Abstract
Clinical and experimental evidence supports that chronic oxidative stress is a primary contributing factor to numerous retinal degenerative diseases, such as age-related macular degeneration (AMD). Eyes obtained postmortem from AMD patients have extensive free radical damage to the proteins, lipids, DNA, and mitochondria of their retinal pigment epithelial (RPE) cells. In addition, several mouse models of chronic oxidative stress develop many of the pathological hallmarks of AMD. However, the extent to which oxidative stress is an etiologic component versus its involvement in disease progression remains a major unanswered question. Further, whether the primary target of oxidative stress and damage is photoreceptors or RPE cells, or both, is still unclear. In this review, we discuss the major functions of RPE cells with an emphasis on the oxidative challenges these cells encounter and the endogenous antioxidant mechanisms employed to neutralize the deleterious effects that such stresses can elicit if left unchecked.
Keywords: Retinal pigment epithelium, Age-related macular degeneration, Oxidative stress, Mitochondria, Nrf2, Ubiquitin proteolytic system
1. Introduction
The retinal pigment epithelium (RPE) is a single layer of epithelial cells lining the posterior segment of the eye. It is located between the light-sensing photoreceptor cells and the choriocapillaris. Similar to other epithelial cell types, RPE cells are polarized. The apical processes are interdigitated with the outer segments of the photoreceptors, whereas the basolateral side of each cell is aligned along a specialized membrane called Bruch’s membrane (BM) underlying the fenestrated endothelium of the choriocapillaris.
The anatomical positioning of the RPE layer situates these cells for their numerous support functions as guardian and caretaker of the photoreceptors (Strauss, 2005). In conjunction with the endothelium of the retinal vessels, the RPE layer forms the blood–retinal barrier. A primary function of this barrier is to mediate the uptake of ions, water, and nutrients while simultaneously removing metabolic waste products from the subretinal space. These exchange processes are central for maintaining overall metabolic homeostasis and sustenance of the photoreceptor cells. A complementary function of the RPE involves retinoid storage and metabolism. In the classical visual cycle associated with rod photoreceptors, RPE cells convert all-trans-retinol (vitamin A) into 11-cis-retinal and then deliver the 11-cis-retinal to the photoreceptors for phototransduction. 11-cis-Retinal is a chromophoric derivative of vitamin A that binds opsin to generate rhodopsin in photoreceptor outer segments. Coincident with the absorption of a photon of light by rhodopsin and initiation of the phototransduction cascade, 11-cis-retinal is photoisomerized into all-trans-retinal. Upon release from opsin, the all-trans-retinal is reduced in the cytoplasm to all-trans-retinol by all-trans retinol dehydrogenase and subsequently exported to the RPE for recycling back into 11-cis-retinal. The regeneration of 11-cis-retinal in the RPE occurs via an enzymatic cascade consisting of lecithin retinol acyltransferase, RPE65, and 11-cis retinol dehydrogenase. Vitamin A from the circulation also enters RPE cells from the basal side and is likewise processed by these enzymes to produce 11-cis-retinal.
RPE cells are enriched in numerous pigments, such as melanin, lipofuscin, and flavins, which absorb excess light and thereby function to protect the neuroretina from phototoxicity. Paradoxically, these same moieties can underlie photochemical damage to the RPE and retina (Boulton et al., 2001). An additional key function performed by RPE cells is the maintenance of photoreceptor outer segment length. Each day, the RPE ingests the distal tips of the outer segments and, in doing so, balances the growth of these segments that occurs at the proximal end where new membrane stacks are generated. This trimming function of the RPE ensures that a relatively constant outer segment length is maintained, which is essential for proper photoreceptor function (Bok and Hall, 1971; Edwards and Szamier, 1977; LaVail, 1983; Nandrot et al., 2004). RPE cells also secrete growth factors in a directional fashion. Most notably, they release vascular endothelial growth factor basolaterally to the choriocapillaris and pigment epithelial-derived growth factor apically to the subretinal space. Additional immunosuppressive factors are also produced and released by RPE cells to impart immune privilege to ocular tissues (Ishida et al., 2003).
2. Oxidative Stress and Damage in RPE
2.1. Sources of oxidative stress in RPE
The panoply of functions carried out by RPE highlights its central role as guardian and caretaker of the neural retina. It is no coincidence then that impairment of one or more of the above RPE processes can have dire consequences for ocular health and vision. A growing body of clinical and experimental data strongly implicate oxidative stress, and, in particular, chronic intracellular oxidative stress, as a constant threat to the structural and functional integrity of the RPE.
The sources of this stress are diverse. For example, RPE is subjected to very high O2 tension due to its juxtaposition to the blood supply of the choriocapillaris (Alder and Cringle, 1985). Flow rates in the choriocapillaris have been measured at 1400 ml/min per 100 g of tissue, and the venous blood from this vascular bed reportedly has 90% O2 saturation (Alm and Bill, 1970, 1972a, b, 1973). Together, the high flow rate and O2 saturation levels contribute to high O2 tension at the RPE layer. Long-term sunlight exposure is also a significant source of free radical stress to the RPE. In particular, blue light (475 nm) causes photooxidation of RPE biomolecules (including pigments) and can be especially detrimental (Bressler and Bressler, 1995; Cruickshanks et al., 1993; Dorey et al., 1990; Rozanowska et al., 1995).
The enriched mitochondrial population of the RPE exhibits robust metabolic activity to meet the high-energy needs of these cells. This amplified oxidative phosphorylation produces large amounts of ATP but in the process also generates high local concentrations of reactive oxygen species (ROS). As the RPE ages, the capacity to utilize and/or neutralize these mitochondrial-derived ROS likely diminishes. The result is increased collisions of free radicals with DNA, proteins, and lipids within mitochondria and in the cytoplasm. This damaged material can wreak havoc on mitochondrial function and integrity as well as impact cytoplasmic processes and overall cellular health.
Another source of intracellular oxidative stress in RPE is the age-dependent accumulation of lipofuscin (aka aging pigment) (Sparrow et al., 2012). This heterogeneous, autofluorescent complex of lipid–protein aggregates is derived largely from photoreceptor outer segments that are phagocytosed by RPE cells and presumably also contain remnants of triaged RPE organelles (Boulton et al., 2001). Young, healthy, RPE cells efficiently dispose lipofuscin by targeting it to lysosomes for degradation. However, the autophagy and lysosomal pathways within the RPE diminish as we age and thus, so do the capacity to eliminate lipofuscin (Kaarniranta et al., 2010). As a consequence, this waste material builds up to toxic levels in the cytoplasm of RPE cells and produces free radicals. In addition, the components of lipofuscin can be photooxidized and, in doing so, become toxic to the RPE. The best studied of these components is produced by the reaction of two molecules of all-trans-retinal with ethanolamine and is called A2E (N-retinyl-N-retinylidene ethanolamine, 2-[2,6-dimethyl-8-(2,6,6-trimethyl-1-cyclohexen-1-yl)-1E,3E,5E,7E-octatetraenyl]-1-(2-hydroxyethyl)-4-[4-methyl-6-(2,6,6-trimethyl-1-cyclohexen-1-yl)-1E,3E,5E-hexatrienyl]-pyridinium). When oxidized by 430-nm light, various A2E epoxides are generated that arguably become detrimental to RPE health and function. Studies in cultured RPE cells indicate that A2E epoxides have pleiotropic effects, ranging from destabilizing mitochondrial and/or lysosomal membrane integrity to reducing the capacity of RPE cells to process phagocytosed photoreceptor outer segments (Bergmann et al., 2004; Holz et al., 1999; Liu et al., 2008; Vives-Bauza et al., 2008). Photodegradation of A2E can release methylglyoxal, a dicarbonyl that reacts with proteins to produce advanced glycation end products (Wu et al., 2010). In addition to A2E and its derivatives, other hydrophobic components of lipofuscin have been invoked as major contributors to the photoexcitatory-induced free radical production and damage caused to RPE cells (Pawlak et al., 2002, 2003). Collectively, photoexcitation of these outer segment-derived bisretinoids can lead to the generation of singlet oxygen, superoxide anion, and hydroxyl radicals, all of which set off chain reactions of lipid peroxidations (Dillon et al., 1996).
2.2. Targets of oxidative damage in RPE
In an age-dependent manner, these various sources of intracellular free radical stress can converge to elicit compounding deleterious effects. They do so by triggering large-scale damage to RPE biomolecules (i.e., proteins, lipids, and DNA) and by promoting destruction of the RPE mitochondrial network. Mitochondria are an especially susceptible target of oxidative damage because they are a rich source of ROS, the mitochondrial genome lacks protection from histones, and the system for repairing damage to mtDNA is not as proficient and robust as its nuclear counterpart. Furthermore, because RPE cells are postmitotic, damaged mitochondria are not readily eliminated as a function of cell division (Cai et al., 2000). These events, in turn, presumably exacerbate further increases in free radical damage and stress. mtDNA damage due to oxidative stress increases with age and is enriched in the RPE cells of the macula region as compared to the peripheral retina. In age-related macular degeneration (AMD) eyes, this damage parallels disease severity (Lin et al., 2011). These increases in mtDNA damage correlate with decreases in OGG1, a marker of mtDNA repair capacity (Lin et al., 2011), and this damage is distributed throughout the mtDNA genome, as measured by long-extension polymerase chain reaction (Karunadharma et al., 2010). In contrast, normal aging eyes have progressively increasing levels of mtDNA that is concentrated in the common deletion region of the mtDNA. Further, proteomic tracking of RPE mitochondrial protein levels during different stages of AMD revealed changes in the ATP synthase subunits (α, β, δ), mitochondrial import factors, and translation factors (Nordgaard et al., 2008). The exact mechanisms underlying these particular changes in protein levels are unknown, but the data are consistent with the notion that chronic oxidative insult alters fundamental aspects of mitochondrial function, integrity, and viability.
A mechanistic model explaining this self-perpetuating cycle of debilitation remains to be elucidated, but this area of investigation is likely to be a rich source of therapeutic targets. Any unifying explanation will have to incorporate several key contributors. One such factor is that many of the enzymes responsible for handling and disposing of oxidatively damaged proteins contain active site cysteines and, as a consequence, are themselves predicted to be highly susceptible to inactivation by free radicals (Kriegenburg et al., 2011). For example, inactivation of protective enzymes could occur via the addition of molecular oxygen(s) to a cysteine sulfhydryl, leading to the formation of cysteine sulfenic acid, sulfinic acid, or sulfonic acid. Enzymatically inactivating oxidations of this nature as well as protective glutathionylation of active site cysteines in response to redox stress have been demonstrated for components of the ubiquitin proteolytic system (UPS) (Jahngen-Hodge et al., 1997; Obin et al., 1998) and may well apply to enzymes of the autophagy system. The UPS and autophagy systems are the two primary means for triaging and eliminating damaged, intracellular material. The susceptibility of these enzymatic cascades to inactivation by oxidative stress coupled to the induction of these systems during such stress is likely to be an expanding area of research focus in the coming years in light of the notion that the capacity of cells to counter the accumulation of damaged materials declines as a function of age. This decline is consistent with the buildup of potentially toxic aggregates such as lipofuscin, a hallmark of the aging cell.
Mitochondrial DNA damage is a second factor contributing to the cycle of self-perpetuating debilitation induced by chronic oxidative stress in the RPE. Analysis of postmortem AMD eyes (Feher et al., 2006; Karunadharma et al., 2010) has clearly demonstrated an accumulation of modifications and mutations to RPE mitochondrial DNA that are readily attributable to free radical stress. In all likelihood, such stress has both cytoplasmic and mitochondrial origins that together culminate in irreversible changes to mitochondrial DNA, additional free radical production, declining mitochondrial respiratory function, and a growing need for increased flux through the autophagy pathway to dispose of dysfunctional mitochondria. This “domino” effect may well represent one, if not the, precipitating event in AMD development.
A third contributory factor that, to date, has received only minimal appreciation is the impact that chronic oxidative stress in the RPE has on mitochondrial network dynamics (i.e., the equilibrium between mitochondrial fusion and fission). The molecules governing this equilibrium are described in more detail below (Section 4). Briefly, these synchronized processes regulate a dynamic balance in which individual mitochondria physically associate (fusion) or dissociate (fission) from the reticular mitochondrial network. Multiple GTPases mediate fusion and fission, and the stability, function, and/or localization of several of these proteins are controlled by the UPS (Neutzner et al., 2008). Yet, these same UPS components harbor active site cysteines and, as mentioned above, are potentially susceptible to inactivation upon oxidation. The consequences of such events on fusion and fission, particularly over the longer term in RPE cells subjected to chronic oxidative stress, remain to be fully elaborated, but the prediction would be an age-dependent debilitation of mitochondrial network integrity and function. Such mitochondrial deterioration has been noted in postmortem eyes from AMD patients (Feher et al., 2006).
3. Preservation of RPE Integrity and Function
In light of the challenges that oxidative stress poses to RPE health, function, and survival, it is typically not until the later years of life (i.e., age 60–65) that pathological hallmarks in this cell layer begin to manifest, as in the case of AMD. We speculate that two major mechanisms likely account for this. One is the collective workings of the endogenous antioxidant defense system. The second is RPE regeneration.
3.1. Endogenous antioxidant defenses
3.1.1. Central role of Nrf2
The endogenous antioxidant defense system is comprised of an array of factors including enzymes such as the superoxide dismutases (SODs) and catalase, carotenoids such as lutein, zeaxanthin, and β-carotene, vitamins such as l-ascorbate and α-tocopherol, pigments such as melanin, and various other molecules (e.g., glutathione (GSH)) that function in redox maintenance and homeostasis. Additionally, most if not all cells in the body (including RPE cells) express a panel of cytoprotective antioxidant and detoxifying genes commonly referred to as phase II genes. Expression of phase II genes is driven by nuclear factor-related factor 2, a transcription factor better known as Nrf2. Nrf2 belongs to a family of cap-N-collar transcription factors that includes two related transcriptional activators, Nrf1 and Nrf3. Expression profiling in adult mice has demonstrated that Nrf1 is expressed in a range of tissues including heart, muscle, liver, kidney, salivary glands, and prostate (Biswas and Chan, 2010). Nrf2 is ubiquitously expressed throughout the body with highest levels in muscle, kidney, and lung in adults (Moi et al., 1994), and Nrf3 expression has been reported in liver, placenta, and other tissues (Zhang et al., 2009). Nrf2 knockout mice live normal life spans but are more susceptible to tumor formation when challenged (Ramos-Gomez et al., 2001). Embryonic loss of Nrf2 has also been reported to interfere with adipocyte development although the authors concluded that this effect was independent of the antioxidant actions of the transcription factor (Pi et al., 2010).
During redox homeostasis, cells suppress Nrf2 levels primarily through posttranslational mechanisms involving Ub-mediated degradation by the 26S proteasome. This degradation is mediated by a UPS E3 ligase called CUL3Keap1 (Kobayashi et al., 2004; Zhang et al., 2004). CUL3Keap1 consists of three primary proteins, but the complex is modulated by a large panel of secondary factors (Bosu and Kipreos, 2008). The three primary proteins are the cullin 3 (CUL3) scaffold protein, a really interesting new gene (RING)-finger containing protein, ROC1, and the substrate adaptor, Keap1. Keap1 recruits Nrf2 to a CUL3–ROC1 complex for polyubiquitylation. The tagged transcription factor is subsequently delivered to the 26S proteasome for degradation. Upon sensing of an oxidative stress, however, the CUL3Keap1 complex dissociates and Nrf2 turnover is inhibited. The stabilized transcription factor translocates to the nucleus, heterodimerizes with one of several Maf proteins, and binds to the antioxidant response elements in the promoters of phase II genes. This constellation of genes codes for multiple antioxidant and detoxifying enzymes including GSH-S-transferases, NAD(P)H:quinone oxidoreductases, heme oxygenase (HO)-1, and thioredoxin as well as chaperones and components of the proteasome. Collectively, the induction of these cytoprotective proteins enables cells to neutralize free radical stress and restore redox homeostasis (Kensler et al., 2007).
Work in cultured RPE cells suggests a central role for this transcription factor in protecting the RPE from phototoxic stress. For example, Gao and Talalay found that treatment of aRPE19 cells with the Nrf2 activator, sulforaphane, induced the expression of Nrf2 target genes and effectively protected the cells from phototoxicity caused by blue light-activated all-trans-retinaldehyde (Gao and Talalay, 2004). Sulforaphane is an isothiocyanate found in cruciferous vegetables (Zhang et al., 1992) that directly modifies and inhibits the machinery mediating Nrf2 turnover (Dinkova-Kostova et al., 2002). The all-trans-retinaldehyde treatment was used to reconstitute the age-dependent accumulation of this retinoid in RPE cells as a by-product of photoreceptor phototransduction. In complementary studies, the Sparrow laboratory found that sulforaphane treatment protected cultured RPE cells from death induced by photooxidized A2E. The protection was attributed to sulforaphane-induced, Nrf2-mediated increases in GSH levels as well as increases in the phase II enzymes, NAD(P)H:quinone reductase, and GSH-S-transferases (Zhou et al., 2006).
Complementary in vivo and in vitro studies have further highlighted the central role of Nrf2-induced expression and maintenance of thioredoxin and GSH levels in protecting RPE cells from a variety of oxidative challenges (Ha et al., 2006; Nelson et al., 1999, 2002; Tanito et al., 2005; Yoon et al., 2011a). Additionally, multiple laboratories have been delineating the relationship between cigarette smoke, Nrf2, and RPE atrophy (Cano et al., 2010; Chan, 1998; Seddon et al., 1996; Smith et al., 2001). In pilot studies, Handa and colleagues found that the RPE cells of genetically engineered Nrf2 knockout mice have an increased incidence of DNA damage, vacuolization, dilated basolateral infoldings, overall cellular degeneration, and apoptosis following prolonged (6 month) exposure to cigarette smoke. Furthermore, the combination of cigarette smoke and Nrf2 deficiency resulted in evidence consistent with complement factor deposition in BM (Cano et al., 2010). These data bolster findings from other organ systems directly linking Nrf2 to proper function and regulation of the innate immune response (Braun et al., 2002; Thimmulappa et al., 2006a,b). Perhaps most interestingly from a clinical perspective, the RPE cells of mice treated with the synthetic triterpenoid, 2-cyano-3,12-dioxooleana-1,9-dien-28-imidazolide were largely protected from the harmful effects of chronic cigarette smoke exposure. In summary, this work underscores the central role of the Nrf2-driven, antioxidant system in protecting the RPE from chronic oxidative insult.
Zhao et al. recently published the first description of the age-dependent consequences of Nrf2 loss on retinal morphology and function. This study was done without the addition of any exogenous stressors (e.g., cigarette smoke). As predicted by the widely accepted notion that chronic oxidative stress is an underlying trigger of AMD, these investigators detected an age-dependent development of the cardinal pathologies of human AMD (Zhao et al., 2011b). Nrf2 knockout mice developed drusen-like deposits, lipofuscin accumulation, subretinal deposition of inflammatory proteins, and choroidal neovascularization. Electron microscopic analysis revealed that, similar to human AMD eyes (Wang et al., 2009), the RPE cells of Nrf2-deficient mice have an accumulation of phagocytosed photoreceptor outer segments, autophagosomes, autophagolysosomes, and polyubiquitin-positive aggregates (Zhao et al., 2011b). This led the authors to conclude that the absence of Nrf2 resulted in a disabled autophagy–lysosomal degradation system. Interestingly, but perhaps not surprisingly, these various autophagic intermediates were often juxtaposed, or near, BM irregularities. Although a molecular link between Nrf2 and the autophagy defect(s) in the RPE remains to be defined, the authors posit a logical model in which increased cellular stress brought on by the lack of Nrf2 target gene expression increases the load of oxidatively damaged macromolecules and organelles entering the autophagy pathway for degradation. This increased cargo load, combined with the inhibitory effects of lipofuscin on the efficiency of the lysosomal system (Liu et al., 2008; Sparrow and Boulton, 2005), ultimately overwhelms the autophagic capacity of the RPE cell. The result is an accumulation of autophagic intermediates, and the exocytosis of this cellular “garbage” into the sub-RPE space and BM. The exocytosed material, in the form of drusen, elicits the recruitment and deposition of inflammatory and complement factors characteristic of AMD sub-RPE lesions (Zhao et al., 2011b).
Related human studies indicate that Nrf2 inducibility and cytoprotection decrease with age (Suzuki et al., 2008). This decrease has been observed as a marked reduction in detectable Nrf2 mRNA in the lungs of aged smokers versus their nonsmoking counterparts. Curiously, young smokers and young nonsmokers had comparable Nrf2 levels and responsiveness. As this analysis was done in alveolar macrophages, it will be essential to confirm these results at the protein level in human RPE cells considering that such an age-dependent decrease in protection from oxidative challenge could significantly contribute toward a comprehensive molecular explanation for the relatively late onset of AMD. Further, such analyses could offer potential therapeutic strategies aimed at increasing and sustaining Nrf2 levels in the RPE by either pharmacological means and/or by antagonizing Keap1 and other negative regulators of Nrf2 stability. Toward this goal, a large panel of pharmacological Nrf2 activators have been identified and tested in various model systems (Hayes et al., 2010). Future work in this field will require determining, among other things, which of these activators is optimal for protecting RPE cells, which at-risk patient populations would benefit from this activator(s), and the best methods for delivery and sustained release of the agent. Recent advances in nanotechnology and microencapsulation technologies with retinal applications (Birch and Liang, 2007; Kalishwaralal et al., 2010; Zhang et al., 2011) will likely play a central role in the success of these strategies.
Attempts to assess age-dependent changes in the endogenous neuroretinal and RPE antioxidant systems have used different methodologies and, not surprisingly, yielded a range of results, as recently discussed by Cano et al. (2010). Briefly, comparisons of peripheral versus macular retina isolated from fresh cadavers (non-AMD) found that although there was significant variability among individuals, particularly with increasing age, the expression of the major antioxidant enzymes was largely sustained over all ages. GSH peroxidase was an exception as it decreased with age (De La Paz et al., 1996). Five years earlier, an independent study found an age-dependent decrease in catalase activity in both peripheral and macular retina (Liles et al., 1991). Decreases in HO-1 and -2 as well as in catalase were observed in AMD maculas in a 1999 study (Frank et al., 1999), but this was contradicted in a 2004 report (Miyamura and Ogawa, 2004). Interestingly, Miyamura and colleagues found that within a given RPE monolayer, the expression of HO-1 and catalase was variable (i.e., mosaic). In situ hybridization to label HO-1 and catalase mRNAs revealed clumps of positive RPE cells situated near unlabeled RPE cells and a higher proportion of positive cells (as a function of total RPE cells) in the macula versus the periphery. These in situ results were corroborated by processing serial sections for immunohistochemical staining. From this study, the authors concluded that although HO-1 mRNA decreases in both the macula and periphery as a function of age, HO-1 and catalase protein levels are in fact sustained during aging. Mosaic patterns of HO-1 and catalase expression were not observed in ganglion cells, which had uniform staining for both enzymes, implying that the ability of individual RPE cells to manage acute and chronic oxidative insults may vary. It is tempting to speculate that such variability may underlie or contribute to the geographic nature of dry AMD.
In a follow-up work, the Handa laboratory used microarray analysis of laser-captured microdissected RPE cells to perform a global analysis of gene expression patterns in older (nondiseased) human eyes. Perhaps unexpectedly, dramatic differences were not found between macular and peripheral RPE cells, but interestingly from the standpoint of redox homeostasis and protection, GSH-S-transferase M1 was relatively underexpressed in macular RPE (Ishibashi et al., 2004). From this analysis of healthy older eyes, the authors concluded that there is not a wholesale, age-dependent diminution of the antioxidant system in macular RPE cells. Collectively, these data support the notion that there is not an age-dependent, precipitous drop in the Nrf2 circuit in RPE cells, but more likely there may be a dampening or dysregulation of one or more signaling pathways linked to Nrf2-mediated cytoprotection of the RPE. Further, key antioxidant enzymes may be irreversibly inactivated or mis-activated and trigger a redox crisis.
3.1.2. Glutathione
Substantial clinical and experimental evidence demonstrates that GSH makes a major contribution to the endogenous antioxidant defense of RPE cells (Cai et al., 2000). GSH is a tripeptide (l-γ-glutamyl-l-cysteinyl-glycine) that interconverts between a reduced form, designated as “GSH,” and an oxidized form, GSH disulfide, designated as “GSSG.” Physiologically, this conversion often involves GSH molecules neutralizing lipid peroxides as well as hydrogen peroxide (H2O2) in a reaction catalyzed by any one of the family of enzymes referred to collectively as GSH peroxidases. In the case of H2O2, the products of the reaction are GSSG and two molecules of water. GSSG is reduced back to GSH by the action of GSH reductase in a reaction that utilizes nicotinamide adenine dinucleotide phosphate as a cofactor to supply electrons. GSH can also be nonenzymatically or enzymatically (via GSH-S-transferase) conjugated to small molecules, proteins, or lipids in a process referred to as glutathionylation. GSH is at millimolar concentrations in cells and this abundance reflects its central importance to redox homeostasis. Most cell types maintain intracellular GSH at 1–10 mM, whereas GSSG and conjugated GS are typically each below 0.1 mM(Ballatori et al., 2009b). Moreover, age-dependent decreases in the ratio of GSH:GSSG have been detected in human plasma from the elderly, as compared to younger individuals, implying potential links into age-dependent pathologies, including AMD and diabetes (Samiec et al., 1998).
The levels of intracellular GSH are determined by a balance between synthesis and degradation. Interestingly, synthesis takes place intracellularly, whereas GSH catabolism is exclusively an extracellular event. The first and rate-limiting step of GSH synthesis is the ligation of glutamate to cysteine by the enzyme, γ-glutamyl cysteine synthetase (GCS) (aka glutamate cysteine ligase) (Majerus et al., 1971; Minnich et al., 1971). GCS is a heterodimer of a catalytic (GCLC) and a modulatory (GCLM) subunit, and the expression of these two subunits is driven by Nrf2 (Kwak et al., 2003). The second step in the biosynthetic pathway adds a glycine residue to complete the tripeptide and is catalyzed by GSH synthetase. GSH synthesis rates are highly sensitive to the availability of free cysteine (Ballatori et al., 2009a; Meister and Tate, 1976). The extracellular hydrolysis of GSH and GSH-containing molecules (e.g., GSH-S-conjugates) is performed by γ-glutamyl transpeptidase, an ectoenzyme enriched for on the apical surface of many epithelial cell types involved in transport (e.g., kidney, intestine, and epididymis).
The protective function of GSH in RPE cells has been primarily gleaned from cultured cell studies in which efforts to increase the GSH content have yielded protection from oxidative insult. For example, Sternberg and colleagues demonstrated that feeding cultured human RPE cells a mixture of glutamate, cysteine, and glycine was sufficient to boost intracellular GSH levels and protect cells from oxidant-induced toxicity by t-butylhydroperoxide (t-BHP) (Sternberg et al., 1993). Similarly, Nelson et al. stimulated GSH synthesis using the phase II inducer, DMF, to confer resistance to t-BHP (Nelson et al., 1999). In vitro work by Sparrow and colleagues demonstrated that GSH directly reacts with photooxidized A2E and methylglyoxal, a photocleavage product of A2E, but not with nonphotooxidized A2E (Yoon et al., 2011a). Using electrospray ionization mass spectrometry, these investigators showed that GSH can donate a hydrogen atom to photooxidized A2E and its photoproducts as well as form adducts with these modified bisretinoids. These data prompted the authors to suggest that GSH may in fact play an important role in limiting the reactivity and toxicity of bisretinoid photoproducts and facilitate their elimination from RPE cells. This work is consistent with previous findings from this laboratory showing that the Nrf2 activator, sulforaphane, could protect against damage caused by photooxidized A2E (Zhou et al., 2006). It is reasonable to speculate that this protection was conferred by Nrf2 induction of GCS gene expression and subsequent stimulation of GSH synthesis, but other Nrf2 target genes may well have contributed.
3.1.3. Other antioxidants
Similar to photoreceptors, RPE cells may also employ antioxidants including α-tocopherol (vitamin E), l-ascorbic acid (vitamin C), and β-carotene to mitigate the impact of chronic oxidative exposure and stress. Briefly, α-tocopherol functions as a quencher and scavenger of photogenerated singlet oxygen whereby a single molecule of α-tocopherol can deactivate 120 molecules of singlet oxygen (Fahrenholtz et al., 1974). In vivo, vitamin E gets oxidized to an α-chromanoxyl radical and, in doing so, limits the oxidation of polyunsaturated fatty acids. l-ascorbic acid reduces the α-chromanoxyl radical back to regenerate α-tocopherol. This reduction can also be performed by quinones, thiols such as lipoic acid, and GSH. β-Carotene, a precursor of vitamin A, is a membrane antioxidant that can physically deactivate singlet oxygen. Susceptibility of the RPE to mounting oxidative stress and damage in the elderly may be exacerbated by the age-dependent reduction in plasma levels of l-ascorbic acid (Rikans and Moore, 1988) and α-tocopherol (Vandewoude and Vandewoude, 1987).
3.2. RPE regeneration
In addition to these antioxidant defense mechanisms, RPE regeneration may contribute to sustaining retinal health and function during the early and mid-life years. The extent and optimal physiological conditions of such regeneration are areas of intense research (Sugino et al., 2011), but multiple RPE transplant studies have convincingly established that an aged and diseased BM is largely incompatible with regeneration of a functional RPE monolayer (Binder et al., 2002; Gullapalli et al., 2005; Lee and Maclaren, 2011). Thus, prior to the onset of AMD, it is possible that sick RPE cells can be replaced through endogenous regeneration in so long as the integrity of the underlying BM is suitably healthy (Binder et al., 2002). However, following the initiation of retinal degenerative disorders like AMD and compromise of BM integrity, the endogenous recovery of the RPE monolayer is remote without surgical and/or therapeutic intervention. Toward this end, Sugino and colleagues have demonstrated that aged submacular human BM seeded with cell-derived extracellular matrix proteins is an improved substrate for RPE cell survival (Sugino et al., 2011). Thus, the future of BM and/or RPE transplantation could hold great promise for patients with intermediate to advanced AMD.
4. Mitochondrial Network Dynamics
As briefly discussed in Sections 2.1 and 2.2, mitochondria are both a major source and target of ROS in RPE cells. As such, this section elaborates on the proteins that regulate mitochondrial dynamics with the idea that a comprehensive understanding of these dynamics will potentially facilitate drug development efforts to abrogate RPE atrophy. Mitochondria have historically been depicted as individual, round organelles that act independently of one another, but in fact these organelles form a dynamic reticular network, the morphology of which is determined by the relative rates of fission and fusion events (Karbowski and Youle, 2003). The dynamic nature of this network is crucial for equivalent segregation of mitochondria during cell division (Taguchi et al., 2007), quality control of the mitochondrial genome (mtDNA), metabolic regulation, and response to apoptotic stimuli (Liesa et al., 2009).
Fusion of mitochondria involves a membrane potential-dependent melding of both the outer and inner mitochondrial membranes on adjacent mitochondria (Legros et al., 2002), thus decreasing the total number of individual organelles while maintaining total mitochondrial mass. Conversely, during fission, the mitochondria are clamped and divided at determined fission sites, thus increasing the total number of free organelles. Mitochondrial networks driven by fission appear extremely vesicular and disconnected, whereas those driven by fusion appear highly reticular and elongated (e.g., Fig. 4.1). The consequences of disrupting mitochondrial network equilibrium are still being elucidated, but it has been demonstrated that excessive fission of mitochondrial networks is associated with decreased mitochondrial volume and decreased mitochondrial membrane potential (Park et al., 2008). Equilibria shifted toward fusion are associated with mtDNA depletion and deficits in electron transport chain (ETC) activity, likely through ETC assembly defects (Parone et al., 2008). The most aberrant phenotypes are manifested when either fusion is excessively inhibited or fission excessively stimulated (Scott and Youle, 2010).
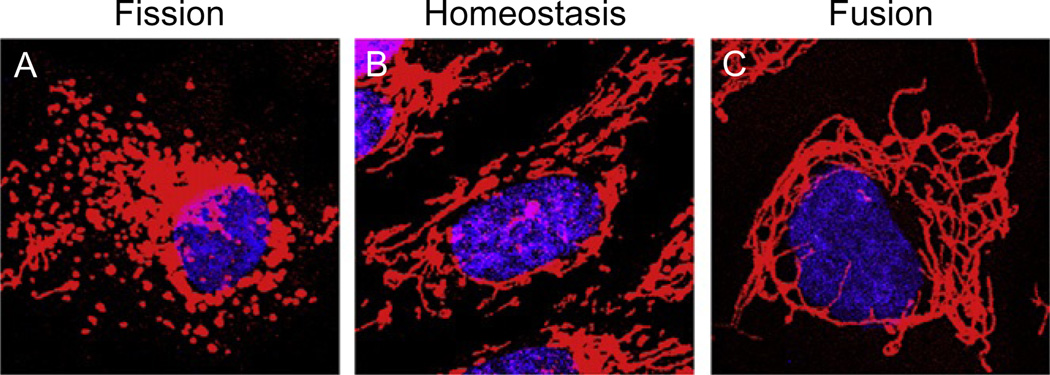
Figure 4.1.
The morphology of the mitochondrial network is dynamic. Confocal images of the various states of the mitochondrial network in telomerase-immortalized, human RPE-1 cells. (A) Treatment with 50 µM of the proteasome inhibitor, MG132, causes fragmentation of the network (i.e., fission). (B) Cells during homeostasis have a reticular network consisting of different size mitochondrial filaments. (C) Cells knocked down for the profission factor, Drp1, exhibit a fused phenotype characterized by interconnected tubular mitochondria. Mitochondria are red and nuclei are blue.
4.1. Drp1 and fission
Most attempts to experimentally disrupt the mitochondrial network have targeted the handful of proteins established to regulate fission and fusion events. Fission events are primarily driven by a large, dynamin-related GTPase (Drp1 in mammals, Dnm1 in yeast). Drp1 is an 80-kDa protein recruited from the cytoplasm to future sites of fission on the mitochondrial outer membrane (MOM). Dnm1p, the Saccharomyces cerevisiae homologue of Drp1, was first discovered to regulate mitochondrial morphology as budding yeast with a null Dnm1 allele have mitochondria clustered on one side of the nucleus. The loss of Dnm1p did not, however, affect the morphologies of other membrane-bound organelles such as the endoplasmic reticulum, Golgi apparatus, or vacuoles (Otsuga et al., 1998). These findings suggest that Dnm1p is not only important for dividing mitochondria but also in determining the subcellular distribution of the mitochondrial network. Several years later, a similar role for mammalian Drp1 was elaborated. Overexpression of a dominant-negative point mutant (K38A) of Drp1 in COS-7 cells, an African green monkey-derived cell line, was found to induce a perinuclear clustering of mitochondrial tubules (Smirnova et al., 1998). These perinuclear clusters were shown to consist of highly interconnected mitochondrial tubules that could be dispersed by treatment with the microtubule depolymerizing agent, nocodazole. These data indicated a role for the microtubule network in either delivery of mitochondria to the ER or in Drp-1 recognition (Smirnova et al., 2001).
Drp1 is primarily a cytoplasmic protein and it must be recruited to mitochondrial sites of fission. As such, overexpression of recombinant Drp1 reportedly did not enhance mitochondrial fission (Smirnova et al., 2001), suggesting that Drp1 recruitment is a highly regulated process intended to restrict fission activity and thereby preserve mitochondrial network equilibrium. Indeed, the recruitment, retention, and activity of Drp1 involve a host of MOM proteins (e.g., Mff; Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010) and posttranslational modifications (e.g., phosphorylation, ubiquitylation, SUMOylation, and redox-dependent thiol modification) (Braschi et al., 2009; Chang and Blackstone, 2007; Cho et al., 2009; Han et al., 2008; Wang et al., 2011; Yonashiro et al., 2006). Once localized to the mitochondria, Drp1 forms an oligomeric, helical structure around the mitochondria (Ingerman et al., 2005; Smirnova et al., 2001). This structure mediates mitochondrial fission through a putative scission-like mechanism (Smirnova et al., 2001; Yoon et al., 2001). Until recently, it was difficult to reconcile how Drp1 helices circumscribe mitochondria to exercise the necessary mechano-scission chemical force in light of the fact that the circumference of a mitochondria is estimated to be 190–200 nm (Friedman et al., 2011) and the maximum circumference of a Drp1 oligomeric helix is approximated at 110–130 nm (Ingerman et al., 2005; Mears et al., 2011). This conundrum has now been apparently resolved as Friedman and colleagues discovered that ER tubules constrict mitochondria to facilitate Drp1-mediated fission. This clamping action by the ER reduces the circumference of the mitochondria sufficiently to permit encompassment by the Drp1 oligomer (Friedman et al., 2011).
The exact MOM proteins and mechanism(s) for recruitment of Drp1 to sites of fission remain controversial. For example, the single-pass transmembrane protein, Fis1, has been implicated (Yoon et al., 2003) based on the observation that overexpression of Fis1 induces mitochondrial fragmentation while simultaneous overexpression of (K38A) Drp1 dampens this effect (James et al., 2003). Additionally, Dnm1p and the yeast homologue of Fis1 (Fis1p) bind to each other and this complex can incorporate a third protein, Mdv1p. All three proteins are required for yeast mitochondrial division (Mozdy et al., 2000; Tieu and Nunnari, 2000; Tieu et al., 2002). The interaction between Fis1 and Drp1 was confirmed in mammalian cells ( James et al., 2003), although no human orthologue for Mdv1p has been identified to date. Mutations targeted at the N-terminal tetratricopeptide repeats of Fis1 prevent Drp1 association and exert a dominant-negative effect on mitochondrial fission (Yu et al., 2005). These data support the notion that Fis1 directly recruits Drp1 to sites of fission.
However, independent studies concluded that the presence of Fis1 is not sufficient to catalyze fission. The most compelling evidence comes from the finding that conditional knockout of Fis1 from colon carcinoma cells is dispensable for fission and for the physical recruitment of Drp1 to the mitochondria (Otera et al., 2010). Additionally, the GTPase activity of Drp1 was not stimulated by Fis1 addition in vitro. Moreover, Suzuki and colleagues did not detect a disruption of the mitochondrial network or the mitochondrial accumulation of Drp1 in cells overexpressing Fis1 (Suzuki et al., 2003). Interestingly, knockdown of Fis1 induced mitochondrial elongation, but the knockdown cells were also larger than the control cells (Otera et al., 2010), indicating that Fis1 may play an indirect role in maintaining mitochondrial dynamics and/or may contribute to cell growth through a mitochondria-independent pathway. These conflicting observations suggest that Fis1 may function as a low affinity receptor for Drp1 but that the recruitment and activation of Drp1 requires other factors.
Mitochondrial fission factor (Mff) is a tail-anchored MOM protein that, similar to Fis1, induces elongation of the mitochondrial network upon depletion by RNA interference (Gandre-Babbe and van der Bliek, 2008). Additionally, knockdown of Mff abolishes the punctate, mitochondrial focal accumulation of Drp1, and this effect is reversed by simultaneous overexpression of Mff (Otera et al., 2010). These data suggest that Mff either recruits and/or anchors Drp1 to the MOM. The finding that overexpression of Mff alone induces a more robust mitochondrial fission phenotype than Fis1 overexpression (Otera et al., 2010) is consistent with the idea that Mff has a higher affinity for Drp1. The cytoplasm-exposed, N-terminal domain of Mff binds Drp1, and this interaction is retained in the face of Mff mislocalization to the plasma membrane (Otera et al., 2010), strongly indicating that Mff is a primary determinant for targeting Drp1 to mitochondria. Additionally, Drp1 oligomerization appears to be required for its mitochondrial recruitment as a Drp1 mutant (A395D) defective in higher order assembly fails to both coimmunoprecipitate with Mff in an in vitro assay and to localize to mitochondria in vivo (Otera et al., 2010).
Although most evidence indicates that Drp1 recruitment to mitochondria promotes fission, recent findings indicate that Drp1 recruitment to the MOM can also inhibit fission. This inhibitory effect is mediated by a vertebrate-specific, MOM protein called mitochondrial elongation factor 1 (MIEF1; also known as MiD51) (Palmer et al., 2011; Zhao et al., 2011a). Overexpression of this novel Drp1 receptor induces elongated mitochondria, whereas MIEF1 depletion fragments the mitochondrial network. MIEF1 was also shown to recruit Drp1 to mitochondria independent of Fis1,Mff, and the fusion factor, mitofusin 2 (Mfn2). Accordingly, Drp1 and MIEF1 coimmunoprecipitate, and this interaction inhibits the GTP-binding capacity of Drp1, as assessed by GTP-agarose pull down (Zhao et al., 2011a). Given the abundance of Drp1 in the cytoplasm, the negative regulatory function of MIEF1 likely plays a critical role in restricting Drp1 activity and thereby preserving mitochondrial network equilibrium.
4.2. Proteins that regulate mitochondrial fusion
While proteins like Drp1 may have a dual role in fission and fusion, most of the machinery that regulates mitochondrial membrane fusion is distinct from the fission machinery, despite the obligate coordination between the two processes. Similar to fission, fusion is also controlled by large GTPases. Mitofusins (Mfn) 1 and 2 are each two-pass transmembrane proteins in the MOM, and the Drosophila/yeast orthologue for both mitofusins is called Fuzzy onions 1 (Fzo1). Fzo1 is a transmembrane protein that passes through the MOM twice such that a short loop of the protein makes contact with the intermembrane space. This intermembrane space segment is required for efficient fusion (Fritz et al., 2001; Legros et al., 2002), underlying not only the importance of fusing both the outer and inner membranes, but also the observation that mitochondria must have an intact membrane potential to undergo fusion (Legros et al., 2002). The mammalian mitofusins are similarly oriented; they project their N-terminal GTPase domains and C-terminal coiled-coil domains out into the cytoplasm and mediate a SNARE-like docking mechanism in trans with mitofusins on adjacent mitochondria (Koshiba et al., 2004; Santel and Fuller, 2001). It was independently demonstrated that only one of the two mitofusins is necessary for fusion, although in the absence of a full complement, fusion proceeds with decreased kinetics. Additionally, heterotypic interactions between Mfn1 and Mfn2 were shown to be more efficient in stimulating mitochondrial fusion than either homotypic interaction (Hoppins et al., 2011). In contrast, depletion of both mitofusins completely halts all detectable fusion and results in fragmented mitochondria (Koshiba et al., 2004). It was further demonstrated that the free “tips” of mitochondria are the most likely regions to undergo fusion, as connections in the middle of preexisting threads are rare, although how this reconciles with the even distribution of Mfn1/2 on the mitochondria is unclear (Yoon et al., 2011b). Mutations in the Mfn2 gene can cause the neurodegenerative disease, Charcot–Marie–Tooth neuropathy type 2A, highlighting the importance of balancing steady-state fission activity with robust and flexible fusion activity (Kijima et al., 2005). The presence of the two mitofusin isoforms presumably imparts a cytoprotective redundancy for the fusion machinery, but the localization of the mitofusins to the MOM implies the existence of a counterpart(s) fusion mediator to account for structural maintenance of the mitochondrial inner membrane (MIM).
Maintenance of the MIM is mediated by a large, dynamin-related GTPase called optical atrophy factor 1 (OPA1). OPA1 is capable of binding Mfn1 and can serve as a link between the two distinct, but synchronized inner and outer fusion events (Cipolat et al., 2004). OPA1 was originally found to have antiapoptotic properties (Olichon et al., 2003), and mutations in one allele of OPA1 lead to dominant optic atrophy, a vision-robbing disease affecting the retinal ganglion cells (Cohn et al., 2007). Contrary to the mitofusins, studying OPA1 is complicated by the presence of eight isoforms in the human genome and by multiple patterns of alternative splicing and proteolytic cleavage. Depending on the variant, nascent OPA1 can either be cleaved by the YME1L protease, generating the short form (S-OPA1) or left unmodified as the long form (L-OPA1) (Song et al., 2007). Of note, this proteolytic regulation of OPA1 is dependent on mitochondrial membrane potential (Guillery et al., 2008). Both populations of OPA1 are essential for proper fusion to occur, demonstrating the need for functional cleavage machinery in the MIM. Initially, it was thought that the entire L-OPA1 population was not proteolyzed, but more recently, it was discovered that the mitochondrial protease OMA1 can cleave L-OPA1 in response to a loss of mitochondrial membrane potential. Notably, OMA1 is itself subject to constitutive proteolytic cleavage by a currently uncharacterized protease. Upon loss of membrane potential, cleavage of OMA1 is inhibited, allowing OMA1 to cleave OPA1, and this proteolysis deactivates OPA1 and inhibits fusion (Head et al., 2009). Head and colleagues further demonstrated that this mechanism is important for the onset of apoptosis.
4.3. Fission, fusion, and apoptosis
Mitochondria are known to be important mediators of apoptosis, or programmed cell death, and this cell fate is a distinguishing pathological hallmark of RPE cells in AMD patients (Kinnunen et al., 2011). Mitochondrial permeabilization and concomitant cytochrome c release are terminal signaling events in the onset of apoptosis. Permeabilization of the mitochondria is accomplished by pore-forming Bcl-2-type proteins called Bak and Bax (Narita et al., 1998). These proteins are inhibited under survival conditions by the Bcl-2-like, antiapoptotic proteins Bcl-xL and Mcl-1 (Willis et al., 2005). Under apoptotic conditions, p53 upregulates Bax and concomitantly downregulates the antiapoptotic factors resulting in the release of cytochrome c and apoptosis-inducing factor and subsequent downstream caspase activation (Culmsee and Landshamer, 2006).
Mounting evidence implicates the fission and fusion machinery in apoptotic signaling. For example, it is widely held that when mitochondrial network equilibrium is shifted toward fusion, cells are more resistant to death signals (Mazure et al., 2011). In addition, the mitochondria become extensively fragmented during apoptosis (Youle and Karbowski, 2005) and accordingly, protection from apoptosis can be conferred by inhibiting the fission machinery via knockdown of either Drp1 or hFis1 (Lee et al., 2004). Drp1 accumulates on mitochondria early during the induction of apoptosis but before caspase activation, suggesting that Drp1 and other members of the fission and fusion machinery may cooperate with the Bcl-2 family proteins to dismantle the mitochondria and amplify the proapoptotic signals. Drp1 normally cycles between the cytoplasm and mitochondria, but Bak/Bax stimulates modification of Drp1 with the ubiquitin (Ub)-like molecule, SUMO. Drp1 SUMOylation correlates with stable association of Drp1 at the mitochondria during apoptotic cell death (Wasiak et al., 2007). Additional work demonstrated that mitochondrial fragmentation induced by nitrosative stress was upstream of Bax foci formation, as overexpression of Mfn1 or dominant-negative Drp1 prevented foci formation.
These observations are consistent with fusion providing protection against apoptosis. It has been proposed that fission of the mitochondria is somehow necessary for insertion of Bax/Bak into the mitochondrial membrane (Yuan et al., 2007). The details of the molecular interactions between the fission/fusion machinery and the apoptotic proteins are still being investigated, but clearly the fields of mitochondrial dynamics and apoptosis are integrated. As our understanding of this relationship grows, so too will the number of potential therapeutic avenues for limiting RPE apoptosis.
4.4. Free radicals influence mitochondrial dynamics
Free radicals are produced both as by-products of exogenous agents, such as sunlight and carcinogens, and as a result of oxygen consumption by mitochondria. The ETC of mitochondria receives reducing equivalents (NADH, FADH2) from the glycolytic pathway and Krebs cycle and serves to generate a proton gradient to drive ATP synthesis. The final electron acceptor in this chain is molecular oxygen (O2), the reduction of which produces water. Stochastically, superoxide () is formed instead of water and can collide with, oxidize, and thereby damage proteins, lipids, and DNA. While superoxide is the chief free radical produced by mitochondria, other types of free radicals can be generated, such as peroxynitrite (ONOO−), through the action of nitric oxide synthase enzymes. It is well established that these various free radical species can damage biomolecules, and the accumulation of this damaged material can be toxic to cells. In this context, it is generally thought that the capacity to triage or sequester such damaged biomolecules serves as a measure of cellular fitness and that this capacity declines as a function of age (Dmitriev and Titov, 2010; Martinez-Vicente and Cuervo, 2007; Zerovnik, 2010).
In addition to damaging biomolecules and organelles, free radical stress can impact mitochondrial dynamics. This was directly demonstrated using high fluence-low power laser irradiation (HF-LPLI) to induce photooxidative stress in ASTC-1 cells (derived from a human lung adenocarcinoma). Wu and colleagues found that mitochondrial fusion was inhibited, whereas Drp1 recruitment to the mitochondria was stimulated, resulting in a profission phenotype (Wu et al., 2011). This effect was not reversed by concomitant overexpression of the fusion factor, Mfn2. HF-LPLI-induced oxidative stress also increased Bax association with the mitochondria and cytochrome c release, indicating that unchecked free radical stress can directly activate the intrinsic apoptotic pathway. Similarly, induction of transient oxidative stress in a hyperglycemia model yielded robust mitochondrial fission. Treatment with p-(trifluoromethoxy)phenylhydrazone to disrupt the mitochondrial membrane potential could prevent this ROS accumulation but did not abrogate the morphological disruption of the mitochondrial network. Furthermore, overexpression of dominant-negative Drp1 was sufficient to prevent hyperglycemia-induced ROS production implying that fragmented mitochondria can be a source of ROS production during hyperglycemic conditions (Yu et al., 2006). These findings highlight just a few of the links between free radical stress and mitochondrial dynamics.
4.5. Regulation of mitochondrial dynamics and protein quality control by Ub E3 ligases
Multiple enzymes of the UPS modulate mitochondrial dynamics. (A more thorough treatment of the UPS is given in Section 5.) Paramount among these UPS components is membrane-associated RING finger (C3HC4) 5 (MARCH5 or MITOL), an E3 Ub ligase embedded in the MOM. MARCH5 is related to a family of E3 ligases found in several herpesviruses that reduce the expression of major histocompatibility complex class I molecules (Bartee et al., 2004). Based on its E3 ligase activity and subcellular localization, MARCH5 has been proposed to be an important regulator of mitochondrial dynamics. MARCH5 has four transmembrane domains with its N and C-termini exposed to the cytoplasm.
The N-terminal domain contains a RING finger domain, which is crucial for both ubiquitylation of substrates and autoubiquitylation of MARCH5 (Yonashiro et al., 2006). As the membrane-associated RING-CH family of Ub ligases have been posited to target the cytoplasmic domains of other membrane proteins for Ub modification (Mansouri et al., 2003), an attractive model positions MARCH5 as a primary regulator of the Ub-dependent modulation of multiple mitochondrial fission and fusion factors. Consistent with this idea, the Youle group demonstrated that the RING-dependent ubiquitylation activity of MARCH5 is necessary for Drp1-mediated fission in HeLa cells (Karbowski et al., 2007) and that ubiquitylated Drp1 coimmunoprecipitates with MARCH5 (Nakamura et al., 2006). It has also been shown that MARCH5 RING mutants form submitochondrial aggregates, to which Drp1 but not Fis1 colocalizes, suggesting that MARCH5’s role in Drp1 recruitment occurs downstream of Fis1 (Karbowski et al., 2007). Remarkably, MARCH5 does not appear to target either Drp1 or Fis1 for proteasomal degradation despite ubiquitylating both in in vitro assays. Furthermore, the RING activity (i.e., ubiquitylating activity) of MARCH5 appears to be crucial for maintaining the mitochondrial network, as overexpression of various RING mutants of MARCH5 induces aberrant mitochondrial interconnections, decreases the total number of individual organelles, and increases the total mitochondrial network volume (Karbowski et al., 2007). These data implicate a nonproteasomal role for MARCH5-mediated ubiquitylation in mitochondrial fission with ubiquitylation of Drp1, Fis1, or both being a prerequisite for fission.
MARCH5 has been most thoroughly characterized as a key regulator of mitochondrial fission, but whether and how it regulates fusion is less clear. MARCH5 coimmunoprecipitates with Mfn2, but not with Mfn1 (Nakamura et al., 2006), indicating that there is some selectivity for the actions of the two mitofusins. It has been established that the elongated, interconnected mitochondria observed when MARCH5 expression or function is silenced result in large part to an inhibition of fission (Karbowski et al., 2007), but it is also possible that wild-type MARCH5 negatively regulates the activities of a fusion factor(s), in particular, Mfn2. Thus, removal of MARCH5 activity from a cell would relieve such inhibition and result in excessive fusogenic activity(s). Using Mfn2-deficient mouse embryonic fibroblasts, the Youle group measured the rates of fusion before and after the introduction of a MARCH5 RING mutant and wild-type Mfn2. While the rate of fusion slightly increased after the introduction of mutant MARCH5, it was well below the rate increase seen when wild-type Mfn2 was reintroduced into the cells, indicating that MARCH5 may have a subtle regulatory influence on Mfn2. Mfn1, however, is a major in vivo ubiquitylation substrate of MARCH5, and ubiquitylated Mfn1 is targeted to the proteasome for degradation. These observations suggest that MARCH5 regulates the fission machinery through nondegradative Ub signals and regulates the fusion machinery in part by negative allosteric regulation of Mfn2 and by controlling the levels of Mfn1. This would classify MARCH5, by all accounts, as a profission factor, but additional studies will be required to fully elaborate the functions of this ligase within mitochondrial network dynamics and regulation.
Although MARCH5 has generally been associated with the maintenance and modulation of the mitochondrial fission and fusion factors, this E3 ligase also contributes a protein quality control function at the MOM. Specifically, MARCH5 has been shown to mediate the Ub-dependent degradation of a mutant of superoxide dismutase 1 (SOD1) that accumulates at the mitochondria and generates toxic levels of ROS (Yonashiro et al., 2009). SOD1 is one of a constellation of antioxidant enzymes that are crucial for neutralizing intracellular free radicals. The SOD isoforms differ in their localization and the metals they chelate, but all catalyze the conversion of superoxide to hydrogen peroxide and molecular oxygen (Fridovich, 1978a,b). The enzyme, catalase, then converts the hydrogen peroxide to water. When SOD1 is mutated or misfolded, as it is in a familial form of amyotrophic lateral sclerosis, it aggregates on the mitochondria and induces mitochondrial dysfunction (Noor et al., 2002). The Yanagi group demonstrated that MARCH5 ubiquitinates mutant SOD1 (mSOD1) and targets it for degradation, thus confirming a role for MARCH5 in protein quality control (Yonashiro et al., 2009). Additionally, knockdown of MARCH5 (or overexpression of a catalytically inactive enzyme) decreased mSOD1 ubiquitination, enhanced mSOD1 accumulation at mitochondria, and increased mSOD-1-derived ROS production. These results suggest that MARCH5 plays a protective role within the cell, but whether this protection is targeted against misfolded proteins in general or is specific to SOD1 remains to be established.
Other E3 ligases localized in the MOM include the dual Ub and SUMO ligase, MULAN/MAPL/GIDE (Braschi et al., 2009; Li et al., 2008; Zhang et al., 2008), and RNF185 (Tang et al., 2011). The precise roles and contributions of these proteins to mitochondrial dynamics largely remain to be delineated. In contrast, the largely cytoplasmic E3 ligase, Parkin, appears to be an especially important regulator of mitochondrial network dynamics. Mutant forms of Parkin are causative of autosomal recessive juvenile Parkinson’s disease (Kitada et al., 1998), a finding consistent with the notion that perturbations of mitochondrial network equilibrium can underlie neurodegeneration (Schon and Przedborski, 2011). Parkin and its cofactor, PTEN-induced kinase 1 (PINK1), which is also genetically linked to familial Parkinson’s disease (Valente et al., 2004), function in quality control of the mitochondria by shuttling damaged organelles to the autophagy/lysosome system for degradation (Dagda and Chu, 2009; Dagda et al., 2009; Vives-Bauza et al., 2010). This pathway of autophagy as it applies to mitochondria is termed, “mitophagy” (Wang and Klionsky, 2011).
In response to cellular stresses that result in a loss of membrane potential, Parkin is selectively recruited to damaged mitochondria by PINK1. PINK1 functions as a sensor of mitochondrial membrane potential. It is proteolytically cleaved and degraded on bioenergetically active mitochondria but stabilized by collapse of the MIM potential. The ensuing stabilization of PINK1, coupled to the catalytic activity of the kinase (Geisler et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010), recruits Parkin to the mitochondria (Narendra et al., 2010) resulting in ubiquitylation of mitochondrial substrates including Mfn1 (Glauser et al., 2011), Marf, a Drosophila orthologue of the mitofusins (Poole et al., 2010; Ziviani et al., 2010), and the voltage-dependent anion-selective channel protein 1 (VDAC1) (Geisler et al., 2010), the most abundant MOM protein. In cultured cells, Parkin-mediated polyubiquitylation of VDAC1 does not detectably alter the stability of VDAC1 (albeit, a direct analysis of half-life was not conducted) (Geisler et al., 2010), implying a nonproteasomal role for the Ub tagging, such as demarcation of the mitochondria for an autophagic fate (Narendra and Youle, 2011).
Mechanistically, it has been suggested that a second major function of Parkin is to induce mitochondrial fission, a prerequisite for mitophagy. Parkin presumably accomplishes this by marking fusion factors such as Mfn1 and Mfn2 for Ub-mediated degradation (Tanaka et al., 2010). Shifting the equilibrium of the mitochondrial network toward fission prevents damaged mitochondria from reassociating with healthy mitochondria and enables the compromised organelles to be effectively segregated and triaged for mitophagy (Narendra and Youle, 2011). Data from studies of the Drosophila orthologues of Parkin, PINK1, and Mfn1 (aka Marf) support this model as depletion of Marf or overexpression of a fission factor partially rescues the phenotypes of muscle-specific knockouts of Parkin or PINK1 (Deng et al., 2008; Poole et al., 2008; Yang et al., 2008).
Minimal work has been done to analyze the expression and/or function of most of the above-described mitochondrial factors in the RPE, despite the abundance of mitochondria in these cells and the critical roles of this organelle in both RPE function and disease (e.g., AMD). Most importantly, such studies could potentially unveil new avenues of therapeutic pursuit. For example, evidence from the Youle laboratory strongly supports the conclusion that PINK1 and Parkin suffice to triage mitochondria for autophagic death (Suen et al., 2010). The authors created heteroplasmic cybrids (cells containing a mixture of mitochondria) with mitochondria having either a wild-type genome or a genome harboring a deleterious mutation of the CoxIV subunit. This mutant of CoxIV disrupts mitochondrial function. Overexpression of Parkin (2–6-fold above endogenous levels) was shown to be sufficient to (1) cull out the dysfunctional mitochondria via mitophagy, (2) promote repopulation by the healthy, wild-type mitochondria, and (3) restore cytochrome c oxidase activity (Suen et al., 2010). The implications of this work for RPE cells could be profound as Parkin displayed an ability to discriminate between healthy and compromised mitochondria and to then selectively target the compromised mitochondria for mitophagy. If such an approach could be applied to spare the mitochondrial failure that besets RPE cells in diseases like AMD, it could provide a means of halting, or at least slowing, disease onset and progression. One could envision delivery of parkin being mediated by subretinal injection of adeno-associated viruses encoding the E3 ligase, or alternatively, of nanoparticles coated with a parkin expression plasmid.
5. Ubiquitin Proteolytic System
5.1. Overview of the UPS system
Intracellular proteins that become damaged and/or misfolded by oxidative stress are typically destined for one of three fates. They can be repaired and refolded, sequestered in aggregates, or targeted for degradation. The UPS plays a major role in targeting and degrading such damaged proteins. The central player of this system is Ub, a highly conserved, 76-amino acid polypeptide that is posttranslationally attached to lysine residues on target proteins. The conjugation of Ub to a target protein is accomplished by an enzyme cascade consisting (minimally) of a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub protein ligase (E3) (Fang and Weissman, 2004). Over the past decade, additional factors have been identified that can facilitate and further increase the efficiency and specificity of Ub conjugation to substrates, but the E1–E2–E3 axis constitutes the core machinery. Akin to phosphorylation and dephosphorylation by kinases and phosphatases, respectively, the ubiquitylation of substrates by E1–E2–E3 is countered by the action of deubiquitylating enzymes, more commonly known as DUBs. At the most basic level, these enzymes, which are either thiol proteases or metalloenzymes, deconstruct polymers of Ub and thereby counter the work of the E1–E2–E3 conjugation machinery. The human genome is estimated to encode 95 DUBs and an excellent survey of these enzymes can be found in Nijman et al. (2005).
Extensive biochemical and biophysical experimentation initiated in the 1980s and continuing to this day has elaborated the general mechanism by which E1–E2–E3 cooperate to ubiquitylate substrates. The initial activation of Ub involves an ATP-consuming reaction catalyzed by E1. This activation is a two-step process (Haas and Rose, 1982; Haas et al., 1982). In the first step, E1 hydrolyzes ATP to adenylate the carboxy-terminal glycine of Ub and release pyrophosphate. The resulting Ub-AMP is tightly bound to E1. In the second step, the active site cysteine of the E1 attacks the Ub-adenylate giving rise to a thioester bond between the cysteine and the carboxy-terminal glycine of Ub and releasing the AMP. This activated Ub is then transferred in a transesterification reaction from the active site cysteine of E1 to the active site cysteine of an E2. A third enzymatic component, an E3 protein ligase, cooperates with the activated E2 to transfer the Ub to substrates. Crystallographic and corroborating biochemical studies have demonstrated that the charging of an E2 with Ub by E1 is mutually exclusive from the association of that E2 with its cognate E3(s). This is attributable to E1 and E3s have an overlapping interaction surface on E2. Substrate selection and specificity are primarily conferred through the pairing of particular E2–E3 combinations, and it is estimated that the human genome encodes 40–60 different E2s and 600–1000 distinct E3s. In vitro and in vivo evidence clearly show that a single E2 can partner with multiple E3s and vice versa. E3s can be single proteins or multi-subunit complexes. In both cases, their primary function is to recruit and facilitate transfer of Ub to substrates.
The E3s can be broadly classified into two main types: those containing a RING finger domain or a U-box domain and those with a homologous to E6-AP carboxy-terminal (HECT) domain (Ye and Rape, 2009). In addition to the structural differences between these E3 classes, a primary distinction lies in the mechanism by which their respective substrates are modified with Ub. The RING finger/U-box E3s do not contain a catalytic cysteine residue, and as a consequence, they function as scaffolds to bring together a Ub-charged E2 and a substrate. The Ub is then transferred directly from the active site cysteine of the E2 to the substrate. In contrast, HECT domain E3s harbor an active site cysteine and receive Ub from the E2 via a trans-thiolation reaction. These E3s then catalytically transfer the Ub to the substrate. This distinction is important particularly as it pertains to the topology of polyUb chains that get attached to substrates.
In the case of the RING finger/U-box E3s, this topology is primarily dictated by the E2 although the E3 can enforce or constrain the formation of particular topologies (David et al., 2011). In contrast, HECT domain E3s appear to govern this process irrespective of the E2 that they partner with to receive the Ub (Kim et al., 2007). Further mechanistic nuance comes into play because some HECT E3s transfer a single Ub at a time when building a polyUb chain on their cognate substrate(s) whereas others prebuild a chain on their active site cysteine and transfer the chain en bloc to the substrate (Wang and Pickart, 2005).
PolyUb chain topology is believed to be a major factor in determining the fate of ubiquitylated substrates (Pickart and Fushman, 2004). Substrates are initially modified on one or more lysine residues with a single Ub in a process referred to as monoubiquitylation. Monoubiquitylation in and of itself can function to recruit new binding partners, alter cellular localization, regulate substrate function and activity and, in select instances, target a substrate for degradation (Ye and Rape, 2009). Alternatively, monoubiquitylation can prime a substrate for the addition of subsequent Ubs; these are attached sequentially to the previously added Ub(s) to form a polyUb chain. Ub has seven lysines, and each can function as an acceptor for polyUb chain synthesis (Kirkpatrick et al., 2006). As a result, polyUb chains can be homogeneous or heterogeneous. Homogeneous polyUb chains are synthesized utilizing a common acceptor lysine on the proximal Ub of the growing chain. For example, polyUb chains can consist of Ub–Ub linkages exclusively between K48 of the proximal Ub and the carboxy-terminal glycine of the distal Ub. Such K48-linked chains on substrates target these proteins to the 26S proteasome for degradation (Chau et al., 1989; Finley et al., 1994; Thrower et al., 2000). Alternatively, K63-linked chains were initially discovered and characterized for their nonproteolytic role in recruiting factors to sites of DNA damage (Hoege et al., 2002; Spence et al., 1995). Recent evidence, however, indicates that K63-linked polyUb chains can, in particular contexts, also function as degradation signals (Xu et al., 2009). Heterogeneous polyUb chains are comprised of a composite of Ub–Ub linkages with different lysine residues serving as acceptor sites on the proximal Ub. Depending on the specific configuration and length of these chains, substrates may be targeted for either proteolytic or nonproteolytic outcomes. Heterogeneous polyUb chains can also have a forked structure and such structures are readily produced in vitro (Kim et al., 2007) and in vegetative yeast cultures (Peng et al., 2003). In this scenario, polyUb chains extend from two or more acceptor lysines on a proximal Ub. In mammalian cells, factors such as S5a/Rpn10 block the synthesis of forked chains produced by the E2, UbcH5, in cooperation with particular RING and U-box E3s (Kim et al, 2009). These forked chains are resistant to deubiquitylation and degradation by the proteasome and by blocking their synthesis; S5a/Rpn10 effectively promotes the degradation of particular substrates (Kim et al., 2009).
The best-studied consequence of polyubiquitylation is to target substrates to the 26S proteasome for degradation. The 26S proteasome is a macromolecular assembly of proteases. These proteases line the interior face of a barrel-shaped, 20S core particle consisting of four stacks of seven-membered rings. The proteases belong to the N-terminal nucleophile hydrolase family. Specifically, there are three proteolytic activities referred to as β1, β2, and β5, and each harbors a catalytic, N-terminal threonine active site. β1 has a preference for cleaving on the carboxyl-side of acidic residues (caspase-like activity), whereas β2 cleaves after lysines and arginines (tryptic-like activity), and β5 after hydrophobic residues (chymotrypsin-like activity). The core particle is capped at one or both ends with a 19S regulatory particle (RP) comprised of a base and a lid structure. The asymmetrically distributed proteins of the RP collectively function to recruit polyubiquitylated substrates, clip off the polyUb chains, unfold the substrates, open an axial channel in the 20S chamber, and translocate the denatured substrates into this chamber for proteolytic reduction to peptides. The resulting peptide fragments are then either cleaved by cytoplasmic peptidases into amino acids or consumed en bulk for hydrolysis by the lysosome. Disassembly of the polyUb chains prior to substrate delivery to the 20S chamber enables the individual Ub molecules to be recycled by reentering the free pool of Ub for conjugation to new substrates (Finley, 2009).
An extensive discussion of the proteins localized to the 19S RP is beyond the scope of this chapter, and the reader is referred to a comprehensive treatment of this topic (Finley, 2009). However, several key players of the 19S RP warrant mentioning. Although the human protein names are used here, much of the seminar work on these factors was done in yeast. S5A and hRpn13 are integral constituents of the RP base that function as polyUb receptors by recruiting Ub-modified substrates for proteasome-mediated degradation (Lam et al., 2002; Qiu et al., 2006). Also of note are the polyUb receptors hHR23/Rad23, hPLIC, and Ddi1 (Kaplun et al., 2005; Kleijnen et al., 2000; Wang et al., 2003). These, and several other proteins, are not integral constituents of the proteasome but rather, are weakly and reversibly associated with the RP and as a result, are commonly referred to as shuttling Ub receptors. They bind Ub polymers through their respective Ub-associated domains and escort them to the proteasome.
Once at the lid, disassembly of the polyUb chain is achieved by three proteasomal DUBs, called Poh1, Uch37, and Usp14 (Borodovsky et al., 2001; Yao and Cohen, 2002; Yao et al., 2006). Poh1, a DUB originally characterized in S. cerevisiae and called Rpn11, likely carries out the lion’s share of this activity as it can remove chains en bloc from tagged substrates (Yao and Cohen, 2002). In contrast, Uch37 and Usp14 function to trim polyUb chains. Uch37, in fact, has been proposed to serve an editing function, removing Ub from “lightly” ubiquitylated substrates and thus preventing their degradation (Lam et al., 1997). Together, the trimming and editing functions of Uch37 and Usp14 play critical roles in preparing substrates for proteasome-mediated turnover (Hanna et al., 2006; Lam et al., 1997; Thrower et al., 2000), triaging suboptimally ubiquitylated substrates from proteasomal processing (Lam et al., 1997), and maintaining the necessary pool of free Ub (Finley, 2009; Koulich et al., 2008). Unfolding of substrates destined for proteasomal degradation is an energy-consuming process performed by a ring-like structure at the RP base consisting of six ATPase subunits of the AAA-ATPase family (Braun et al., 1999; Lee et al., 2001; Navon and Goldberg, 2001).
5.2. UPS components and function in RPE and retina
Although the UPS functions in all cells of the body, efforts to characterize specific components of the system have revealed enrichment of particular E2 and E3 enzymes in certain tissues, cells, and anatomical niches. Evidence to date in the retina indicates that different retinal cell types appear to express distinct subsets of UPS components. Such expression profiling data must always be interpreted bearing in mind that the conclusions gleaned are derived from a compilation of analyses (e.g., mRNA vs. protein) from multiple species (e.g., human, mouse, and bovine) and are potentially limited by the specificity and sensitivity of the reagents (e.g., antibodies) used. Accepting this caveat, four different Ub-conjugating enzymes (E214K, E220K, E225K, and E235K) have been identified in bovine rod outer segments (Obin et al., 1996). Several E3 ligases have been detected including parkin, which is expressed in photoreceptors as well as across the retina (Esteve-Rudd et al., 2010), and KLHL7, which is expressed in ganglion cells and the inner nuclear layer (Friedman et al., 2009). PGP 9.5, a Ub carboxy-terminal hydrolase, is only present in retinal ganglion and horizontal cells (Bonfanti et al., 1992), but the Ub hydrolase, UCH-L3, is enriched in photoreceptor inner segments (Sano et al., 2006) and Ub carboxy-terminal hydrolase-1 (UCH-L1) was detected in the RPE layer of human retinal sections (Glenn et al., 2011) as well as in horizontal, bipolar, amacrine, and ganglion cells (Esteve-Rudd et al., 2010). Recently, it has been demonstrated that Ub-specific protease 2 (USP2) is rhythmically expressed in the retina and controls the circadian clock at the evening light to dark transition (Scoma et al., 2011).
5.3. Mechanisms for handling oxidatively damaged proteins
We have reported that a subset of highly conserved metazoan Ub-conjugating enzymes, the class III E2s, are differentially expressed in the mouse retina (Mirza et al., 2010). The mouse versions of the class III E2s are called UbcM2, UbcM3, and UBE2E2, and each is identical to its human counterpart. These enzymes are distinguished from one another by unique N-terminal extensions of 40–60 residues (Matuschewski et al., 1996). Using rabbit polyclonal antibodies we generated against these extensions, we found that UbcM2 is expressed in ganglion cells, the inner nuclear layer, a subpopulation of photoreceptors, and RPE cells. UBE2E2 was detected in retinal ganglion cells and a subpopulation of nuclei of the inner nuclear layer. In contrast, unequivocal detection of UbcM3 in retinal cells was not observed (Mirza et al., 2010). Retinal E3s and cognate substrates for the class III E2s remain to be determined, and conditional knockouts that deplete each from the RPE will be needed to rigorously determine which, if any, of these enzymes functions to eliminate oxidatively damaged proteins. It is, however, tempting to speculate that the class III E2s fulfill this protective function. These enzymes are functional homologues of a pair of yeast E2s, Ubc4, and Ubc5 that play essential roles in mediating the degradation of misfolded and oxidatively damaged proteins (Kaganovich et al., 2008; Matuschewski et al., 1996; Medicherla and Goldberg, 2008) and are required for viability (Stoll et al., 2011). Additionally, each of the enzymes interacts with carboxy-terminal Hsp70 interacting protein (CHIP) (Xu et al., 2008). CHIP is an E3 ligase that directly binds to heat-shock proteins 70 and 90 (Hsp70 and Hsp90, respectively) to mediate the ubiquitylation of their cognate client proteins. A subset of these ubiquitylated clients are targeted for degradation, whereas others are destined for nonproteolytic outcomes. Furthermore, we recently reported that the active site cysteine and the activity of UbcM2 are preserved in photooxidatively stressed mouse retinas (Mirza et al., 2010), although similar experiments with cultured retinal cells have revealed that other E2s are catalytically inactivated following hydrogen peroxide exposure (Dudek et al., 2005; Shang et al., 2001). These data imply that UbcM2 function is preserved in the face of retinal photooxidative challenge, which nicely positions the enzyme to facilitate the clearance of damaged proteins.
Although the UPS mediates the disposal of oxidatively damaged proteins to prevent their toxic accumulation, Medicherla and Goldberg have demonstrated that in S. cerevisiae, only newly synthesized proteins that have incurred oxidative damage are targeted for degradation by the UPS. In contrast, long-lived proteins (≥60 min postsynthesis) that become oxidatively damaged are largely not triaged (Medicherla and Goldberg, 2008). The authors believe that these findings can be explained by the idea that nascently synthesized proteins subjected to oxidative damage are prevented from folding properly which thereby triggers their UPS-mediated degradation. In contrast, because “older” proteins are already folded and incorporated into macromolecular complexes, oxidative damage does not drive their unfolding. The “older” proteins are thus more resistant to being denatured by oxidants and consequently less susceptible to degradation by the UPS. It remains to be determined whether this scenario holds true for human cells but if so, it could provide new insights into the etiology of AMD and other neurodegenerative disorders. For example, it implies that the protein aggregates that accumulate in AMD patients are primarily derived from freshly synthesized, oxidant-damaged proteins. As a corollary, it suggests that enhancing the capacity of the UPS to degrade this class of compromised proteins may be a legitimate therapeutic strategy for treating AMD. Therefore, it will be of great importance to validate the studies of Medicherla and Goldberg in mammalian retinas, and likewise to determine the fate of oxidatively damaged “older” proteins, many of which are likely being inactivated, though not denatured, in the highly oxidizing environment of the retina and RPE.
Because the retina and RPE are highly susceptible to oxidative damage, sequestration and/or degradation of damaged biomolecules is required to prevent the toxic accumulation of these species. Their buildup is in fact a hallmark of numerous neurodegenerative disorders including AMD (Sas et al., 2007). Yet, the identification and characterization of UPS enzymes that protect the retina is only the “tip of the iceberg” with respect to understanding how the various retinal cell types neutralize and eliminate damaged molecules. In this regard, work from the Frydman laboratory has identified two novel, cellular “compartments” for sequestering misfolded proteins (Kaganovich et al., 2008). One is a quality control compartment, named JUNQ, which is enriched with chaperone proteins (e.g., Hsp104) and 26S proteasomes. The JUNQ accumulates soluble misfolded proteins that can either be processed for Ub-dependent degradation or alternatively, refolded by chaperones. The second compartment is the IPOD (insoluble protein deposit), and it functions to sequester insoluble, aggregated proteins.
These compartments are further distinguished by their intracellular localizations. Whereas the JUNQ is juxtanuclear and associated with the cytoplasmic face of the endoplasmic reticulum, the IPOD is situated peripherally (perivacuolar in yeast) and quarantines aggregates that cannot be salvaged. Numerous autophagic marker proteins, such as Atg8, colocalize with the IPOD, consistent with the idea that the constituents of the IPOD are destined for autophagy-mediated elimination. Importantly, the IPOD is the site of accumulation of disease-associated, amyloidogenic proteins such as prion proteins and Huntington’s protein. Intriguingly, polyubiquitylation is a critical factor in determining the solubility of a misfolded protein and thus whether the protein gets sorted to the JUNQ or to the IPOD. These findings imply that the UPS could potentially be harnessed (e.g., overexpression of particular UPS enzymes) to direct oxidatively damaged and misfolded proteins to the JUNQ for destruction. This could impart multiple benefits to the retinal cells of AMD patients including decreasing the kinetics with which toxic aggregates accumulate in IPODs, reducing the clogging of proteasomes, and preventing the saturation and sequestration of chaperones (Chen et al., 2011). Clearly, much more work needs to be done to establish and characterize the full complement of mechanisms that confer protection from the chronic free radical stress imposed on the retina and RPE.
6. Concluding Remarks
Based on the functions of RPE cells and their susceptibility to chronic oxidative challenge, we posit the following model for the events that trigger RPE atrophy in maculopathies such as AMD (Fig. 4.2). As we age, A2E and related bisretinoids compromise the capacity of the RPE lysosomal system to efficiently and completely degrade the phospholipids of ingested photoreceptor outer segments (Finnemann et al., 2002). This leads to bisretinoid accumulation as outer segments are continually phagocytosed by the RPE but cannot be efficiently degraded. The accumulating photooxidized products react with and are reduced by GSH (Yoon et al., 2011a) but, in the process, deplete this major antioxidant defense molecule from RPE cells. The cells attempt to compensate for this reduction in GSH levels by increasing GSH synthesis but are largely unsuccessful at sustaining such efforts because of dysregulated Nrf2 activity (Cano et al., 2010). Nrf2 transcriptional activity drives expression of γ-glutamylcysteine ligase, the first and rate-limiting step in GSH synthesis. The ensuing decline of ROS-neutralizing capacity along with the concomitant buildup of bisretinoid photoproducts leads to an increase in unchecked cytoplasmic free radical production. These excess free radicals damage proteins, lipids, DNA, and organelles, especially mitochondria. The levels of these damaged biomolecules breach a survival threshold for the RPE by overwhelming the UPS, autophagy, and chaperone defense systems and by compromising mitochondrial integrity, which propagates additional ROS production and ultimately triggers RPE apoptosis.
Figure 4.2.
Model of the steps that initiate dry AMD onset.
ACKNOWLEDGMENTS
Work in the Plafker laboratory is supported in part by grant 1R01GM092900-A1 from NIH/NIGMS, by a Karl Kirchgessner Foundation Vision Research Grant, and by monies from the Oklahoma Medical Research Foundation (OMRF). We apologize to any colleagues whose work was inadvertently overlooked or not cited.
REFERENCES
- Alder VA, Cringle SJ. The effect of the retinal circulation on vitreal oxygen tension. Curr. Eye Res. 1985;4:121–129. doi: 10.3109/02713688508999977. [DOI] [PubMed] [Google Scholar]
- Alm A, Bill A. Blood flow and oxygen extraction in the cat uvea at normal and high intraocular pressures. Acta Physiol. Scand. 1970;80:19–28. doi: 10.1111/j.1748-1716.1970.tb04765.x. [DOI] [PubMed] [Google Scholar]
- Alm A, Bill A. The oxygen supply to the retina. I. Effects of changes in intraocular and arterial blood pressures, and in arterial P O2 and P CO2 on the oxygen tension in the vitreous body of the cat. Acta Physiol. Scand. 1972a;84:261–274. doi: 10.1111/j.1748-1716.1972.tb05177.x. [DOI] [PubMed] [Google Scholar]
- Alm A, Bill A. The oxygen supply to the retina. II. Effects of high intraocular pressure and of increased arterial carbon dioxide tension on uveal and retinal blood flow in cats. A study with radioactively labelled microspheres including flow determinations in brain and some other tissues. Acta Physiol. Scand. 1972b;84:306–319. doi: 10.1111/j.1748-1716.1972.tb05182.x. [DOI] [PubMed] [Google Scholar]
- Alm A, Bill A. Ocular and optic nerve blood flow at normal and increased intraocular pressures in monkeys (Macaca irus): a study with radioactively labelled micro-spheres including flow determinations in brain and some other tissues. Exp. Eye Res. 1973;15:15–29. doi: 10.1016/0014-4835(73)90185-1. [DOI] [PubMed] [Google Scholar]
- Ballatori N, Krance SM, Marchan R, Hammond CL. Plasma membrane glutathione transporters and their roles in cell physiology and pathophysiology. Mol. Aspects Med. 2009a;30:13–28. doi: 10.1016/j.mam.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballatori N, Krance SM, Notenboom S, Shi S, Tieu K, Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biol. Chem. 2009b;390:191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. Down-regulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J. Virol. 2004;78:1109–1120. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann M, Schutt F, Holz FG, Kopitz J. Inhibition of the ATP-driven proton pump in RPE lysosomes by the major lipofuscin fluorophore A2-E may contribute to the pathogenesis of age-related macular degeneration. FASEB J. 2004;18:562–564. doi: 10.1096/fj.03-0289fje. [DOI] [PubMed] [Google Scholar]
- Binder S, Stolba U, Krebs I, Kellner L, Jahn C, Feichtinger H, et al. Transplantation of autologous retinal pigment epithelium in eyes with foveal neovascularization resulting from age-related macular degeneration: a pilot study. Am. J. Ophthalmol. 2002;133:215–225. doi: 10.1016/s0002-9394(01)01373-3. [DOI] [PubMed] [Google Scholar]
- Birch DG, Liang FQ. Age-related macular degeneration: a target for nanotechnology derived medicines. Int. J. Nanomedicine. 2007;2:65–77. doi: 10.2147/nano.2007.2.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol. Appl. Pharmacol. 2010;244:16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok D, Hall MO. The role of the pigment epithelium in the etiology of inherited retinal dystrophy in the rat. J. Cell Biol. 1971;49:664–682. doi: 10.1083/jcb.49.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfanti L, Candeo P, Piccinini M, Carmignoto G, Comelli MC, Ghidella S, et al. Distribution of protein gene product 9.5 (PGP 9.5) in the vertebrate retina: evidence that immunoreactivity is restricted to mammalian horizontal and ganglion cells. J. Comp. Neurol. 1992;322:35–44. doi: 10.1002/cne.903220104. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton M, Rozanowska M, Rozanowski B. Retinal photodamage. J. Photochem. Photobiol. B. 2001;64:144–161. doi: 10.1016/s1011-1344(01)00227-5. [DOI] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, et al. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- Braun S, Hanselmann C, Gassmann MG, Aufdemkeller U, Born-Berclaz C, Chan K, et al. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol. Cell. Biol. 2002;22:5492–5505. doi: 10.1128/MCB.22.15.5492-5505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler NM, Bressler SB. Preventative ophthalmology. Age-related macular degeneration. Ophthalmology. 1995;102:1206–1211. doi: 10.1016/s0161-6420(95)30889-5. [DOI] [PubMed] [Google Scholar]
- Cai J, Nelson KC, Wu M, Sternberg P, Jr, Jones DP. Oxidative damage and protection of the RPE. Prog. Retin. Eye Res. 2000;19:205–221. doi: 10.1016/s1350-9462(99)00009-9. [DOI] [PubMed] [Google Scholar]
- Cano M, Thimmalappula R, Fujihara M, Nagai N, Sporn M, Wang AL, et al. Cigarette smoking, oxidative stress, the anti-oxidant response through Nrf2 signaling, and age-related macular degeneration. Vision Res. 2010;50:652–664. doi: 10.1016/j.visres.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D. Cigarette smoking and age-related macular degeneration. Optom. Vis. Sci. 1998;75:476–484. doi: 10.1097/00006324-199807000-00015. [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb. Perspect. Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn AC, Toomes C, Potter C, Towns KV, Hewitt AW, Inglehearn CF, et al. Autosomal dominant optic atrophy: penetrance and expressivity in patients with OPA1 mutations. Am. J. Ophthalmol. 2007;143:656–662. doi: 10.1016/j.ajo.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Cruickshanks KJ, Klein R, Klein BE. Sunlight and age-related macular degeneration. The Beaver Dam Eye Study. Arch. Ophthalmol. 1993;111:514–518. doi: 10.1001/archopht.1993.01090040106042. [DOI] [PubMed] [Google Scholar]
- Culmsee C, Landshamer S. Molecular insights into mechanisms of the cell death program: role in the progression of neurodegenerative disorders. Curr. Alzheimer Res. 2006;3:269–283. doi: 10.2174/156720506778249461. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Chu CT. Mitochondrial quality control: insights on how Parkinson’s disease related genes PINK1, parkin, and Omi/HtrA2 interact to maintain mitochondrial homeostasis. J. Bioenerg. Biomembr. 2009;41:473–479. doi: 10.1007/s10863-009-9255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Y, Ternette N, Edelmann MJ, Ziv T, Gayer B, Sertchook R, et al. E3 ligases determine ubiquitination site and conjugate type by enforcing specificity on E2 enzymes. J. Biol. Chem. 2011;286:44104–44115. doi: 10.1074/jbc.M111.234559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Paz MA, Zhang J, Fridovich I. Antioxidant enzymes of the human retina: effect of age on enzyme activity of macula and periphery. Curr. Eye Res. 1996;15:273–278. doi: 10.3109/02713689609007621. [DOI] [PubMed] [Google Scholar]
- Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon J, Gaillard ER, Bilski P, Chignell CF, Reszka KJ. The photochemistry of the retinoids as studied by steady-state and pulsed methods. Photochem. Photobiol. 1996;63:680–685. doi: 10.1111/j.1751-1097.1996.tb05673.x. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev LF, Titov VN. Lipid peroxidation in relation to ageing and the role of endogenous aldehydes in diabetes and other age-related diseases. Ageing Res. Rev. 2010;9:200–210. doi: 10.1016/j.arr.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Dorey CK, Delori FC, Akeo K. Growth of cultured RPE and endothelial cells is inhibited by blue light but not green or red light. Curr. Eye Res. 1990;9:549–559. doi: 10.3109/02713689008999595. [DOI] [PubMed] [Google Scholar]
- Dudek EJ, Shang F, Valverde P, Liu Q, Hobbs M, Taylor A. Selectivity of the ubiquitin pathway for oxidatively modified proteins: relevance to protein precipitation diseases. FASEB J. 2005;19:1707–1709. doi: 10.1096/fj.05-4049fje. [DOI] [PubMed] [Google Scholar]
- Edwards RB, Szamier RB. Defective phagocytosis of isolated rod outer segments by RCS rat retinal pigment epithelium in culture. Science. 1977;197:1001–1003. doi: 10.1126/science.560718. [DOI] [PubMed] [Google Scholar]
- Esteve-Rudd J, Campello L, Herrero MT, Cuenca N, Martin-Nieto J. Expression in the mammalian retina of parkin and UCH-L1, two components of the ubiquitin-proteasome system. Brain Res. 2010;1352:70–82. doi: 10.1016/j.brainres.2010.07.019. [DOI] [PubMed] [Google Scholar]
- Fahrenholtz SR, Doleiden FH, Trozzolo AM, Lamola AA. On the quenching of singlet oxygen by alpha-tocopherol. Photochem. Photobiol. 1974;20:505–509. doi: 10.1111/j.1751-1097.1974.tb06610.x. [DOI] [PubMed] [Google Scholar]
- Fang S, Weissman AM. A field guide to ubiquitylation. Cell. Mol. Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feher J, Kovacs I, Artico M, Cavallotti C, Papale A, Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiol. Aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnemann SC, Leung LW, Rodriguez-Boulan E. The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3842–3847. doi: 10.1073/pnas.052025899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank RN, Amin RH, Puklin JE. Antioxidant enzymes in the macular retinal pigment epithelium of eyes with neovascular age-related macular degeneration. Am. J. Ophthalmol. 1999;127:694–709. doi: 10.1016/s0002-9394(99)00032-x. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide dismutases: defence against endogenous superoxide radical. Ciba Found. Symp. 1978a:77–93. doi: 10.1002/9780470715413.ch6. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Superoxide radicals, superoxide dismutases and the aerobic lifestyle. Photochem. Photobiol. 1978b;28:733–741. doi: 10.1111/j.1751-1097.1978.tb07009.x. [DOI] [PubMed] [Google Scholar]
- Friedman JS, Ray JW, Waseem N, Johnson K, Brooks MJ, Hugosson T, et al. Mutations in a BTB-Kelch protein, KLHL7, cause autosomal-dominant retinitis pigmentosa. Am. J. Hum. Genet. 2009;84:792–800. doi: 10.1016/j.ajhg.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S, Rapaport D, Klanner E, Neupert W, Westermann B. Connection of the mitochondrial outer and inner membranes by Fzo1 is critical for organellar fusion. J. Cell Biol. 2001;152:683–692. doi: 10.1083/jcb.152.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Talalay P. Induction of phase 2 genes by sulforaphane protects retinal pigment epithelial cells against photooxidative damage. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10446–10451. doi: 10.1073/pnas.0403886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Glauser L, Sonnay S, Stafa K, Moore DJ. Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 2011;118:636–645. doi: 10.1111/j.1471-4159.2011.07318.x. [DOI] [PubMed] [Google Scholar]
- Glenn JV, Mahaffy H, Dasari S, Oliver M, Chen M, Boulton ME, et al. Proteomic profiling of human retinal pigment epithelium exposed to an advanced glycation-modified substrate. Graefes Arch. Clin. Exp. Ophthalmol. 2011;250(3):349–359. doi: 10.1007/s00417-011-1856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery O, Malka F, Frachon P, Milea D, Rojo M, Lombes A. Modulation of mitochondrial morphology by bioenergetics defects in primary human fibroblasts. Neuromuscul. Disord. 2008;18:319–330. doi: 10.1016/j.nmd.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Gullapalli VK, Sugino IK, Van Patten Y, Shah S, Zarbin MA. Impaired RPE survival on aged submacular human Bruch’s membrane. Exp. Eye Res. 2005;80:235–248. doi: 10.1016/j.exer.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Ha KN, Chen Y, Cai J, Sternberg P., Jr Increased glutathione synthesis through an ARE-Nrf2-dependent pathway by zinc in the RPE: implication for protection against oxidative stress. Invest. Ophthalmol. Vis. Sci. 2006;47:2709–2715. doi: 10.1167/iovs.05-1322. [DOI] [PubMed] [Google Scholar]
- Haas AL, Rose IA. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J. Biol. Chem. 1982;257:10329–10337. [PubMed] [Google Scholar]
- Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J. Biol. Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, et al. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J. Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Hayes JD, McMahon M, Chowdhry S, Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 2010;13:1713–1748. doi: 10.1089/ars.2010.3221. [DOI] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Holz FG, Schutt F, Kopitz J, Eldred GE, Kruse FE, Volcker HE, et al. Inhibition of lysosomal degradative functions in RPE cells by a retinoid component of lipofuscin. Invest. Ophthalmol. Vis. Sci. 1999;40:737–743. [PubMed] [Google Scholar]
- Hoppins S, Edlich F, Cleland MM, Banerjee S, McCaffery JM, Youle RJ, et al. The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol. Cell. 2011;41:150–160. doi: 10.1016/j.molcel.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Tian J, Handa JT. Similarity of mRNA phenotypes of morphologically normal macular and peripheral retinal pigment epithelial cells in older human eyes. Invest. Ophthalmol. Vis .Sci. 2004;45:3291–3301. doi: 10.1167/iovs.04-0168. [DOI] [PubMed] [Google Scholar]
- Ishida K, Panjwani N, Cao Z, Streilein JW. Participation of pigment epithelium in ocular immune privilege. 3. Epithelia cultured from iris, ciliary body, and retina suppress T-cell activation by partially non-overlapping mechanisms. Ocul. Immunol. 2003;11:91–105. doi: 10.1076/ocii.11.2.91.15914. [DOI] [PubMed] [Google Scholar]
- Jahngen-Hodge J, Obin MS, Gong X, Shang F, Nowell TR, Jr, Gong J, et al. Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 1997;272:28218–28226. doi: 10.1074/jbc.272.45.28218. [DOI] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Kaarniranta K, Hyttinen J, Ryhanen T, Viiri J, Paimela T, Toropainen E, et al. Mechanisms of protein aggregation in the retinal pigment epithelial cells. Front. Biosci. (Elite Ed.) 2010;2:1374–1384. doi: 10.2741/e198. [DOI] [PubMed] [Google Scholar]
- Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalishwaralal K, BarathManiKanth S, Pandian SR, Deepak V, Gurunathan S. Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surf. B Biointerfaces. 2010;79:340–344. doi: 10.1016/j.colsurfb.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, Raveh D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol. Cell. Biol. 2005;25:5355–5362. doi: 10.1128/MCB.25.13.5355-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunadharma PP, Nordgaard CL, Olsen TW, Ferrington DA. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2010;51:5470–5479. doi: 10.1167/iovs.10-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, et al. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum. Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 2009;28:1867–1877. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen K, Petrovski G, Moe MC, Berta A, Kaarniranta K. Molecular mechanisms of retinal pigment epithelium damage and development of age-related macular degeneration. Acta Ophthalmol. 2011;90(4):299–309. doi: 10.1111/j.1755-3768.2011.02179.x. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- Koulich E, Li X, DeMartino GN. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegenburg F, Poulsen EG, Koch A, Kruger E, Hartmann-Petersen R. Redox control of the ubiquitin-proteasome system: from molecular mechanisms to functional significance. Antioxid. Redox Signal. 2011;15:2265–2299. doi: 10.1089/ars.2010.3590. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, Cohen RE. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- LaVail MM. Outer segment disc shedding and phagocytosis in the outer retina. Trans. Ophthalmol. Soc. U.K. 1983;103(Pt 4):397–404. [PubMed] [Google Scholar]
- Lee E, Maclaren RE. Sources of retinal pigment epithelium (RPE) for replacement therapy. Br. J. Ophthalmol. 2011;95:445–449. doi: 10.1136/bjo.2009.171918. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell. 2002;13:4343–4354. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- Liles MR, Newsome DA, Oliver PD. Antioxidant enzymes in the aging human retinal pigment epithelium. Arch. Ophthalmol. 1991;109:1285–1288. doi: 10.1001/archopht.1991.01080090111033. [DOI] [PubMed] [Google Scholar]
- Lin H, Xu H, Liang FQ, Liang H, Gupta P, Havey AN, et al. Mitochondrial DNA damage and repair in RPE associated with aging and age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:3521–3529. doi: 10.1167/iovs.10-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lu W, Reigada D, Nguyen J, Laties AM, Mitchell CH. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4(−/−) mice: pharmacologic approaches and functional recovery. Invest. Ophthalmol. Vis. Sci. 2008;49:772–780. doi: 10.1167/iovs.07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerus PW, Brauner MJ, Smith MB, Minnich V. Glutathione synthesis in human erythrocytes. II. Purification and properties of the enzymes of glutathione biosynthesis. J. Clin. Invest. 1971;50:1637–1643. doi: 10.1172/JCI106652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M, Bartee E, Gouveia K, Hovey Nerenberg BT, Barrett J, Thomas L, et al. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 2003;77:1427–1440. doi: 10.1128/JVI.77.2.1427-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- Matuschewski K, Hauser HP, Treier M, Jentsch S. Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J. Biol. Chem. 1996;271:2789–2794. doi: 10.1074/jbc.271.5.2789. [DOI] [PubMed] [Google Scholar]
- Mazure NM, Brahimi-Horn MC, Pouyssegur J. Hypoxic mitochondria: accomplices in resistance. Bull. Cancer. 2011;98:40–46. doi: 10.1684/bdc.2011.1360. [DOI] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Tate SS. Glutathione and related gamma-glutamyl compounds: biosynthesis and utilization. Annu. Rev. Biochem. 1976;45:559–604. doi: 10.1146/annurev.bi.45.070176.003015. [DOI] [PubMed] [Google Scholar]
- Minnich V, Smith MB, Brauner MJ, Majerus PW. Glutathione biosynthesis in human erythrocytes. I. Identification of the enzymes of glutathione synthesis in hemolysates. J. Clin. Invest. 1971;50:507–513. doi: 10.1172/JCI106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza S, Plafker KS, Aston C, Plafker SM. Expression and distribution of the class III ubiquitin-conjugating enzymes in the retina. Mol. Vis. 2010;16:2425–2437. [PMC free article] [PubMed] [Google Scholar]
- Miyamura N, Ogawa T, Boylan S, Morse LS, Handa JT, Hjelmeland LM. Topographic and age–dependent expression of heme oxygenase–1 and catalase in the human retinal pigment epithelium. Invest. Ophthalmol. Vis. Sci. 2004;45(5):1562–1565. doi: 10.1167/iovs.02-0761. [DOI] [PubMed] [Google Scholar]
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. U.S.A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Kim Y, Brodie SE, Huang X, Sheppard D, Finnemann SC. Loss of synchronized retinal phagocytosis and age-related blindness in mice lacking alphavbeta5 integrin. J. Exp. Med. 2004;200:1539–1545. doi: 10.1084/jem.20041447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Youle RJ. Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 2011;14:1929–1938. doi: 10.1089/ars.2010.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Shimizu S, Ito T, Chittenden T, Lutz RJ, Matsuda H, et al. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14681–14686. doi: 10.1073/pnas.95.25.14681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol. Cell. 2001;8:1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- Nelson KC, Carlson JL, Newman ML, Sternberg P, Jr, Jones DP, Kavanagh TJ, et al. Effect of dietary inducer dimethylfumarate on glutathione in cultured human retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 1999;40:1927–1935. [PubMed] [Google Scholar]
- Nelson KC, Armstrong JS, Moriarty S, Cai J, Wu MW, Sternberg P, Jr, et al. Protection of retinal pigment epithelial cells from oxidative damage by oltipraz, a cancer chemopreventive agent. Invest. Ophthalmol. Vis. Sci. 2002;43:3550–3554. [PubMed] [Google Scholar]
- Neutzner A, Benard G, Youle RJ, Karbowski M. Role of the ubiquitin conjugation system in the maintenance of mitochondrial homeostasis. Ann N. Y. Acad. Sci. 2008;1147:242–253. doi: 10.1196/annals.1427.012. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Noor R, Mittal S, Iqbal J. Superoxide dismutase—applications and relevance to human diseases. Med. Sci. Monit. 2002;8:RA210–RA215. [PubMed] [Google Scholar]
- Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of agerelated macular degeneration. Invest. Ophthalmol. Vis. Sci. 2008;49:2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obin MS, Jahngen-Hodge J, Nowell T, Taylor A. Ubiquitinylation and ubiquitin-dependent proteolysis in vertebrate photoreceptors (rod outer segments). Evidence for ubiquitinylation of Gt and rhodopsin. J. Biol. Chem. 1996;271:14473–14484. doi: 10.1074/jbc.271.24.14473. [DOI] [PubMed] [Google Scholar]
- Obin M, Shang F, Gong X, Handelman G, Blumberg J, Taylor A. Redox regulation of ubiquitin-conjugating enzymes: mechanistic insights using the thiolspecific oxidant diamide. FASEB J. 1998;12:561–569. doi: 10.1096/fasebj.12.7.561. [DOI] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, et al. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D, Keegan BR, Brisch E, Thatcher JW, Hermann GJ, Bleazard W, et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Wiederkehr A, Kirkpatrick C, Mattenberger Y, Martinou JC, Marchetti P, Demaurex N, Wollheim CB. Selective actions of mitochondrial fission/fusion genes on metabolism–secretion coupling in insulin–releasing cells. J. Biol. Chem. 2008;283(48):33347–33456. doi: 10.1074/jbc.M806251200. Epub 2008 Oct 2. PMID: 18832378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak A, Rozanowska M, Zareba M, Lamb LE, Simon JD, Sarna T. Action spectra for the photoconsumption of oxygen by human ocular lipofuscin and lipofuscin extracts. Arch. Biochem. Biophys. 2002;403:59–62. doi: 10.1016/S0003-9861(02)00260-6. [DOI] [PubMed] [Google Scholar]
- Pawlak A, Wrona M, Rozanowska M, Zareba M, Lamb LE, Roberts JE, et al. Comparison of the aerobic photoreactivity of A2E with its precursor retinal. Photochem. Photobiol. 2003;77:253–258. doi: 10.1562/0031-8655(2003)077<0253:cotapo>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, et al. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- Pi J, Leung L, Xue P, Wang W, Hou Y, Liu D, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3410–3415. doi: 10.1073/pnas.051618798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikans LE, Moore DR. Effect of aging on aqueous-phase antioxidants in tissues of male Fischer rats. Biochim. Biophys. Acta. 1988;966:269–275. doi: 10.1016/0304-4165(88)90076-1. [DOI] [PubMed] [Google Scholar]
- Rozanowska M, Jarvis-Evans J, Korytowski W, Boulton ME, Burke JM, Sarna T. Blue light-induced reactivity of retinal age pigment. In vitro generation of oxygenreactive species. J. Biol. Chem. 1995;270:18825–18830. doi: 10.1074/jbc.270.32.18825. [DOI] [PubMed] [Google Scholar]
- Samiec PS, Drews-Botsch C, Flagg EW, Kurtz JC, Sternberg P, Jr, Reed RL, et al. Glutathione in human plasma: decline in association with aging, age-related macular degeneration, and diabetes. Free Radic. Biol. Med. 1998;24:699–704. doi: 10.1016/s0891-5849(97)00286-4. [DOI] [PubMed] [Google Scholar]
- Sano Y, Furuta A, Setsuie R, Kikuchi H, Wang YL, Sakurai M, et al. Photoreceptor cell apoptosis in the retinal degeneration of Uchl3-deficient mice. Am. J. Pathol. 2006;169:132–141. doi: 10.2353/ajpath.2006.060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Sas K, Robotka H, Toldi J, Vecsei L. Mitochondria, metabolic disturbances, oxidative stress and the kynurenine system, with focus on neurodegenerative disorders. J. Neurol. Sci. 2007;257:221–239. doi: 10.1016/j.jns.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: the next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoma HD, Humby M, Yadav G, Zhang Q, Fogerty J, Besharse JC. The deubiquitinylating enzyme, USP2, is associated with the circadian clockwork and regulates its sensitivity to light. PLoS One. 2011;6:e25382. doi: 10.1371/journal.pone.0025382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I, Youle RJ. Mitochondrial fission and fusion. Essays Biochem. 2010;47:85–98. doi: 10.1042/bse0470085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Willett WC, Speizer FE, Hankinson SE. A prospective study of cigarette smoking and age-related macular degeneration in women. JAMA. 1996;276:1141–1146. [PubMed] [Google Scholar]
- Shang F, Nowell TR, Jr, Taylor A. Removal of oxidatively damaged proteins from lens cells by the ubiquitin-proteasome pathway. Exp. Eye Res. 2001;73:229–238. doi: 10.1006/exer.2001.1029. [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith W, Assink J, Klein R, Mitchell P, Klaver CC, Klein BE, et al. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow JR, Boulton M. RPE lipofuscin and its role in retinal pathobiology. Exp. Eye Res. 2005;80:595–606. doi: 10.1016/j.exer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Sparrow JR, Gregory-Roberts E, Yamamoto K, Blonska A, Ghosh SK, Ueda K, et al. The bisretinoids of retinal pigment epithelium. Prog. Retin. Eye Res. 2012;31:121–135. doi: 10.1016/j.preteyeres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P, Jr, Davidson PC, Jones DP, Hagen TM, Reed RL, Drews-Botsch C. Protection of retinal pigment epithelium from oxidative injury by glutathione and precursors. Invest. Ophthalmol. Vis. Sci. 1993;34:3661–3668. [PubMed] [Google Scholar]
- Stoll KE, Brzovic PS, Davis TN, Klevit RE. The essential Ubc4/Ubc5 function in yeast is HECT E3-dependent, and RING E3-dependent pathways require only monoubiquitin transfer by Ubc4. J. Biol. Chem. 2011;286:15165–15170. doi: 10.1074/jbc.M110.203968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin over-expression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino IK, Sun Q, Wang J, Nunes CF, Cheewatrakoolpong N, Rapista A, et al. Comparison of FRPE and human embryonic stem cell-derived RPE behavior on aged human Bruch’s membrane. Invest. Ophthalmol. Vis. Sci. 2011;52:4979–4997. doi: 10.1167/iovs.10-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J. Mol. Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Betsuyaku T, Ito Y, Nagai K, Nasuhara Y, Kaga K, et al. Down-regulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2008;39:673–682. doi: 10.1165/rcmb.2007-0424OC. [DOI] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Wang B, Li N, Wu Y, Jia J, Suo T, et al. RNF185, a novel mitochondrial ubiquitin E3 ligase, regulates autophagy through interaction with BNIP1. PLoS One. 2011;6:e24367. doi: 10.1371/journal.pone.0024367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanito M, Masutani H, Kim YC, Nishikawa M, Ohira A, Yodoi J. Sulforaphane induces thioredoxin through the antioxidant-responsive element and attenuates retinal light damage in mice. Invest. Ophthalmol. Vis. Sci. 2005;46:979–987. doi: 10.1167/iovs.04-1120. [DOI] [PubMed] [Google Scholar]
- Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest. 2006a;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, et al. Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem. Biophys. Res. Commun. 2006b;351:883–889. doi: 10.1016/j.bbrc.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WDrepeat protein that interacts with the dynaminrelated GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Vandewoude MF, Vandewoude MG. Vitamin E status in a normal population: the influence of age. J. Am. Coll. Nutr. 1987;6:307–311. doi: 10.1080/07315724.1987.10720192. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Anand M, Shirazi AK, Magrane J, Gao J, Vollmer-Snarr HR, et al. The age lipid A2E and mitochondrial dysfunction synergistically impair phagocytosis by retinal pigment epithelial cells. J. Biol. Chem. 2008;283:24770–24780. doi: 10.1074/jbc.M800706200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. U.S.A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Pickart CM. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 2005;24:4324–4333. doi: 10.1038/sj.emboj.7600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Goh AM, Howley PM, Walters KJ. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry. 2003;42:13529–13535. doi: 10.1021/bi035391j. [DOI] [PubMed] [Google Scholar]
- Wang AL, Lukas TJ, Yuan M, Du N, Tso MO, Neufeld AH. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Song P, Du L, Tian W, Yue W, Liu M, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J. Biol. Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yanase E, Feng X, Siegel MM, Sparrow JR. Structural characterization of bisretinoid A2E photocleavage products and implications for age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7275–7280. doi: 10.1073/pnas.0913112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhou F, Zhang Z, Xing D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011;278:941–954. doi: 10.1111/j.1742-4658.2011.08010.x. [DOI] [PubMed] [Google Scholar]
- Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct. Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 2006;8:994–1002. doi: 10.1038/ncb1460. [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R, Sugiura A, Miyachi M, Fukuda T, Matsushita N, Inatome R, et al. Mitochondrial ubiquitin ligase MITOL ubiquitinates mutant SOD1 and attenuates mutant SOD1-induced reactive oxygen species generation. Mol. Biol. Cell. 2009;20:4524–4530. doi: 10.1091/mbc.E09-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KD, Yamamoto K, Zhou J, Sparrow JR. Photo-products of retinal pigment epithelial bisretinoids react with cellular thiols. Mol. Vis. 2011a;17:1839–1849. [PMC free article] [PubMed] [Google Scholar]
- Yoon Y, Galloway CA, Jhun BS, Yu T. Mitochondrial dynamics in diabetes. Antioxid. Redox Signal. 2011b;14:439–457. doi: 10.1089/ars.2010.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Karbowski M. Mitochondrial fission in apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:657–663. doi: 10.1038/nrm1697. [DOI] [PubMed] [Google Scholar]
- Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J. Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- Yu T, Robotham JL, Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Gerencser AA, Liot G, Lipton SA, Ellisman M, Perkins GA, et al. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 2007;14:462–471. doi: 10.1038/sj.cdd.4402046. [DOI] [PubMed] [Google Scholar]
- Zerovnik E. Protein conformational pathology in Alzheimer’s and other neurodegenerative diseases; new targets for therapy. Curr. Alzheimer Res. 2010;7:74–83. doi: 10.2174/156720510790274437. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proc. Natl. Acad. Sci. U.S.A. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redoxregulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Huang J, Li HL, Liu T, Wang YY, Waterman P, et al. GIDE is a mitochondrial E3 ubiquitin ligase that induces apoptosis and slows growth. Cell Res. 2008;18:900–910. doi: 10.1038/cr.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kobayashi A, Yamamoto M, Hayes JD. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J. Biol. Chem. 2009;284:3195–3210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Hopkins JJ, Heier JS, Birch DG, Halperin LS, Albini TA, et al. Ciliary neurotrophic factor delivered by encapsulated cell intraocular implants for treatment of geographic atrophy in age-related macular degeneration. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6241–6245. doi: 10.1073/pnas.1018987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Liu T, Jin S, Wang X, Qu M, Uhlen P, et al. Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 2011a;30:2762–2778. doi: 10.1038/emboj.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Chen Y, Wang J, Sternberg P, Freeman ML, Grossniklaus HE, et al. Age-related retinopathy in NRF2-deficient mice. PLoS One. 2011b;6:e19456. doi: 10.1371/journal.pone.0019456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gao X, Cai B, Sparrow JR. Indirect antioxidant protection against photooxidative processes initiated in retinal pigment epithelial cells by a lipofuscin pigment. Rejuvenation Res. 2006;9:256–263. doi: 10.1089/rej.2006.9.256. [DOI] [PubMed] [Google Scholar]
- Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl. Acad. Sci. U.S.A. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]