural data [20] which indicated that, in the crystal complex of PPIP5K2KD, the InsP8 product was under considerable steric strain. It had seemed reasonable to conclude that the increase in entropy upon product release would discourage re-binding of InsP8 for dephosphorylation. Thus, we soaked InsP8 into a crystal complex of PPIP5K2KD and ADP. We found the conformation and catalytic environments of this complex to be very similar (results not shown) to those that we described previously when InsP8 was synthesized inside the crystals [20]. We therefore have no evidence that the mechanism of InsP8 dephosphorylation proceeds any differently than by a reversal of 5-InsP7 phosphorylation.
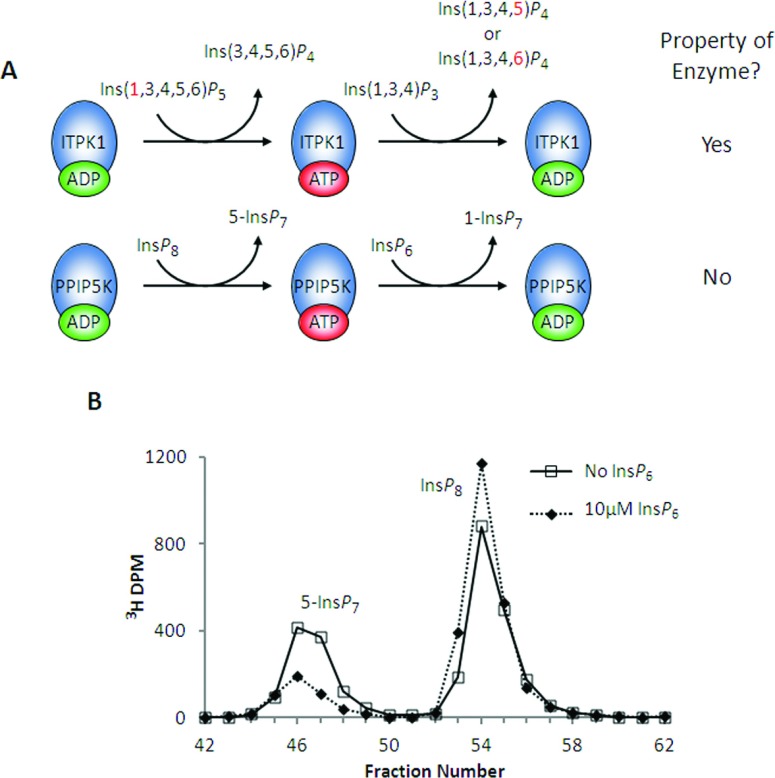
One of the other inositol phosphate kinases that is reversible, ITPK1, is PPIP5K's closest structural homologue [20]. Both PPIP5K2 and ITPK1 envelop ATP tightly within two sets of antiparallel β-sheets that comprise an ATP-grasp fold; in each case, less than 10% of the nucleotide is solvent exposed [20,47]. In the case of ITPK1, the bound nucleotide promotes a phosphotransferase activity by which the dephosphorylation of Ins(1,3,4,5,6)P5 to Ins(3,4,5,6)P4 is stimulated by the phosphorylation of Ins(1,3,4)P3 [47]. The analogous reaction for PPIP5K would be the stimulation of InsP8 dephosphorylation by the presence of InsP6 (Figure 5A). To investigate if that were possible, we assayed the ‘reverse’ (i.e., InsP8 dephosphorylation) activity of PPIP5K2KD in the presence of physiological [InsP6] (Figure 5B). Unlike ITPK1, the phosphorylation of one substrate (InsP6) by PPIP5K2KD merely inhibited (by >70%) the dephosphorylation of another (InsP8) (Figure 5B). We conclude that even though both ITPK1 and PPIP5K are ATP-grasp kinases, only ITPK1 can act as a phosphotransferase.
Figure 5. PPIP5K2KD is not a phosphotransferase: InsP6 inhibits InsP8 dephosphorylation by PPIP5K2KD.
(A, top) This graphic depicts how Ins(1,3,4)P3 stimulates Ins(1,3,4,5,6)P5 dephosphorylation by ITPK1 [47]. The 1-phosphate group (shown in red) that is removed from Ins(1,3,4,5,6)P5 is not released into the bulk phase but is retained by the nucleotide and donated to Ins(1,3,4)P3; this returns the enzyme to its ADP state, ready for another cycle of Ins(1,3,4,5,6)P5 dephosphorylation. (A, bottom) The analogous potential reaction sequence for PPIP5K. (B) Either no enzyme (broken line) or 0.63 μg/ml PPIP5K2KD (solid line; dotted line) was incubated with 10 μM [3H]InsP8 and 10 mM ADP for 10 min as indicated in the Materials and methods section. Where indicated (dotted line) incubations also contained 10 μM InsP6. The reactions were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section. Representative HPLC data are shown.
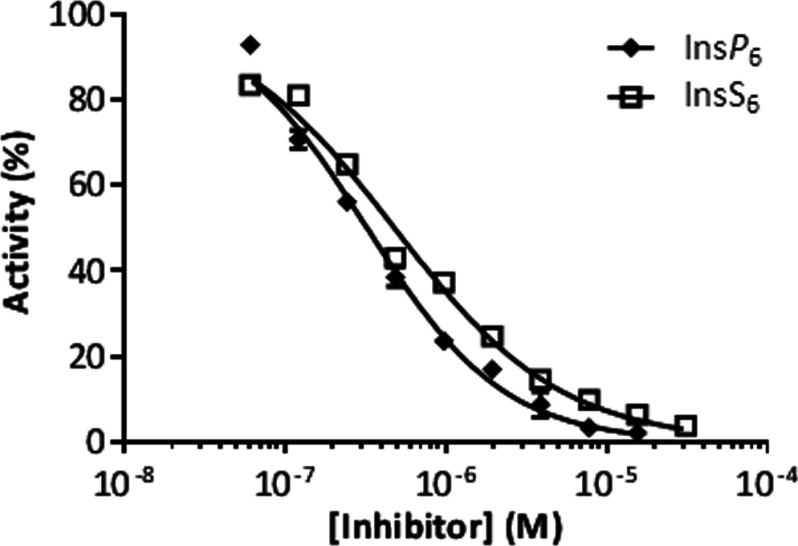
The inhibition of InsP8 dephosphorylation by InsP6 was also demonstrated by using a luciferase-based ATP-detection method (IC50=0.32 μM; Figure 6). The latter result indicates that the luciferase assay could be employed as a high-throughput tool to screen for inhibitors of this enzyme. For example, by using this method we found that InsS6 also inhibited the enzyme (IC50=0.48 μM; Figure 6).
Figure 6. InsP6 and InsS6 inhibit InsP8 dephosphorylation by PPIP5K2KD; a luciferase-based ATP assay.
A total of 0.3 μg/ml PPIP5K2KD was incubated at 37°C with 1.4× reaction buffer (ATP Determination Kit, Molecular Probes), 650 μM ADP and 250 nM InsP8 for 15 min as described in the Materials and methods section. InsS6 and InsP6 were included in the assays as indicated. Then 16 μl of either an ATP standard solution or reaction solution was added into 100 μl Standard Reaction Solution (ATP Determination Kit, Molecular Probes) and luminescence was recorded after 15 s using a BioTek Synergy 2. Data are presented as the means±S.E.M. (n≥3). A dose–response curve was fit using GraphPad Prism v5.03. The IC50 values for InsS6 and InsP6 were 0.48 and 0.32 μM, respectively.
Equilibrium and rate studies of PPIP5K2KD under varied [ATP] and [ADP]
Tables 1 and 2 show that PPIP5K2KD is reversible in vitro and also InsP8 dephosphorylation was associated with a Km value for ADP that has potential physiological relevance. We therefore next investigated to what extent changes in the initial ATP/ADP ratios would influence either the initial reaction rate or equilibrium point of the kinase reaction.
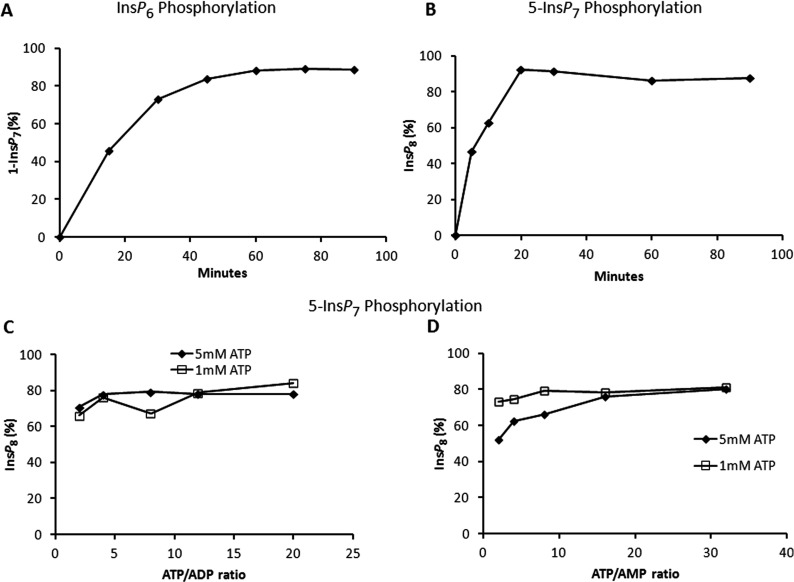
In planning these experiments, we noted that total cellular [ATP] generally varies between 1 and 5 mM, with total cellular ATP/ADP ratios generally in the range of 2–12 [46,49,50]. At physiologically relevant [ATP] and [ADP] concentrations of 5 and 0.5 mM, respectively, the PPIP5K2KD-catalysed equilibrium point was attained at near complete (80–90%) phosphorylation of either InsP6 or 5-InsP7 (Figures 7A–7C). Furthermore, the PPIP5K equilibrium point for the phosphorylation of 5-InsP7 to InsP8 was neither affected by varying total ATP (1 or 5 mM) nor by changing the ATP/ADP ratio from 2 to 20 (Figure 7C). We also assayed PPIP5K2KD activity under physiological extremes of ATP/ADP ratio (2 against 20), using both 1 and 5 mM total [ATP] (results not shown). The changes that we made to the ATP/ADP ratios did not alter reaction rates. These data indicate that the reversibility of PPIP5Ks is not biologically significant in vivo, in contrast to a previous proposal [25]. Although the Vmax for InsP8 dephosphorylation is relatively high, the enzyme's relatively low affinity for ADP may in part explain why that reaction is not favoured, as long as physiological levels of ATP are also present. Our data also demonstrate that the synthesis of both 1-InsP7 and InsP8 is likely well-protected from the consequences of the deteriorating [ATP]/ [ADP] ratio that cells experience when undergoing bioenergetic stress.
Figure 7. Effect of ATP/ADP and AMP upon the equilibrium condition for PPIP5K2KD.
(A) and (B) show a time course of InsP6 and 5-InsP7 phosphorylation, respectively. In (A), 20 μg/ml enzyme was incubated with reaction buffer (see the Materials and methods section) containing 15 μM InsP6, 5 mM ATP and 0.5 mM ADP. In (B), 0.33 μg/ml enzyme was incubated with reaction buffer (see the Materials and methods section) containing 1 μM 5-InsP7 and 5 mM ATP. Assays were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section. (C) and (D) show the equilibrium point for the phosphorylation of 5-InsP7 to InsP8 in which 0.33 μg/ml enzyme was incubated as described in the Materials and methods section under initial conditions of 1 μM 5-InsP7 and either 1 (squares) or 5 (diamonds) mM ATP; either the ATP/ADP ratio (C) or the ATP/AMP ratio (D) was varied as indicated. Assays were conducted for 60 min. The reactions were then quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section.
Equilibrium and rate studies of PPIP5K2KD under varied [AMP]
InsP8 levels in vivo have been found to decrease in response to mild bioenergetic stress that is reflected more in an increase in [AMP] rather than a decrease in [ATP]/[ADP] [15]. However, we did not find any effect of AMP upon the equilibrium condition catalysed by PPIP5K2KD (Figure 7D). We further found that these same changes in [ATP]/[AMP] ratios did not affect PPIP5K2KD reaction rate (results not shown). Thus the decrease in InsP8 in relation to elevated [AMP] in vivo [15] likely proceeds through a signalling pathway, rather than through direct effects on InsP8 synthesis by PPIP5K2. We [15] have previously excluded AMPK (AMP-activated protein kinase) from being involved, so we will need to consider other mechanisms.
The stoichiometry of PPIP5K2KD
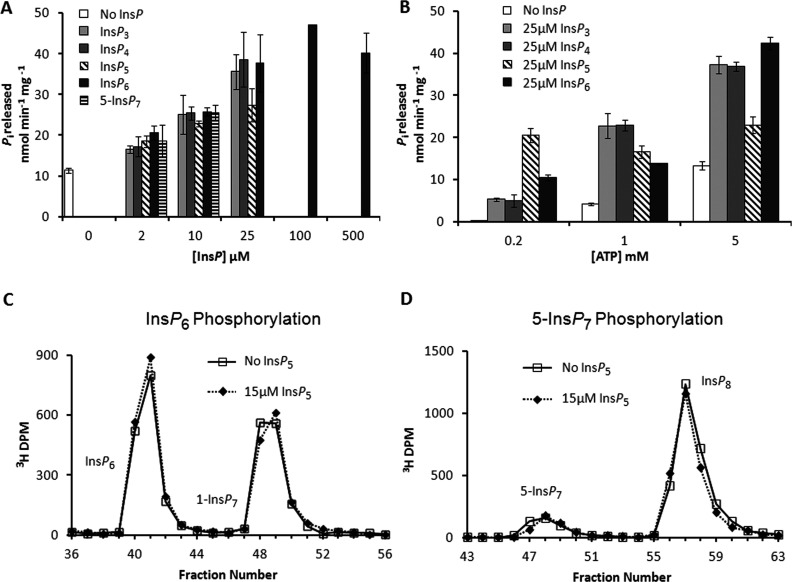
Reaction stoichiometry is pertinent to the regulation of metabolic pathways [16]. Moreover, since PPIP5K activity consumes ATP, stoichiometry is also highly relevant to the potential interface of PP-InsPs with cellular bioenergetic status [5,15]. We [20] previously demonstrated that PPIP5K2 showed a slow ATPase activity (0.5 nmol min−1 mg−1) when it was incubated with 0.1 mM ATP in the absence of inositol phosphate substrate. That did not seem significant in relation to the Vmax values for InsP6 and 5-InsP7 phosphorylation (43 and 190 nmol min−1 mg−1; Table 1). However, in the current study we found that at physiological [ATP] (5 mM), the inositol phosphate-independent ATPase activity increased to approximately 12 nmol min−1 mg−1 (Figures 8A and 8B). Moreover, the rate of ATP hydrolysis was further elevated up to 4-fold as increasing concentrations of either InsP6 or 5-InsP7 were added, with a maximally effective concentration of 25 μM (Figure 8A).
Figure 8. ATPase activity of PPIP5K2KD.
(A) and (B) Orthophosphate released from the hydrolysis of ATP was measured using a Malachite Green detection method [55]. The 27–270 μg/ml PPIP5K2KD was incubated at 37°C for 120 min in 50 μl reactions containing 20 mM Tris/HCl, pH 7.5, 10 mM ATP (unless otherwise indicated), 100 mM KCl, 0.1 mM EDTA and 5 mM MgCl2. (A) Stimulation of PPIP5K2KD ATPase activity by InsP3, InsP4, InsP5, InsP6 and 5-InsP7 (concentrations as indicated, [ATP]=5 mM). (B) Effect of ATP concentration (0.2, 1 or 5 mM) on basal (absence of InsP) and InsP-stimulated (25 μM) ATPase activity. Results are presented as the means±S.D. (n=3). (C) and (D) provide representative HPLC data indicating that 15 μM InsP5 (dotted lines) does not inhibit phosphorylation of either InsP6 or 5- InsP7, respectively. In (C), 20 μg/ml PPIP5K2KD was incubated for 15 min with reaction buffer (see the Materials and methods section) containing 15 μM [3H]InsP6, 5 mM ATP and 0.5 mM ADP. In (D), 0.41 μg/ml PPIP5K2KD was incubated for 5 min with reaction buffer (see the Materials and methods section) containing 1 μM 5-[3H]InsP7, 5 mM ATP and 0.5 mM ADP. The reactions were quenched and neutralized and analysed by Partisphere SAX HPLC as described in the Materials and methods section.
InsP3 (inositol 1,3,4-trisphosphate), InsP4 (inositol 1,3,4,5-tetrakisphosphate) and InsP5 also stimulated ATPase activity (Figures 8A and 8B). These InsPs elevated ATPase activity even at low (0.2 μM) [ATP] (Figure 8B). It is surprising that this ATPase activity can be stimulated by an inositol phosphate that is neither a substrate [20] nor an inhibitor (Figures 8C and 8D). Nevertheless, such data (Figures 8C and 8D) lead us to conclude that even the relatively high levels of InsP5 that occur in cells cannot access the enzyme's active site to stimulate ATPase activity when InsP6 and 5-InsP7 are available.
A comparison of the rates of the ATPase activity (Figures 8A and 8B) in relation to the kinase activity (Table 1) indicates that approximately two molecules of ATP are consumed for every molecule of InsP6 that is phosphorylated. As the Vmax is higher for 5-InsP7 phosphorylation compared with InsP6 phosphorylation (Table 1), there is a much smaller impact of the ATPase activity upon that reaction stoichiometry (1.2 molecules of ATP are consumed for every molecule of 5-InsP7 that is phosphorylated). In other words, the bioenergetic cost to the cell is much smaller for 5-InsP7 phosphorylation (17% ATP ‘wastage’) than it is for InsP6 phosphorylation (50% ATP ‘wastage’). Thus, the synthesis of InsP8 by pathway I is more energetically efficient than is pathway II (Figure 1).
An analogous ATPase reaction has been observed in ATP-grasp ligases [51], but to our knowledge this is the first demonstration that this is a property of an ATP-grasp kinase. We propose that the dynamic nature of the active site and the partly associative reaction mechanism make it inevitable that there will be some non-productive ATPase activity. We [20] have also previously determined that InsP6 is more mobile within the active site than is 5-InsP7. That may explain why the phosphorylation of InsP6 is less efficient.
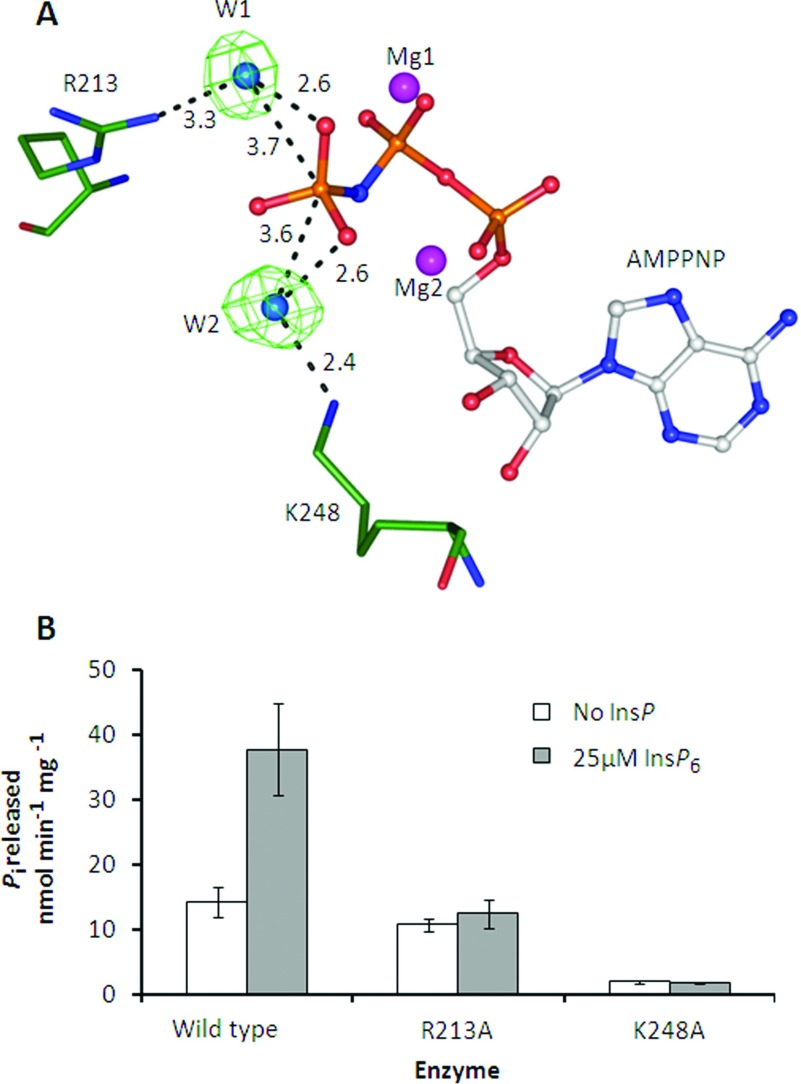
We studied the mechanism of this ATPase activity through structural and mutagenesis studies. Previous analysis [20] of the crystal structure of the PPIP5K2KD indicated that there are two water molecules (W1 and W2; Figure 9A) that are candidates to carry out nucleophilic attack on the γ-phosphate of ATP (Figure 9A). The two amino acid residues that are candidates for activating these putative nucleophiles are Arg213 and Lys248, respectively (Figure 9A). We therefore assayed ATPase activities of R213A and K248A mutants of PPIP5K2KD. Both mutants showed a reduction in InsP6-stimulated ATPase activity, as expected since Arg213 and Lys248 are required for binding of InsP6 [20]. In contrast, only the K248A mutant showed a significant reduction in the rate of inositol phosphate-independent ATPase activity (Figure 9B). We therefore conclude that W2 is the water molecule that is activated for nucleophilic attack upon the γ-phosphate of ATP.
Figure 9. The effects of R213A and K248A mutations upon ATPase activity of PPIP5K2KD.
(A) Structure of a portion of the wild-type PPIP5K2KD catalytic centre [20] showing water molecules bridging the interaction between the ATP analogue AMPPNP (adenosine 5′-[β,γ-imido]triphosphate) and R213 and K248. (B) The effect of single-site mutants of PPIP5K2KD on basal and InsP6-stimulated (25 μM) ATPase activity. ATPase activity was measured as in Figure 8.
CONCLUDING COMMENTS
Our new kinetic information concerning PPIP5K stoichiometry, rates, substrate affinities and equilibrium conditions assists our understanding of the dynamic behaviour of this signalling enzyme in vivo and the role it has in PP-InsP generation. Since the catalytic and structural features of the active sites of PPIP5K1 and PPIP5K2 are so similar [18,20], we believe the conclusions that we have made are equally applicable to both enzymes. Furthermore, our studies with recombinant protein have led us to conclude that contrary to an earlier report [25], the kinetic characteristics of PPIP5Ks do not match the synthesis of InsP8 to fluctuations in adenine nucleotide levels. Instead, the opposite is true; the kinetic properties of PPIP5Ks ensure that InsP8 synthesis is protected against cellular bioenergetic stress. The features of this enzyme stand in contradiction to the reduction in InsP8 levels that accompanies experimental manipulations in cultured cells that are generally thought to invoke mild bioenergetic stress [15]. It will be important to uncover the signalling pathways that are involved and their biological relevance.
AUTHOR CONTRIBUTION
Jeremy Weaver, Huanchen Wang and Stephen Shears designed the experiments, analysed the results and wrote the paper. Jeremy Weaver and Huanchen Wang conducted the experiments.
FUNDING
This work was supported by the Intramural Research Program of the National Institutes of Health/National Institute of Environmental Health Sciences.
References
- 1.Wundenberg T., Mayr G. W. Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol. Chem. 2012;393:979–998. doi: 10.1515/hsz-2012-0133. [DOI] [PubMed] [Google Scholar]
- 2.Saiardi A. Cell signalling by inositol pyrophosphates. Subcell. Biochem. 2012;59:413–443. doi: 10.1007/978-94-007-3015-1_14. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty A., Kim S., Snyder S. H. Inositol pyrophosphates as mammalian cell signals. Sci. Signaling. 2011;4:re1. doi: 10.1126/scisignal.2001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsui M. M., York J. D. Roles of inositol phosphates and inositol pyrophosphates in development, cell signaling and nuclear processes. Adv. Enzyme Regul. 2010;50:324–337. doi: 10.1016/j.advenzreg.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shears S. B. Diphosphoinositol polyphosphates: metabolic messengers? Mol. Pharmacol. 2009;76:236–252. doi: 10.1124/mol.109.055897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker C. J., Illies C., Gaboardi G. C., Berggren P. O. Inositol pyrophosphates: structure, enzymology and function. Cell. Mol. Life Sci. 2009;66:3851–3871. doi: 10.1007/s00018-009-0115-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y. S., Mulugu S., York J. D., O’Shea E. K. Regulation of a cyclin-cdk-cdk inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty A., Koldobskiy M. A., Bello N. T., Maxwell M., Potter J. J., Juluri K. R., Maag D., Kim S., Huang A. S., Dailey M. J., et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azevedo C., Burton A., Ruiz-Mateos E., Marsh M., Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21161–21166. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhandari R., Saiardi A., Ahmadibeni Y., Snowman A. M., Adam R. C., Kristiansen T. Z., Molina H., Pandey A., Werner J. K., Jr, Juluri K. R., et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saiardi A., Bhandari R., Resnick A. C., Snowman A. M., Snyder S. H. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 12.Choi K., Mollapour E., Shears S. B. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Pesesse X., Choi K., Zhang T., Shears S. B. Signaling by higher inositol polyphosphates: synthesis of bisdiphosphoinositol tetrakisphosphate (‘InsP8’) is selectively activated by hyperosmotic stress. J. Biol. Chem. 2004;279:43378–43381. doi: 10.1074/jbc.C400286200. [DOI] [PubMed] [Google Scholar]
- 14.Onnebo S. M. N., Saiardi A. Inositol pyrophosphates modulate hydrogen peroxide signalling. Biochem. J. 2009;423:109–118. doi: 10.1042/BJ20090241. [DOI] [PubMed] [Google Scholar]
- 15.Choi K., Mollapour E., Choi J. H., Shears S. B. Cellular energetic status supervises the synthesis of bis-diphosphoinositol tetrakisphosphate independently of AMP-activated protein kinase. Mol. Pharmacol. 2008;74:527–536. doi: 10.1124/mol.107.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohwer J. M. Kinetic modelling of plant metabolic pathways. J. Exp. Bot. 2012;63:2275–2292. doi: 10.1093/jxb/ers080. [DOI] [PubMed] [Google Scholar]
- 17.Padmanabhan U., Dollins D. E., Fridy P. C., York J. D., Downes C. P. Characterization of a selective inhibitor of inositol hexakisphosphate kinases. Use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fridy P. C., Otto J. C., Dollins D. E., York J. D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- 19.Choi J. H., Williams J., Cho J., Falck J. R., Shears S. B. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Falck J. R., Hall T. M. T., Shears S. B. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 2012;8:111–116. doi: 10.1038/nchembio.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulugu S., Bai W., Fridy P. C., Bastidas R. J., Otto J. C., Dollins D. E., Haystead T. A., Ribeiro A. A., York J. D. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 22.Gokhale N. A., Zaremba A., Shears S. B. Receptor-dependent compartmentalization of PPIP5K1, a kinase with a cryptic polyphosphoinositide binding domain. Biochem. J. 2011;434:415–426. doi: 10.1042/BJ20101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szijgyarto Z., Garedew A., Azevedo C., Saiardi A. Influence of inositol pyrophosphates on cellular energy dynamics. Science. 2011;334:802–805. doi: 10.1126/science.1211908. [DOI] [PubMed] [Google Scholar]
- 24.Nagel A., Barker C. J., Berggren P. O., Illies C. Diphosphosinositol polyphosphates and energy metabolism: assay for ATP/ADP ratio. Methods Mol. Biol. 2010;645:123–131. doi: 10.1007/978-1-60327-175-2_8. [DOI] [PubMed] [Google Scholar]
- 25.Huang C. F., Voglmaier S. M., Bembenek M. E., Saiardi A., Snyder S. H. Identification and purification of diphosphoinositol pentakisphosphate kinase, which synthesizes the inositol pyrophosphate bis(diphospho)inositol tetrakisphosphate. Biochemistry. 1998;37:14998–15004. doi: 10.1021/bi981920l. [DOI] [PubMed] [Google Scholar]
- 26.Voglmaier S. M., Bembenek M. E., Kaplin A. I., Dormán G., Olszewski J. D., Prestwich G. D., Snyder S. H. Purified inositol hexakisphosphate kinase is an ATP synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens L., Radenberg T., Thiel U., Vogel G., Khoo K. H., Dell A., Jackson T. R., Hawkins P. T., Mayr G. W. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J. Biol. Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- 28.Menniti F. S., Miller R. N., Putney J. W., Jr, Shears S. B. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- 29.Knight Z. A., Shokat K. M. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Shears S. B. How versatile are inositol phosphate kinases? Biochem. J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillippy B. Q., Ullah A. H., Ehrlich K. C. Purification and some properties of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from immature soybean seeds. J. Biol. Chem. 1994;269:28393–28399. [PubMed] [Google Scholar]
- 32.Raboy V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009;177:281–296. [Google Scholar]
- 33.Losito O., Szijgyarto Z., Resnick A. C., Saiardi A. Inositol pyrophosphates and their unique metabolic complexity: analysis by gel electrophoresis. PLoS ONE. 2009;4:e5580. doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher S. K., Novak J. E., Agranoff B. W. Inositol and higher inositol phosphates in neural tissues: homeostasis, metabolism and functional significance. J. Neurochem. 2002;82:736–754. doi: 10.1046/j.1471-4159.2002.01041.x. [DOI] [PubMed] [Google Scholar]
- 35.Loss O., Azevedo C., Szijgyarto Z., Bosch D., Saiardi A. Preparation of quality inositol pyrophosphates. J. Vis. Exp. 2011;55:e3027. doi: 10.3791/3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perelló J., Isern B., Muñoz J. A., Valiente M., Grases F. Determination of phytate in urine by high-performance liquid chromatography-mass spectrometry. Chromatographia. 2004;60:265–268. [Google Scholar]
- 37.Hoenig M., Lee R. J., Ferguson D. C. A microtiter plate assay for inorganic-phosphate. J. Biochem. Biophys. Methods. 1989;19:249–251. doi: 10.1016/0165-022x(89)90031-6. [DOI] [PubMed] [Google Scholar]
- 38.Brooks H. B., Geeganage S., Kahl S. D., Montrose C., Sittampalam S., Smith M. C., Weidner J. R. In: Basics of enzymatic assays for HTS. In Assay Guidance Manual. Sittampalam G. S., Weidner J., Auld D., Glicksman M., Arkin M., Napper A., Inglese J., editors. Bethesda, MD: Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004. pp. 5–7. [PubMed] [Google Scholar]
- 39.Patton C., Thompson S., Epel D. Some precautions in using chelators to buffer metals in biological solutions. Cell Calcium. 2004;35:427–431. doi: 10.1016/j.ceca.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H., Thompson J., Prestwich G. D. A scalable synthesis of the IP7 isomer, 5-PP-Ins(1,2,3,4,6)P5. Org. Lett. 2009;11:1551–1554. doi: 10.1021/ol900149x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin H., Fridy P. C., Ribeiro A. A., Choi J. H., Barma D. K., Vogel G., Falck J. R., Shears S. B., York J. D., Mayr G. W. Structural analysis and detection of biological inositol pyrophosphates reveal that the family of VIP/diphosphoinositol pentakisphosphate kinases are 1/3-kinases. J. Biol. Chem. 2009;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albert C., Safrany S. T., Bembenek M. E., Reddy K. M., Reddy K. K., Falck J. R., Bröcker M., Shears S. B., Mayr G. W. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem. J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barker C. J., Wright J., Hughes P. J., Kirk C. J., Michell R. H. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem. J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caffrey J. J., Safrany S. T., Yang X., Shears S. B. Discovery of molecular and catalytic diversity among human diphosphoinositol-polyphosphate phosphohydrolases. An expanding Nudt family. J. Biol. Chem. 2000;275:12730–12736. doi: 10.1074/jbc.275.17.12730. [DOI] [PubMed] [Google Scholar]
- 45.Safrany S. T., Caffrey J. J., Yang X., Bembenek M. E., Moyer M. B., Burkhart W. A., Shears S. B. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soboll S., Scholz R., Heldt H. W. Subcellular metabolite concentrations. Dependence of mitochondrial and cytosolic ATP systems on the metabolic state of perfused rat liver. Eur. J. Biochem. 1978;87:377–390. doi: 10.1111/j.1432-1033.1978.tb12387.x. [DOI] [PubMed] [Google Scholar]
- 47.Chamberlain P. P., Qian X., Stiles A. R., Cho J., Jones D. H., Lesley S. A., Grabau E. A., Shears S. B., Spraggon G. Integration of inositol phosphate signaling pathways via human ITPK1. J. Biol. Chem. 2007;282:28117–28125. doi: 10.1074/jbc.M703121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P., Snyder S. H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 49.Zhao S., Snow R. J., Stathis C. G., Febbraio M. A., Carey M. F. Muscle adenine nucleotide metabolism during and in recovery from maximal exercise in humans. J. Appl. Physiol. 2000;88:1513–1519. doi: 10.1152/jappl.2000.88.5.1513. [DOI] [PubMed] [Google Scholar]
- 50.Daniel J. L., Robkin L., Molish I. R., Holmsen H. Determination of the ADP concentration available to participate in energy metabolism in an actin-rich cell, the platelet. J. Biol. Chem. 1979;254:7870–7873. [PubMed] [Google Scholar]
- 51.Fawaz M. V., Topper M. E., Firestine S. M. The ATP-grasp enzymes. Bioorg. Chem. 2011;39:185–191. doi: 10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saiardi A., Nagata E., Luo H. R., Snowman A. M., Snyder S. H. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- 53.Draškovič P., Saiardi A., Bhandari R., Burton A., Ilc G., Kovačevič M., Snyder S. H., Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Schneider C. A., Rasband W. S., Eliceiri K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Meth. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Veldhoven P. P., Mannaerts G. P. Inorganic and organic phosphate measurements in the nanomolar range. Anal. Biochem. 1987;161:45–48. doi: 10.1016/0003-2697(87)90649-x. [DOI] [PubMed] [Google Scholar]