Abstract
Chlorophyllous pigments are essential for photosynthesis. Bacteriochlorophyll (BChl) b has the characteristic C8-ethylidene group and therefore is the sole naturally occurring pigment having an absorption maximum at near-infrared light wavelength. Here we report that chlorophyllide a oxidoreductase (COR), a nitrogenase-like enzyme, showed distinct substrate recognition and catalytic reaction between BChl a- and b-producing proteobacteria. COR from BChl b-producing Blastochloris viridis synthesized the C8-ethylidene group from 8-vinyl-chlorophyllide a. In contrast, despite the highly conserved primary structures, COR from BChl a-producing Rhodobacter capsulatus catalyzes the C8-vinyl reduction as well as the previously known reaction of the C7 = C8 double bond reduction on 8-vinyl-chlorophyllide a. The present data indicate that the plasticity of the nitrogenase-like enzyme caused the branched pathways of BChls a and b biosynthesis, ultimately leading to ecologically different niches of BChl a- and b-based photosynthesis differentiated by more than 150 nm wavelength.
Chlorophylls (Chls) and bacteriochlorophylls (BChls) are tetrapyrrole pigments essential for harvesting light energy, driving photosystems, and achieving charge separation. Phototrophic proteobacteria have either BChl a or BChl b, depending on species. Among naturally occurring pigments, BChl b is the sole pigment showing an absorption maximum at near-infrared light wavelength [λmax (Qy) = 795 nm in diethyl ether]. When it is incorporated as a light-harvesting pigment-protein complex, an absorption band of the BChl-b protein occurs at 1020 nm in Blastochloris (previously known as Rhodopseudomonas) viridis, significantly red-shifted from ~860 nm of an orthologous light-harvesting protein with BChl a1,2,3. Difference in the chemical structure between BChl a and BChl b occurs only at the C8 position, the ethyl group (as in compound 4 of Fig. 1) or the ethylidene group (8), respectively. The ethylidene group on BChl b provides a more π-conjugated system than BChl a, and thus causes the red shift of the Qy absorption maximum, making BChl b capable of absorbing near-infrared light even in the monomeric state. However, the enzyme responsible for catalyzing the C8-ethylidene formation has remained unknown.
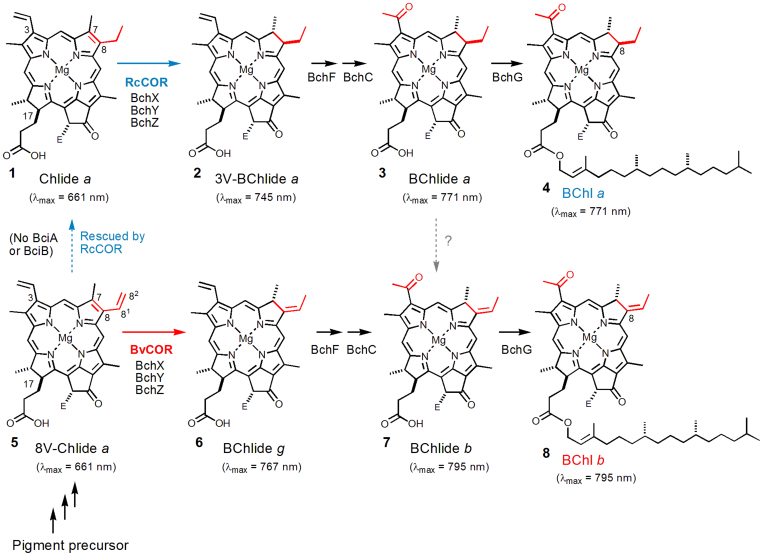
Figure 1. Late steps of BChls a and b biosynthesis.
The presented λmax peak positions are values obtained in diethyl ether. Esterification of a phytyl group to a propionate residue at the C17 position does not alter λmax values. E = COOCH3. It is noteworthy that none of the known photosynthetic proteobacteria producing BChl b produce BChl a, and vice versa.
The closely related structures of BChls a and b suggest a similarity in their biosynthesis. All genes encoding enzymes responsible for BChl a biosynthesis have been identified in the model phototrophic proteobacterium Rhodobacter capsulatus4,5,6. Chlorophyllide a (Chlide a: compound 1 in Fig. 1) is a key “hub” intermediate in biosynthetic pathways of all natural Chls and BChls4. In higher plants and cyanobacteria, a phytyl group is introduced to a propionate residue at the C17 position of Chlide a, producing Chl a. In the biosynthesis of BChl a, two additional steps are required prior to the phytyl esterification. The first step involves conversion of chlorin to a bacteriochlorin ring, i.e., the reduction of the C7 = C8 double bond of Chlide a (1,2-trans-hydrogenation; see 1 to 2 in Fig. 1)7. This step is known to be catalyzed by chlorophyllide a oxidoreductase (COR), which is a nitrogenase-like enzyme8 and a three-subunit complex consisting of BchX, BchY and BchZ in R. capsulatus7. The product of this reaction, 3-vinyl-bacteriochlorophyllide a (3V-BChlide a: compound 2 in Fig. 1) is further modified in the next step: conversion of the C3-vinyl group to an acetyl group by BchF and BchC6,9, producing BChlide a (3).
According to genome information, the bchXYZ and bchFC gene sets are also present in BChl b-producing bacteria. The ethylidene formation, therefore, has been thought to occur on BChlide a to form BChlide b (the gray broken arrow from 3 to 7 in Fig. 1) at the penultimate step of BChl biosynthesis4. However, any attempt to extract prospective genes for the C8-ethylidene synthase, which is assumed to exclusively occur in BChl b-producing bacteria, using comparative genomics of phototrophic bacteria has failed. Instead, we found that photosynthetic bacteria producing BChl b lack a gene encoding divinyl reductase (recently termed BciA or BciB10,11,12). Divinyl reductase is known to make Chlide a from 8-vinyl-Chlide a (8V-Chlide a: 5 in Fig. 1)4,10,11. Thus we hypothesized that COR might react with 8V-Chlide a (not Chlide a) in BChl b-producing bacteria and achieve 1,4-trans-addition of hydrogen at the C7,82 positions of 8V-Chlide a, resulting in the direct formation of the C8-ethylidene group on BChlide g, a precursor to BChl b (see 5 to 6 in Fig. 1). To confirm this hypothesis, two CORs from BChl a- and b-producing proteobacteria were purified and assayed with two substrates, 8V-Chlide a and Chlide a.
Results
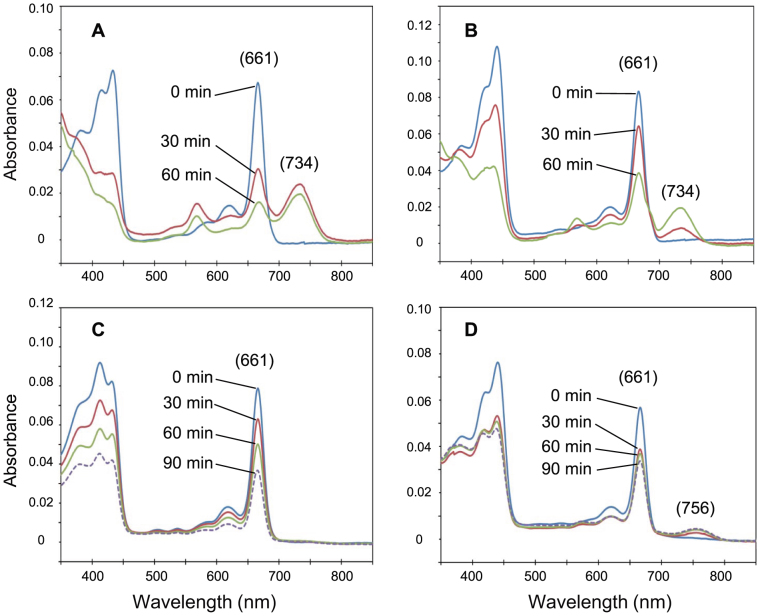
Two sets of COR proteins BchXYZ from the BChl b-producing bacterium B. viridis and the BChl a-producing bacterium R. capsulatus were purified (Fig. S1) and mixed with 8V-Chlide a and Chlide a. COR activities were assayed by absorption spectral changes in 80%-acetone extracts, basically according to the previous report7. As a control, the known function of R. capsulatus COR (RcCOR) was confirmed by mixing with Chlide a. After incubation, the appearance of a peak at 734 nm that corresponded to the product 3V-BChlide a was accompanied by a decrease in the peak of the substrate Chlide a at 661 nm (Fig. 2A). This indicated that the C7 = C8 double bond was reduced by RcCOR. On the other hand, when COR of B. viridis (BvCOR) was mixed with Chlide a, no product peak was observed at the 700–750 nm region (Fig. 2C), suggesting that Chlide a is not a suitable substrate for BvCOR. When BvCOR was mixed with 8V-Chlide a, a new peak at 756 nm appeared (Fig. 2D). This peak position was clearly red-shifted from the 734-nm peak of 3V-BChlide a by more than 20 nm (Fig. 2A vs. 2D), implying that BvCOR converted 8V-Chlide a to BChlide g which has the C8-ethylidene group.
Figure 2. Changes in absorption spectra of enzymatic assay mixtures.
Absorption spectra were recorded in 80% acetone (20% aqueous buffer). R. capsulatus COR was mixed with Chlide a (A) and 8V-Chlide a (B). B. viridis COR was mixed with Chlide a (C) and 8V-Chlide a (D). The Qy absorption peaks of the substrates and products are shown in parentheses.
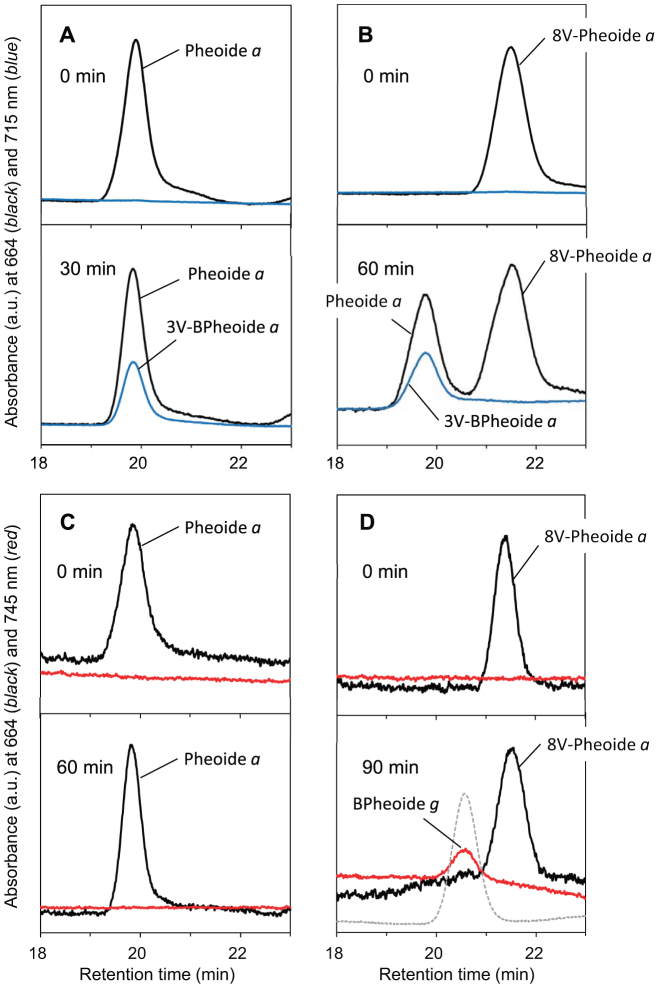
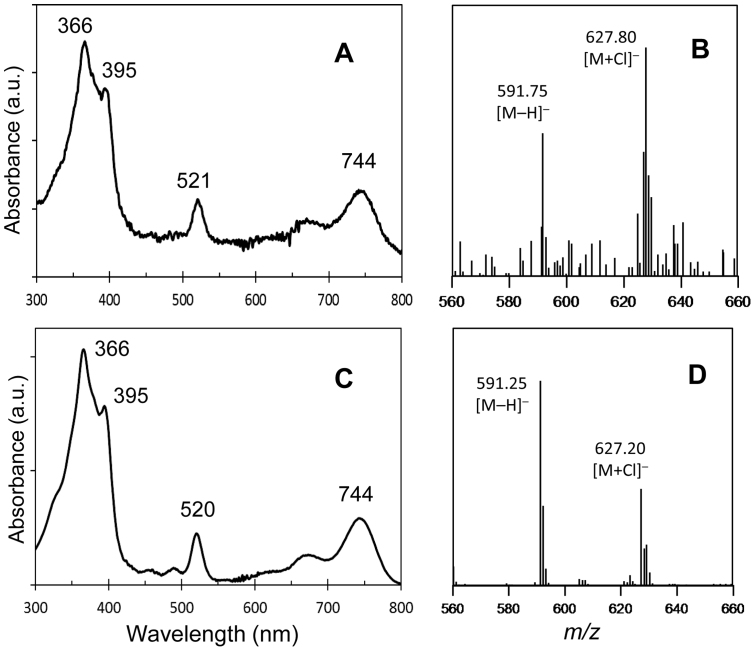
To further identify the product showing the 756-nm peak, the assay mixtures were analyzed by LC-MS. Prior to the sample injection into LC-MS, Chlide and BChlide pigments in the assay mixtures were demetallated by treatment with 10% acetic acid (pheophytinization). As a result of the demetallation, mass spectra were dominated by molecules of the corresponding pheophorbide (Pheoide) and bacteriopheophorbide (BPheoide). The assay mixture of BvCOR and 8V-Chlide a showed a new elution peak at 20.5 min after 90-min incubation (Fig. 3D), which eluted approximately 1 min earlier than the substrate peak of 8V-Pheoide a and at the same time as the BPheoide g standard. In-line absorption spectrum of the new product eluting at 20.5 min was almost identical to that of the BPheoide g standard (Fig. 4A vs. 4C) and clearly different from that of demetallated 3V-BChlide a (Qy/Soret = 715/352 nm in the HPLC solvent). In addition, the molecular-ion mass numbers of the assay product that eluted at 20.5 min were 591.75 Da as [M−H]− and 627.80 Da as [M+Cl]−, which were identical within error to those of the BPheoide g standard (Fig. 4B vs. 4D). These results established that the assay product was BChlide g, and that BvCOR recognizes 8V-Chlide a (not Chlide a) as the substrate and then converts it to BChlide g. Thus, BvCOR is responsible for synthesizing the C8-ethylidene group.
Figure 3. Reverse-phase HPLC analysis of pigments formed in the COR assays after acidic pheophytinization.
R. capsulatus COR was mixed with Chlide a (A) and 8V-Chlide a (B), and B. viridis COR was mixed with Chlide a (C) and 8V-Chlide a (D). Elution profiles were measured at 664 nm (black line) to monitor Chlide pigments, and 715 nm (blue line) and 745 nm (red line) to monitor BChlide pigments. Upper and lower panels of columns represent elution profiles before and after incubation, respectively: incubation time is described in each panel. In (D), an elution profile of the BPheoide g standard is shown as gray dashed line.
Figure 4. Identification of products from the assay mixture of BvCOR and 8V-Chlide a.
In-line absorption (A) and mass spectra (B) of the 20.5-min elution peak in Fig. 3D are shown. Panels (C) and (D) represent in-line absorption and mass spectra of the BPheoide g standard, respectively.
When RcCOR was assayed with the unusual substrate 8V-Chlide a, the product peak at 734 nm was also observed (Fig. 2B). In this assay, it is of interest whether the C8-vinyl group was also reduced. The assay mixture containing RcCOR and 8V-Chlide a showed two HPLC elution peaks when monitored at 664 nm (Fig. 3B, lower panel, black line): one of the peaks at 21.5 min represented the demetallated substrate 8V-Pheoide a (see Fig. 3B upper panel, Fig. S2CD), and the other peak at 19.8 min eluted at the same time as Pheoide a as shown in Fig. 3AC. The in-line absorption spectrum of the elution peak at 19.8 min (Fig. S2A) actually showed that the fraction contained two pigments, namely, a Pheoide derivative showing Qy/Soret bands at 664/409 nm and a BPheoide derivative with Qy/Qx/Soret bands at 714/516/355 nm. The former and latter absorption band positions are identical to those of Pheoide a and 3V-BPheoide a. The in-line mass spectrum of the elution peak at 19.8 min had two masses of 591.20 and 593.25 Da (Fig. S2B, as [M−H]−), which are 2 and 4 mass units larger than that of the demetallated substrate 8V-Pheoide a (Fig. S2D), respectively. These results established that the product peak at 19.8 min in Fig. 3B contained demetallated Chlide a and 3V-BChlide a, strongly suggesting that RcCOR has an additional “latent” activity to reduce the C8-vinyl group of 8V-Chlide a, producing Chlide a, and then performing its known catalytic reaction to reduce the C7 = C8 double bond of Chlide a, finally producing 3V-BChlide a.
Discussion
In the alignments of BchX/BchY/BchZ sequences, no specific conserved region are found in proteins from B. viridis and Thioflavicoccus mobilis, another BChl b-producing bacterium (Fig. S3). Amino acid sequence identities and similarities of each COR proteins between various phototrophic proteobacteria and B. viridis indeed are very high (Table S1, roughly 66–85% in identities and 78–89% in similarities). By considering highly aligned primary structures, the number of amino acid substitutions necessary to convert the known function of COR into an ethylidene synthase is probably very small. To produce BChl b, only small events would be required in the course of evolution: the loss of the gene encoding divinyl reductase and the slight changes in genes encoding COR (bchX, Y, and/or Z).
BChl b-producing bacteria occur in the phylogeny of α- and γ-Proteobacteria and do not form a monophyletic cluster. Phylogenetic trees based on COR proteins (Fig. S4) showed that BchXs from the two BChl b-producing bacteria B. viridis and T. mobilis were not grouped together, and the BchX tree was actually consistent with bacterial phylogeney based on house-keeping markers such as 16S rRNA sequence. On the other hand, in phylogenies of BchY and BchZ, these proteins from the two BChl b-producing bacteria were clustered together, although bootstrap values were low. These implied that amino acid substitutions causing the functional change of COR have occurred in BchY and BchZ, but not in BchX.
COR is a member of nitrogenase-like enzyme family, which includes dark-operative protochlorophyllide oxidoreductase (DPOR)8. The COR subunits BchX, BchY, and BchZ show significant similarities to NifH, NifD, and NifK of nitrogenase, and BchL, BchN, and BchB of DPOR, respectively8. Nitrogenase-like enzymes consist of two separable components; an ATP-dependent reductase component (Fe protein cognates) and a catalytic component (MoFe protein cognates). A recent crystal structure study revealed that the NB-protein, the catalytic component of DPOR, provides the protochlorophyllide-binding site at the inner subsurface region of the two proteins13. In a similar fashion, BchY and BchZ proteins are supposed to provide the Chlide binding site. By analogy to BchL14, BchX forms a homodimer X protein, and functions as an ATP-dependent reductase component catalyzing the electron transfer from ferredoxin to the YZ-protein. We performed control assay experiments in which COR protein(s) was not added to the assay mixtures. Neither of BChlide g or 3V-BChlide a was produced from 8V-Chlide a with dithionite and ATP alone.
This study clearly suggests that a new enzyme specific for formation of the C8-ethylidene group has not been required in the course of evolution, and provides a unique example that orthologous proteins do not always have the same catalytic activity. The C8-ethylidene group is probably synthesized by simple 1,4-trans-hydrogenation of the C7,82 positions of 8V-Chlide a by BvCOR. On the other hand, the previously known function of RcCOR, the C7 = C8 double bond reduction, would be achieved by 1,2-trans-hydrogenation at the C7,8 positions. Using a structural model, the distance between carbons of the C7 = C8 double bond of (8V-) Chlide a was calculated to be 1.37 Å (for 1,2-hydrogenation). In contrast, the distance between C7 and C82 carbons of the cisoid C8−C81 rotamer in 8V-Chlide a was calculated to be 3.16 Å (for 1,4-hydrogenation). The relevant amino acid substitutions occurred in BvCOR probably have extended the distance between two reactive sites in YZ-component by about 1.8 Å, compared to those in RcCOR.
In addition to the reported function of the C7 = C8 double bond reduction, this study revealed that RcCOR appears to have a latent capacity to hydrogenate the C8-vinyl group of 8V-Chlide a, probably through catalysis of the 1,2-addition as well. We performed control experiments in which the 8V-Chlide-a solution in the presence of an excess amount of dithionite was incubated under illuminated condition, in order to see whether photo-reduction occurs especially on the C8-vinyl group without COR protein. Then, no new product was observed, indicating that the additional C8-vinyl reduction was achieved in the fashion of enzymatic reaction. A recent study supports this idea, in which a mutant of the green sulfur bacterium Chlorobaculum tepidum that lacks divinyl reductase accumulated 8-vinyl-Chl a, but still synthesized normal BChl a15. In this mutant, COR probably compensated the function of divinyl reductase in the branch of BChl biosynthesis.
Nitrogenase catalyzes not only di-nitrogen reduction but also reduction of many compounds with multi-bonds such as acetylene and cyanide. In addition, nitrogenase was recently shown to catalyze reductive catenation of carbon monoxide to form a series of hydrocarbon compounds, although the activity was very low16. Thus, flexibility of substrate recognition and enzymatic activities would be inherent characteristics of nitrogenase-like enzymes.
BChl a is recognized as a widely-distributed predominant bacterial pigment in coastal waters of the global ocean17,18. Some of genes encoding enzymes for BChl a biosynthesis have been used as phylogenetic and ecological markers18. It is possible that the ecological significance of BChl b in the global ocean has been underestimated because previously recognized wide distributions of BChl a were revealed based on cultivation-independent genomic analyses using photosynthetic genes other than bchXYZ18,19. Our new observations should help efforts to address global distribution of BChl-mediated photosynthesis, and characterization of poorly understood microbes in marine phytoplankton. Moreover, the mechanism to produce BChl b would aid in the development of artificial photosynthesis systems and also protein engineering.
Methods
Cloning of genes coding for COR from B. viridis and R. capsulatus
B. viridis DSM 133 was obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), and was grown under anaerobic-light conditions at 30°C in the medium described in Ref. 20. Preparation of genomic DNAs from B. viridis was performed basically according to method described previously21. The bchX gene of B. viridis was amplified by PCR using the genomic DNA as a template and primers BvX-f1 (5′- ATGGTAACCTGCATTAGCGCCATGACGTCCCCCGCCAGAGC-3′) and BvX-r1 (5′- ATGGTAACCTGCATTATATCACACCGTGTCGTAAATCACTTCGA-3′; BspMI restriction sites are italicized). The bchY and the flanking bchZ genes of B. viridis were amplified together by PCR using the primers BvY-f2 (ATGGTAGGTCTCAGCGCCATGAGGGCTGCCAGTTACGTTC) and BvZ-r1 (ATGGTAGGTCTCATATCACGCAGCCTGCCCCCCGACA; BsaI restriction sites are italicized). The amplified DNA fragments were excised from agarose gels and purified using a NucleoSpin Extract II kit (Macherey-Nagel, Duren, Germany). Purified DNA fragments containing bchX and bchY-bchZ genes were digested with BspMI and BsaI, respectively, and were cloned into the BsaI restriction sites of the expression vector pASK-IBA5plus (IBA, Göttingen, Germany), yielding plasmids pA5-BvX and pA5-BvYZ, respectively. These plasmids were transformed into the E. coli strain Rosetta-2 (Novagen, Darmstadt, Germany) to overexpress BchX and BchY-BchZ from B. viridis as fusion proteins with an affinity tag (Strep-tag II) under the control of the tet promoter. Entire DNA sequences containing bchX, bchY, and bchZ were deposited into GenBank (Accession No. AB738834).
Two plasmids, pJN1X and pJN1YZ, were also constructed to overexpress BchX and BchY-BchZ from R. capsulatus in E. coli. Two primers, Ask5bchX-f1 (5′- TAGTGAGGTCTCTGCGCCACTGACGCACCCAACCTGAAG-3′) and Ask5bchX-r1 (5′- TAGTGAGGTCTCATATCAGACATCGTCGTAGATCACTTC-3′), were used to amplify the entire coding region for bchX of R. capsulatus by PCR. Another two primers, Ask5bchY-f1 (5′- TAGTGAGGTCTCTGCGCCACCGATCTTCCGCAAGCCGAAG-3′) and Ask5bchZ-r1 (5′- TAGTGAGGTCTCATATCAGTTCCCCCCCTTCCGATCCATG-3′), were used to amplify the chromosomal region covering the entire coding regions of bchY and bchZ of R. capsulatus, which are located contiguously on the chromosome. The amplified fragments, consisting of the entire coding regions for bchX and bchY-bchZ of R. capsulatus, were digested with BsaI (indicated by italics in the sequences) and were ligated into the BsaI sites of pASK-IBA5plus, yielding the overexpression plasmids pJN1X and pJN1YZ. PCR reactions were performed with KOD-plus polymerase (Toyobo, Osaka, Japan) using genomic DNAs from appropriate species as templates.
Purification of BchX and BchY-BchZ proteins
E. coli strains harboring the expression plasmids described above were cultured in 100 mL LB media containing 100 μg/mL ampicillin at 37°C overnight. The cultures were transferred into 1.2 L of fresh LB media containing 100 μg/mL ampicillin, and were incubated at 37°C for 2 h. Protein overexpression was induced by addition of anhydrotetracycline to a final concentration of 200 μg/L, and the cultures were then grown at 20°C for 6 h. Purification of BchX and BchY-BchZ proteins was performed in an anaerobic chamber as described previously7.
Preparation of Chlide a and 8V-Chlide a
Chl a was purified from the cyanobacterium Spirulina sp. as previously described22 and 8V-Chl a was purified from the mutant of Synechocystis sp. PCC6803 which lacks divinyl reductase11 in the same way. Chlide a and 8V-Chlide a were prepared from Chl a and 8V-Chl a, respectively, by using chlorophyllase from Chenopodium album expressed in E. coli. The E. coli strain overexpressing chlorophyllase was a generous gift from Dr. Toru Tsuchiya of Kyoto University23. Five hundred microliters of cell lysates of the E. coli strain (corresponding to ~5 mg total proteins) were suspended with 2.5 mL of Tris-HCl buffer (pH 7.5) containing 0.1% Triton X-100, 5 mM sodium ascorbate, and 3 μL of pyridine. The purified pigments Chl a and 8V-Chl a (~500 nmol in 50 μL acetone) were added to the suspension and incubated at 30°C for 1 h. The reaction was stopped by adding acetone to a final concentration of 80%. The reaction solution in 80% acetone was centrifuged (6000 × g, 15 min), and the supernatant was recovered in a new glass test tube. Half a volume of hexane was added to the supernatant, and the lower acetone phase was collected. Equal volumes of diethyl ether and then water were added to the acetone solution, and the upper ether phase was collected and dried either under a stream of nitrogen gas or by a rotary evaporator. The purity of prepared Chlide pigments was checked by HPLC. If degradation products were observed, further purification of Chlide was performed by preparative HPLC.
(8V-)Chlide a reductase activity assay and LC-MS measurements
Assays of COR activity, and the following measurements of absorption spectra, were performed as described previously7. Purified Chlide a and 8V-Chlide a were dissolved in DMSO and added to assay mixtures to a final concentration of 5 μM for measurements of absorption spectra, and to a final concentration of 10 μM for LC-MS measurements. The LC-MS system consisted of a C18 reverse-phase column (Synergi Hydro-RP 4 μm, 4.6 φ × 150 mm; Shimadzu GLC Ltd., Kyoto, Japan), a photodiode-array spectrophotometer detector (SPD-M20A; Shimadzu, Kyoto) and a LCMS-2010EV quadrupole mass spectrometer equipped with an electronspray ionization (ESI) probe (Shimadzu, Kyoto). The mobile phase was methanol:acetonitrile:mono-ethanolamine (pH 7.0) = 52.5:30.0:17.5 (v/v/v), and the flow rate was isocratic at 1 mL/min. The ESI-MS settings were as follows: capillary temperature; 230°C, sheath gas (N2) pressure; 0.1 MPa, and spray voltage; 2.0 kV (negative-ion ESI). The column oven was set at 40°C. Assay mixtures were treated with 10% acetic acid, which demetallated the Chlide pigments, increased separation capacities for the pigments on the Synergi reverse-phase column, and improved the stability of assay products. The treated assay mixtures were filtered through a 0.45-μm filter prior to injection.
The BPheoide g standard was synthesized as follows: BChl g was extracted from Heliobacterium modesticaldum with acetone:methanol (7:2, v/v) and was purified by normal-phase HPLC. The purified BChl g was treated with 1% (v/v) HCl to produce bacteriopheophytin g. Diethyl ether and then water were added to the solution, and the ether phase containing bacteriopheophytin g was collected and evaporated to dryness under a stream of nitrogen gas. Bacteriopheophytin g was incubated with chlorophyllase, and the resulting BPheoide g was collected as described above.
Phylogenetic analysis
Amino acid sequences of BchX, BchY, and BchZ were aligned using the ClustalW2 program embedded in the MEGA5 software24. Construction of phylogenetic trees was performed by the neighbor-joining method using the p-distance parameter. Bootstrap values (50% or more) are described beside nodes.
Author Contributions
Y.T. and H.T. designed the research. J.H. prepared genomic DNAs of B. viridis for draft genome sequencing and determined the complete sequences of photosynthetic gene cluster of this organism. Y.T., J.N. and M.K. cloned genes and made overexpression strains. Y.T. and H.Y. purified BchXYZ proteins and performed enzymatic assays. Y.T., T.Y. and T.M. prepared substrates for the assay and the pigment standards, established conditions for the LC-MS measurement, and identified assay products by LC-MS. Y.T., Y.F. and H.T. analyzed the data and wrote the manuscript.
Supplementary Material
Supplementary figures and tables
Acknowledgments
We thank Dr. Toru Tsuchiya for his generous gift of the E. coli strain overexpressing Chenopodium album chlorophyllase. This work was partially supported by Grants-in-Aid for Scientific Research (A) (No. 22245030 to H.T.), for Scientific Research (B) (No. 23370020 to Y.F.), for Scientific Research (C) (No. 23570065 to M.K. and No. 24550065 to T.M.), and for Young Scientists (B) (No. 24750169 to J.H. and No. 24770040 to J.N.) from the Japan Society for the Promotion of Science (JSPS) as well as for Scientific Research on Innovative Areas (“Artificial Photosynthesis”, No. 24107002 to H.T.) from the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT). Y.F. was supported by the Japan Science and Technology Agency (JST) [Advanced Low Carbon Technology Research and Development Program (ALCA) and Precursory Research for Embryonic Science and Technology (PRESTO)].
References
- Zubova S. V., Melzer M. & Prokhorenko I. R. Effect of environmental factors on the composition of lipopolysaccharides released from the Rhodobacter capsulatus cell wall. Biol. Bull. 32, 168–173 (2005). [PubMed] [Google Scholar]
- Stomp M., Huisman J., Stal L. J. & Matthijs H. C. P. Colorful niches of phototrophic microorganisms shaped by vibrations of the water molecule. ISME J. 1, 271–282 (2007). [DOI] [PubMed] [Google Scholar]
- Jay F., Lambillotte M., Stark W. & Mühlethaler K. The preparation and characterisation of native photoreceptor units from the thylakoids of Rhodopseudomonas viridis. EMBO J. 3, 773–776 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew A. G. M. & Bryant D. A. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu. Rev. Microbiol. 61,113–129 (2007). [DOI] [PubMed] [Google Scholar]
- Masuda T. & Fujita Y. Regulation and evolution of chlorophyll biosynthesis. Photochem. Photobiol. Sci. 7, 1131–1149 (2008). [DOI] [PubMed] [Google Scholar]
- Bollivar D. W., Suzuki J. Y., Beatty J. T., Dobrowolski J. M. & Bauer C. E. Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J. Mol. Biol. 237, 622–640 (1994). [DOI] [PubMed] [Google Scholar]
- Nomata J., Mizoguchi T., Tamiaki H. & Fujita Y. A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis: reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 281, 15021–15028 (2006). [DOI] [PubMed] [Google Scholar]
- Raymond J., Siefert J. L., Staples C. R. & Blankenship R. E. The natural history of nitrogen fixation. Mol. Biol. Evol. 21, 541–554 (2004). [DOI] [PubMed] [Google Scholar]
- Suzuki J. Y., Bollivar D. W. & Bauer C. E. Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet. 31, 61–89 (1997). [DOI] [PubMed] [Google Scholar]
- Chew A. G. M. & Bryant D. A. Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J. Biol. Chem. 282, 2967–2975 (2007). [DOI] [PubMed] [Google Scholar]
- Ito H., Yokono M., Tanaka R. & Tanaka A. Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J. Biol. Chem. 283, 9002–9011 (2008). [DOI] [PubMed] [Google Scholar]
- Liu Z. & Bryant D. A. Multiple types of 8-vinyl reductases for (bacterio)chlorophyll biosynthesis occur in many green sulfur bacteria. J. Bacteriol. 193, 4996–4998 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraki N. et al. X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114 (2010). [DOI] [PubMed] [Google Scholar]
- Nomata J., Kitashima M., Inoue K. & Fujita Y. Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett. 580, 6151–6154 (2006). [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Harada J. & Tamiaki H. Characterization of chlorophyll pigments in the mutant lacking 8-vinyl reductase of green photosynthetic bacterium Chlorobaculum tepidum. Bioorg. Med. Chem. 20, 6803–6810 (2012). [DOI] [PubMed] [Google Scholar]
- Hu Y., Lee C. C. & Ribbe M. W. Extending the carbon chain: hydrocarbon formation catalyzed by vanadium/molybdenum nitrogenases. Science 333, 753–755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolber Z. S., Van Dover C. L., Niderman R. A. & Falkowski P. G. Bacterial photosynthesis in surface waters of the open ocean. Nature 407, 177–179 (2000). [DOI] [PubMed] [Google Scholar]
- Béjà O. et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415, 630–633 (2002). [DOI] [PubMed] [Google Scholar]
- Selje N., Simon M. & Brinkhoff T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427, 445–448 (2004). [DOI] [PubMed] [Google Scholar]
- Maki H., Matsuura K., Shimada K. & Nagashima K. V. P. Chimeric photosynthetic reaction center complex of purple bacteria composed of the core subunits of Rubrivivax gelatinosus and the cytochrome subunit of Blastochloris viridis. J. Biol. Chem. 278, 3921–3928 (2003). [DOI] [PubMed] [Google Scholar]
- Pospiech A. & Neumann B. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11, 217–218 (1995). [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Harada J. & Tamiaki H. Structural determination of dihydro- and tetrahydrogeranylgeranyl groups at the 17-propionate of bacteriochlorophylls-a. FEBS Lett. 580, 6644–6648 (2006). [DOI] [PubMed] [Google Scholar]
- Tsuchiya T. et al. Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: Finding of a lipase motif and the induction by methyl jasmonate. Proc. Natl. Acad. Sci. USA 96, 15362–15367 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables