Abstract
The nuclear receptor superfamily includes transcription factors that transduce steroid, thyroid and retinoid hormones and other ligands in conjunction with coregulators. To date, over 350 coregulators have been reported in the literature, and advances in proteomic analyses of coregulator protein complexes have revealed that a far greater number of coregulator-interacting proteins also exist. Coregulator dysfunction has been implicated in diverse pathological states, genetic syndromes and cancer. A hallmark of disease related to the disruption of normal coregulator function is the pleiotropic effect on animal physiology, which is frequently manifested as the dysregulation of metabolic and neurological systems. Coregulators have broad physiological and pathological functions that make them promising new drug targets for diseases such as hormone-dependent cancers. Advances in proteomics, genomics and transcriptomics have provided novel insights into the biology of coregulators at a system-wide level and will lead the way to a new understanding of how coregulators can be evaluated in the context of complex and multifaceted genetic factors, hormones, diet, the environment and stress. Ultimately, better knowledge of the associations that exist between coregulator function and human diseases is expected to expand the indications for the use of future coregulator-targeted drugs.
Introduction
The nuclear receptor superfamily is compromised of ligand-activated transcription factors that transduce steroid, thyroid and retinoid hormones and other hormonal signals into distinct physiological responses and ‘orphan’ nuclear receptors for which ligands have not been identified. In the past two decades, numerous studies have demonstrated that nuclear receptors accomplish this role in close collaboration with coregulators, which are integral to the mechanisms by which nuclear receptors elicit their physiological functions. Coregulators include both coactivators that generally associate with agonist-bound nuclear receptors to stimulate gene expression, and co repressors that are usually bound to unliganded or antagonist-bound nuclear receptors to repress gene expression.1 Given the essential role of coregulators in steroid, retinoid and thyroid hormone signalling, it seems inevitable that they would be implicated in a wide range of pathological conditions. Indeed, a growing body of evidence has accumulated that has borne out this prediction, which will be the focus of this Review.
Nuclear receptors are modular proteins that first bind their cognate ligands and then bind to specific sequences in the promoters of their target genes. Their interactions with the RNA polymerase II holocomplex and the chromatin environment that surrounds these genes depend upon, and are modified by, coregulators.2 Coregulators have broad genome-wide effects on gene expression through their capacity to interact with numerous nuclear receptors and other types of transcription factors. Our understanding of coregulators has matured from early work to characterize their mechanism of action to an under standing of their physiological functions and roles in human disease states. Now, this understanding of coregulator biology can be used translationally in the clinic through the development of coregulator-targeting agents.
Coactivators and genetic disorders
To date, over 350 coregulators have been reported in the literature, but proteomic analyses of coregulators have revealed that this number is a gross underestimation of the total number of coregulators. Over 100 genetic mouse models exist that link individual coregulators to distinct physiological functions and pathological states.3 In this Review, the discussion will be restricted to findings in the past decade that connect coregulators to human disease and physiology, focusing on instances that emphasize how coregulators can be distinguished as prominent molecular components of human disease (Table 1).
Table 1.
Coregulators involved in known and putative human genetic disease states
| Coregulator | Human disease state | Relationship between coregulator and disease |
|---|---|---|
| SRC-26 | Von Gierke syndrome | Loss of SRC-2 results in reduced G6Pase in the liver*, phenocopying the glycogen storage disorder of this syndrome‡ |
| SRC-310 | CACT deficiency | Loss of SRC-3* resembles CACT‡ deficiency with hypoketotic hypoglycaemia and muscle weakness |
| PGC-1α13,14 | Genetic predisposition to obesity | Single nucleotide polymorphisms result in increased risk for diabetes mellitus and obesity‡ |
| CBP and p30021,22 | Rubenstein–Taybi syndrome | Heterozygous disruption of CBP or p300 results in mental retardation and other defects due to reduced histone acetylation‡ |
| MTA127 | Parkinson disease | MTA1 forms a complex with DJ1 and Pitx3 to control tyrosine hydroxylase expression§ |
| TAZ28,29 | Thyroid gland dysgenesis | TAZ is required for maintenance of genes responsible for thyroid gland functionठ|
| Lipin-231,32 | Majeed syndrome | A missense mutation in lipin-2 is responsible for this syndrome and the mutation results in a loss of lipin-2 coactivator function‡ |
| MRTF-A33,34 | Cardiac myocyte stress | MRTF-A coactivator function is required for sufficient SRF-mediated gene expression in response to cardiac myocyte mechanical stretching* |
| N-CoR and SMRT39 | Resistance to thyroid hormone | Mutations in the thyroid hormone receptor can result in the retention of N-CoR and SMRT to the receptor when bound to agonist ligands‡ |
| N-CoR and SMRT40 | Gene fusions in leukaemias | N-CoR–SMRT gene fusions result in inappropriate repression of genes required to maintain leukocytes in their differentiated state‡ |
| HDAC444,45 | Brachydactyly mental retardation syndrome | Disruption of HDAC4 is responsible for bone defects observed in this syndrome‡ |
| Multiple HDACs46,47 | Chronic obstructive pulmonary disease | Reduced expression of multiple HDACs results in reduced suppression of inflammation by glucocorticoids‡ |
Mouse studies.
Human studies.
Cell-culture-based studies.
Abbreviations: CACT, carnitine–acylcarnitine translocase; G6Pase, glucose 6-phosphatase; HDAC, histone deacetylase; SRF, serum response factor.
The steroid receptor coactivator family contains three homologous members in mice, SRC-1 (also known as NCoA-1), SRC-2 (also known as NCoA-2), and SRC-3 (also known as NCoA-3). Each member of the SRC family can enhance the transcriptional activities of nuclear receptors and certain other types of transcription factors.4 Mouse genetic studies have demonstrated distinct roles for each SRC in reproduction, cancer and energy metabolism.4,5 These studies have revealed intriguing links between SRCs and human genetic disorders. For example, a striking similarity in the phenotype between Src2−/− mice and humans with Von Gierke disease has been described.6 Mutations in glucose-6-phosphatase —an essential, rate-limiting liver enzyme that serves as a terminal gatekeeper for hepatic glucose release into the plasma—result in Von Gierke syndrome. SRC-2 functions as a key regulator of glucose-6-phosphatase expression, and knockout of SRC-2 in mice results in reduced glucose-6-phosphatase expression leading to a state that resembles Von Gierke disease. Further work has established the role of SRC-2 in regulating fat absorption and whole-body energy accretion.7
In other mouse knockout studies, SRC-1 and SRC-2 have been found to have additional and distinct roles in energy metabolism. Src1−/− mice become obese due to decreased energy expenditure. Conversely, Src2−/− mice are lean due to the reduced transcriptional capacity of PPARγ2, a nuclear receptor involved in adipocyte differentiation.8 In Src2−/− mice, an increase in the interaction between PPARγ co-activator-1α (PGC-1α) and SRC-1 is observed that promotes joint SRC-1-driven and PGC-1α-driven thermogenesis in brown adipose tissue. By contrast, SRC-3 promotes white adipose cell differentiation, and adult Src3−/− mice have decreased adipose tissue mass.9
Another link between SRC family coactivators and a genetic defect in energy metabolism is the finding that in mice, SRC-3 plays a central role in the control of long-chain fatty acid metabolism by directly regulating expression of the gene that encodes carnitine–acylcarnitine translocase.10 Oxidation of lipid substrates is essential for survival in fasting and other catabolic conditions, which spares glucose for use by glucose-dependent tissues. Genetic deficiency of carnitine–acylcarnitine translocase in humans is accompanied by a constellation of metabolic problems and the build up of toxic metabolites, which leads to hypoketotic hypoglycaemia, hyperammonaemia, and impaired neurologic, cardiac and skeletal muscle performance, each of which is apparent in mice lacking SRC-3. Consistent with human cases of carnitine–acylcarnitine translocase deficiency, dietary rescue with short-chain fatty acids drastically attenuates the clinical hallmarks of the disease in mice devoid of SRC-3. Collectively, these findings position SRC-3 as a key regulator of β-oxidation in muscle. Moreover, these data allude to another possible link between what is generally considered to be a solely monogenic syndrome (caused by the loss of a metabolic enzyme) and a coregulator.
Clearly, all three SRC family coactivators subserve important and distinct metabolic functions in healthy, nor mal physiology in mice. Superficially, this fact could seem surprising given that the emphasis on SRC family coregulators has largely been restricted to their roles in cancer biology. However, as discussed below, we speculate that the participation of coregulators in both energy metabolism and cancer is linked to their roles as master regulators that integrate cellular pathways to determine whether a cell will proliferate in response to energy status cues.
Clear examples also exist for other coregulators that are involved in genetic disease states. PGC-1α is another critical coregulator responsible for the regulation of energy metabolism.11,12 Early work demonstrated that PGC-1α is expressed in muscle and brown adipose tissue in mice and that this expression is highly inducible in response to fasting and exposure to cold. In humans, a polymorphism in the gene that encodes PGC-1α and another polymorphism in the promoter of this gene were later found to be associated with an increased risk of type 2 diabetes mellitus.13,14 The related coregulator PGC-1β is also involved in metabolic functions. Lack of PGC-1β in mouse models results in reduced mitochondrial function and other defects in adipose tissue metabolism.15–17 PGC-1β drives the formation of oxidative type IIX muscle fibres, which are important for short-term, anaerobic muscle activity.18 PGC-1α dysfunction has been implicated in the dysregulation of mitochondrial biogenesis and energy metabolism and PGC-1α has, therefore, received attention as a potential drug target. For instance, drugs that are able to stimulate PGC-1α might have potential for the treatment of mitochondrial defects associated with Huntington disease in the brain,19 or in the control of bile acid homeostasis in the liver.20
Other coregulators have been linked to genetic diseases that affect the nervous system, immune response and other biological systems. For instance, Rubenstein–Taybi syndrome results from mutations in the genes that encode CBP or p300 and leads to mental retardation and characteristic morphological defects.21,22 CBP and p300 are both powerful histone acetyltransferases and patients with the Rubenstein–Taybi syndrome possess chromatin with hypoacetylated histones; therefore, histone deacetylase (HDAC) inhibitor therapies have been investigated as potential small molecule therapeutics to treat this syndrome.23 The phenotype of this syndrome is also reminiscent of the pleiotropic character seen for other genetic diseases linked to coregulator function. Although the neurological characteristics of individuals with Rubenstein–Taybi syndrome have received the most attention, these individuals have a variety of other medical issues.24
SRC-1 is also involved in brain development. In the adult cerebellum, Purkinje cells express SRC-1 and a time-course analysis of Purkinje cell development during embryogenesis in mutant mice lacking SRC-1 revealed a delay in the development of these cells.25 Ultimately, this loss of SRC-1 led to moderate motor dysfunction in adult mice. Other studies have implicated SRC-1 and SRC-3 in diverse aspects of brain function consistent with the role that these coregulators have in modulating the actions of sex steroids in distinct regions of the brain.26
Another potential link between a coregulator and neurological function was identified for metastasis-associated protein 1 (MTA1), a coregulator overexpressed in breast and other cancers. The role of this coregulator in mice was in the regulation of dopamine production in the brain.27 MTA1 was found to co-activate expression of tyrosine hydroxylase responsible for dopamine synthesis in neuronal cells, and Mta1−/− mice had reduced levels of tyrosine hydroxylase expression in the striatum and substantial nigra of the brain. MTA1 was found to drive tyrosine hydroxylase expression in conjunction with DJ1 (also known as Parkinson disease 7) and Pitx3 at the bicoid binding element in the tyrosine hydroxylase promoter. Because Pitx3 and DJ1 expression defects have already been linked to Parkinson disease, it is likely that MTA1 contributes to polygenic control of tyrosine hydroxylase expression.
In thyroid development, the coactivator TAZ (also known as WWTR1) has been shown to be a coregulator of Pax8 and other genes necessary for thyroid differentiation in mice, which suggests a role for this coregulator in thyroid dysgenesis.28 The Pax8 and TTF-1 transcription factors are necessary for thyroid gland development and TAZ was identified as a coregulator for these transcription factors. Consistent with its role in thyroid gland function, TAZ overexpression has been linked to thyroid carcinomas in humans.29 In a separate cell-culture-based study that links a coregulator to thyroid gland function, thyroid-receptor-mediated gene expression was shown to depend on the CARM1 and SNF5 coregulators.30
Defects in coregulator biology have been linked to Majeed syndrome through a missense mutation in the lipin-2 protein, manifesting as inflammation, osteomyelitis, fever and anaemia in humans.31 In a previous study, lipin-1 was found to function as a coregulator for PPARγ.32 Further examination revealed that the mutation in lipin-2 in patients with Majeed syndrome is linked to the phosphatidate phosphatase enzymatic function of the protein, which is required for it to function as a coactivator.
Links between coregulators and cardiac disease have also been reported. Myocardin-related transcription factor A (MRTF-A) has been characterized as a potent coregulator that promotes gene expression driven by serum response factor (SRF) in mice.33 Mechanical stretching of cardiomyocytes induces nuclear accumulation of MRTF-A, leading to enhanced SRF-mediated gene expression. In mouse knockout studies, expression of the gene that encodes brain natriuretic peptide (BNP) and other SRF-dependent fetal cardiac genes in response to acute mechanical stress was blunted in mice lacking MRTF-A.34 In relation to cardiac disease, mutation of an SRF-binding site within the promoter of the gene that encodes BNP, or knockdown of MRTF-A, reduced the responsiveness of this promoter to mechanical stretching. Overall, these findings revealed a unique mechanism whereby mechanical stress-regulated nuclear import of a coregulator controls cardiac myocyte gene expression.
Corepressors and genetic disease
SMRT (also known as N-CoR2)35 and N-CoR (also known as N-CoR1)36 were the first corepressors to be identified, and numerous cell culture and animal model studies have demonstrated their biological roles.37 Given the wide range of physiological roles of corepressors, similar to coactivator coregulators, corepressors have been associated with genetic disease and are gaining increased consideration as drug targets.38 The link of these corepressors to human disease was first realized in cases of genetic resistance to thyroid hormones. Individuals with resistance to thyroid hormone often possess point mutations in their thyroid hormone receptors that result in a failure of the receptor to release N-CoR or SMRT upon binding to thyroid hormone, which leads to a spectrum of medical problems.39
In addition to their role in resistance to thyroid hormone, N-CoR and SMRT have been linked to acute pro-myelocytic leukaemia and acute myeloid leukaemia in humans. Genetic translocations that result in the expression of corepressor proteins fused to other proteins that are not normally regulated by N-CoR or SMRT, lead to inappropriate repression of genes required to stop cell growth. In many of these leukaemias, pharmacological inhibition of HDACs associated with N-CoR and SMRT in corepressor protein complexes can be an effective therapy.40 BCL-6 corepressor (BCoR) has been linked to oculofaciocardiodental and Lenz microphthalmia syndromes.41 BCoR is a corepressor of retinoid acid signalling and fusions between this corepressor and the retinoic acid receptor-α have been found in acute pro-myelocytic leukaemias.42 Another corepressor linked to human disease is RIP140 (also known as NRIP1), which subserves a wide range of roles in the regulation of energy metabolism in liver, muscle and adipose tissue.43
Brachydactyly mental retardation syndrome is associated with a deletion at the chromosome 2q37 locus, leading to disabilities, developmental delays, behavioural abnormalities, sleep disturbance, craniofacial and skeletal abnormalities and autism. Finer analysis of this deletion region revealed that HDAC4 mutation is responsible for the phenotype.44 Consistent with the human phenotype, Hdac4−/− mice have bone malformations resulting from premature ossification of developing bones.45
Following observations that HDACs are important for the repression of proinflammatory cytokines in alveolar macrophages, a link was found between this phenomenon and chronic obstructive pulmonary disease (COPD).46 Aggregate HDAC activity in alveolar macrophages from patients with COPD has been found to be inversely correlated with disease severity. In healthy macrophages, glucocorticoid-bound glucocorticoid receptors are able to direct HDACs to the promoters of proinflammatory cytokines to reduce airway swelling, but in patients with COPD, the loss of HDAC activity blunts the anti-inflammatory actions of glucocorticoids. Interestingly, by combining glucocorticoids with the HDAC activator theophylline, restoring the anti-inflammatory effects of glucocorticoids is possible in patients with COPD.47 A similar mechanism has been reported to be responsible for glucocorticoid resistance in patients with asthma and in other inflammatory diseases in the lung.48,49
Coregulators and cancer
By modulating gene expression regulated by hormones, growth factors and cytokines, coregulators can promote pathological processes associated with cancer, including cell proliferation, differentiation, carcinogenesis and metastasis.4 The SRC family of coregulators has been prominently implicated in many cancer types and, therefore, deserve strong consideration as key targets for future generations of anticancer drugs. This recommendation is attested to by the finding that SRC-3 expression is consider ably upregulated in breast cancers and correlates with HER2 positivity, disease recurrence in HER2-positive breast cancers and resistance to endocrine therapy.50,51 SRC-1 is also required for breast cancer metastasis in a mouse model system.52
SRC-3 overexpression has been implicated in a variety of cancers and, in the past 2 years, it has been shown to be highly associated with rapid progression of lung cancers.53,54 Recurrent oncogenic themes in lung cancers have been used to identify several potential therapeutic targets, including epidermal growth factor receptor (EGFR), K-ras, PIK3CA, BRAF and p53.55–59 Although new drug targets for these proteins have been developed (for example, the EGFR inhibitors gefitinib and erlotinib),60,61 their monotherapeutic clinical efficacy has been disappointing. Indeed, a common theme of clinical studies has been the limited capacity of a single therapeutic strategy to block cancer cell growth.20 Possibly, new approaches that combine existing targeted therapy with coregulator-targeting drugs might be used to more effectively treat lung cancers.
Coactivators and resistance to chemotherapy
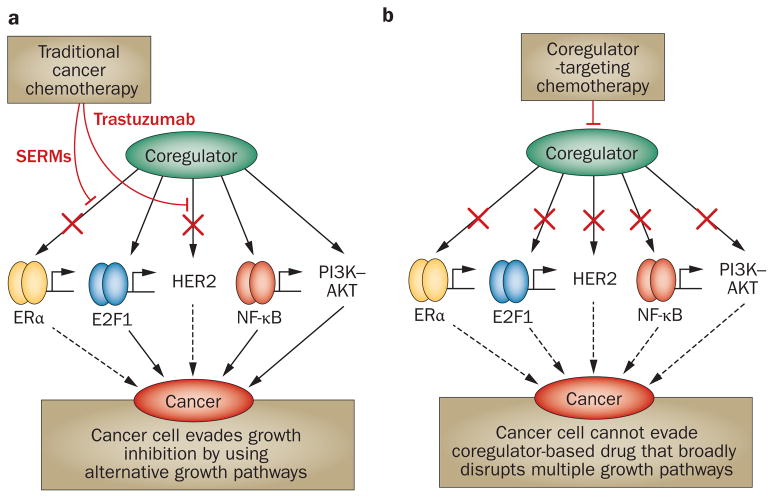
Most cancers are highly adaptable and are frequently able to evade the growth-inhibiting action of individual anti-cancer agents. For instance, growth-promoting pathways, such as those involving HER2, PI3K–AKT or NF-κB, are frequently upregulated in breast cancers in response to treatment with anti-oestrogens. With so many growth-promoting mechanisms available, the cancer cell can evade single chemotherapeutic agents that target discrete growth factor pathways. However, SRC-3 is a central steroid hormone and growth factor signalling integrator;1 therefore, the response of cancer cells to small molecule inhibitors (SMIs) that perturb its coactivator function promises to be different. SRC-3 receives growth signalling information by kinases in the PI3K–AKT,62 NF-κB,63 PKCι, PKCζ64 and other growth factor signalling systems. Phosphorylation of SRC-3 by these kinases enables SRC-3 to function as a coregulator for many transcription factors such as ERα, NF-κB and E2F1.65 Because of the central position of SRC-3 at the hub of multiple growth factor signalling pathways, SMIs that interfere with its coactivator function should simultaneously interfere with the activity of alternative growth signalling pathways that lead to cancer chemotherapy resistance (Figure 1).
Figure 1.
Coregulator-targeted drugs are predicted to overcome cancer cell resistance to chemotherapy. a | Chemotherapeutic agents designed to target ERα, such as selective oestrogen receptor modulators, and HER2 inhibitors such as trastuzumab cannot block coregulator stimulation of other growth promoting pathways driven through E2F1, NF-κB or PI3K–AKT in cancer cells that overexpress coregulators. b | By contrast, a coregulator-targeting drug is predicted to inhibit multiple growth pathways, depriving cancer cells of their capacity to access alternative growth pathways and develop resistance to chemotherapy. Abbreviation: SERMs, selective oestrogen receptor modulators.
Coregulators as cancer drug targets
Although many proteins including nuclear receptors are considered ‘druggable’ targets due to the presence of a high-affinity, high-specificity, ligand-binding site for small lipophilic ligands, coregulators are considered to be hard molecules to target.66 Certain other targeted cancer therapeutic SMIs are typically designed to target the enzyme substrate binding site of kinases.67 Nuclear receptor ligands such as tamoxifen and the EGFR tyrosine kinase inhibitor gefitinib represent examples of these types of SMIs. By contrast, many important proteins involved in cancer cell growth have traditionally been thought to be beyond the reach of SMIs. In the past decade, however, examples of SMIs for such hard targets, which include drugs that are able to target Bcl2, p53, TNF-α, β-catenin, Rac and HIV gp120, have become available.68–72
Thus, although coregulators such as SRC-3 lack a high-affinity ligand-binding pocket or a defined enzyme catalytic surface, given the importance of SRC-3 as a key oncogene, a strong impetus to develop SRC-3 SMIs exists that cannot be ignored. Indeed, high-throughput screens in laboratories have already identified SMIs that are able to interfere with the binding of nuclear receptors to coregulators; for example, binding of SRC family members to ERα, ERβ and PPARγ.73–75 Importantly, we demonstrated that a SMI can directly target SRC-3 or SRC-1 independent of its association with nuclear receptors, leading to coactivator protein degradation.76 In a proof-of-principle study, gossypol was characterized as an SRC-3/SRC-1 SMI that binds directly to the coregulator receptor interacting domain and can block cancer cell growth.
SMIs directed against other coregulators might also have strong therapeutic value. For instance, SMIs that can modulate the activities of metabolic coregulators such as PGC-1α and RIP140 would be expected to have value in the treatment of the metabolic syndrome and type 2 diabetes mellitus. Considering the phenotypes of mouse knockouts of PGC-1α, RIP140, SRC-1 and SRC-3,12,43,77,78 it is predicted that SMIs against these coregulators would be well tolerated.
Coregulator ‘omics’
High-throughput mass spectrometric analyses of coregulator complexes in the past few years have led to the realization that coregulators exist in protein complexes, broadly falling under the category of tight binding coregulator proteins and a large number of loosely interacting protein partners.79 Importantly, if the loosely interacting partners are counted, approximately half of the proteins encoded by the genome can be considered to be transcriptional coregulators. This study also revealed that cancer gene products group together in select protein complexes, which supports the idea that the perturbation of a protein complex as a whole can underlie distinct disease states and that coregulator complexes should be considered as a whole when evaluating the probable response to a given targeted therapeutic agent. This model might also apply to the aetiology of polygenic metabolic or central nervous system diseases, in which mutations in multiple genes may be responsible. If mutations accumulate within two or more proteins that exist in a single coregulator complex, in aggregate, these genetic lesion might lead to serious malfunction of the coregulator complex, resulting in disease.
Ongoing technological advances in mass spectrometry, DNA sequencing and mRNA expression analyses are expected to lead to proteomic, genomic and transcriptomic assessments of coregulators that promise to revolutionize how we understand their biology. For instance, SRC coregulators are known to possess an extensive post-translational modification code that forms an essential part of how SRCs can function as ‘master genes’ to control broad transcriptional programs responsible for cell growth, differentiation and metabolic functions.1,80 Proteomic technologies are expected to open up our ability to understand SRC coregulator function at this level, something that cannot be interrogated through high-throughput sequencing or transcriptomics, which cannot interrogate the state of post-translational modifications on proteins.
Nevertheless, genome-wide association studies have identified a variety of coregulator single nucleotide polymorphisms (SNPs) associated with certain disease risks or human traits.81 SNP risk alleles have linked SRC-1 to type 1 diabetes mellitus.82 A SRC-3 SNP has been identified that predicts response to chemotherapy for lymphoblastic lymphomas and another SRC-3 SNP has been linked to breast cancer risk.83,84 Our speculation is that coregulator dysfunction will not be restricted solely to rare genetic conditions such as highly penetrant monogenic genetic diseases. By contrast, many coregulators, including all three SRC coregulators, are not essential for viability and even their complete knockout is not lethal. Probably, their nonessential nature is related to the roles that coregulators such as SRC-3 have as integrators of diverse signals from the environment.1 Coregulators must be genetically flexible enough to function as integrators of diverse metabolic and environmental stimuli; therefore, the genes that encode them cannot be essential for viability. This coregulator genetic variation and their increased freedom to evolve are possibly linked to the vastly diverse environments and geography that different human population groups are able to inhabit. Indeed, computational searches for genes undergoing strong selective pressure have identified SRC-1 as such a gene in an African population.85 Coregulators are, thus, free to function at the vanguard of adaptive genetic changes necessary for humans to exist in diverse and rapidly changing geographic conditions.
Conclusions
Coregulators represent a large and growing functional class of proteins. Although they are legion in number, a full appreciation of the size of this class of proteins and their pervasive involvement in normal and disease physiology is only beginning to be realized. As discussed above, coregulators are key regulators of reproduction, energy metabolism and cancer and also have roles in human genetic disease and cancer. The genetic basis of coregulator biology will expand further once the effect of environmental stress on human physiology is considered. Indeed, in many instances, mouse genetic models of coregulator dysfunction only display phenotypes when the animals are subject to stress such as a high-fat diet, immune challenge or exposure to mutagens or other toxins. The genetic diversity of coregulators may explain distinct differences between individuals in response to specific environmental and dietary cues. Even aging can be seen as a type of stress and it will be important to evaluate coregulators in this context. Future studies will expand our knowledge of the role of coregulators in pathology and will provide important information on how best to apply emerging coregulator-targeting drugs to treat a wide range of diseases.
Key points.
Coregulators interact with nuclear receptors and other transcription factors to alter chromatin and stimulate (coactivators) or repress (corepressors) gene expression
Over 350 coregulators have been identified in the literature, but proteomic studies indicate that this number is a gross underestimate
Many coregulators have been implicated in the physiology of reproduction, energy metabolism, inherited human genetic diseases and cancer
Coregulators are receiving increasing attention as important drug targets for diseases including cancer, inflammatory disorders and genetic syndromes related to their dysfunction
Proteomic, genomic and transcriptomic characterization of coregulators will enable their physiological and pathological roles to be better realized in the context of diverse endocrine, environmental, dietary and stress conditions
Review criteria.
A PubMed search was performed using the keywords “corepressor”, “coactivator”, “co-repressor”, “co-activator”, “coregulator” and “co-regulator”. Selected original research papers and reviews are discussed in this Review. Articles that were published within the past 17 years were selected, with emphasis given to articles the authors felt were of high originality and impact in the field involving coregulator research primarily linked to the nuclear receptor research field. The authors focused on articles that describe the relationship between coregulators and human disease.
Footnotes
Author contributions
Both authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
References
- 1.Lonard DM, Kumar R, O’Malley BW. Minireview: the SRC family of coactivators: an entrée to understanding a subset of polygenic diseases? Mol Endocrinol. 2010;24:279–285. doi: 10.1210/me.2009-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 3.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 4.Lonard DM, O’Malley BW. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 6.Chopra AR, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra AR, et al. Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell Metab. 2011;13:35–43. doi: 10.1016/j.cmet.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picard F, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 9.Louet JF, et al. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci USA. 2006;103:17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.York B, et al. Ablation of steroid receptor coactivator-3 resembles the human CACT metabolic myopathy. Cell Metab. 2012;15:752–763. doi: 10.1016/j.cmet.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z, Boss O. Targeting PGC-1α to control energy homeostasis. Expert Opin Ther Targets. 2007;11:1329–1338. doi: 10.1517/14728222.11.10.1329. [DOI] [PubMed] [Google Scholar]
- 12.Handschin C. The biology of PGC-1α and its therapeutic potential. Trends Pharmacol Sci. 2009;30:322–329. doi: 10.1016/j.tips.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Andersen G, et al. PGC-1α Gly482Ser polymorphism associates with hypertension among Danish whites. Hypertension. 2005;45:565–570. doi: 10.1161/01.HYP.0000158946.53289.24. [DOI] [PubMed] [Google Scholar]
- 14.Ek J, et al. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 15.Sonoda J, Mehl IR, Chong LW, Nofsinger RR, Evans RM. PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc Natl Acad Sci USA. 2007;104:5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lelliott CJ, et al. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol. 2006;4:e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uldry M, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 19.Quintanilla RA, Jin YN, Fuenzalida K, Bronfman M, Johnson GV. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin-expressing cells: possible role of peroxisome proliferator-activated receptor-gamma (PPARgamma) in the pathogenesis of Huntington disease. J Biol Chem. 2008;283:25628–25637. doi: 10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eloranta JJ, Kullak-Ublick GA. Coordinate transcriptional regulation of bile acid homeostasis and drug metabolism. Arch Biochem Biophys. 2005;433:397–412. doi: 10.1016/j.abb.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Petrij F, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 22.Roelfsema JH, et al. Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am J Hum Genet. 2005;76:572–580. doi: 10.1086/429130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 24.Roelfsema JH, Peters DJ. Rubinstein–Taybi syndrome: clinical and molecular overview. Expert Rev Mol Med. 2007;9:1–16. doi: 10.1017/S1462399407000415. [DOI] [PubMed] [Google Scholar]
- 25.Nishihara E, et al. SRC-1 null mice exhibit moderate motor dysfunction and delayed development of cerebellar Purkinje cells. J Neurosci. 2003;23:213–222. doi: 10.1523/JNEUROSCI.23-01-00213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlier TD. Importance of steroid receptor coactivators in the modulation of steroid action on brain and behavior. Psychoneuroendocrinology. 2009;34 (Suppl 1):S20–S29. doi: 10.1016/j.psyneuen.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Reddy SD, et al. Multiple coregulatory control of tyrosine hydroxylase gene transcription. Proc Natl Acad Sci USA. 2011;108:4200–4205. doi: 10.1073/pnas.1101193108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Palma T, et al. TAZ is a coactivator for Pax8 and TTF-1, two transcription factors involved in thyroid differentiation. Exp Cell Res. 2009;315:162–175. doi: 10.1016/j.yexcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 29.de Cristofaro T, et al. TAZ/WWTR1 is overexpressed in papillary thyroid carcinoma. Eur J Cancer. 2011;47:926–933. doi: 10.1016/j.ejca.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Choi HK, et al. The functional role of the CARM1-SNF5 complex and its associated HMT activity in transcriptional activation by thyroid hormone receptor. Exp Mol Med. 2007;39:544–555. doi: 10.1038/emm.2007.60. [DOI] [PubMed] [Google Scholar]
- 31.Donkor J, et al. A conserved serine residue is required for the phosphatidate phosphatase activity but not the transcriptional coactivator functions of lipin-1 and lipin-2. J Biol Chem. 2009;284:29968–29978. doi: 10.1074/jbc.M109.023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finck BN, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Cen B, et al. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuwahara K, et al. Myocardin-related transcription factor A is a common mediator of mechanical stress- and neurohumoral stimulation-induced cardiac hypertrophic signaling leading to activation of brain natriuretic peptide gene expression. Mol Cell Biol. 2010;30:4134–4148. doi: 10.1128/MCB.00154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 36.Hörlein AJ, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 37.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of corepressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 38.Hsia EY, Goodson ML, Zou JX, Privalsky ML, Chen HW. Nuclear receptor coregulators as a new paradigm for therapeutic targeting. Adv Drug Deliv Rev. 2010;62:1227–1237. doi: 10.1016/j.addr.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoh SM, Chatterjee VK, Privalsky ML. Thyroid hormone resistance syndrome manifests as an aberrant interaction between mutant T3 receptors and transcriptional corepressors. Mol Endocrinol. 1997;11:470–480. doi: 10.1210/mend.11.4.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quintás-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011;25:226–235. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- 41.Ng D, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–416. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto Y, et al. BCOR as a novel fusion partner of retinoic acid receptor α in a t(X;17) (p11;q12) variant of acute promyelocytic leukemia. Blood. 2010;116:4274–4283. doi: 10.1182/blood-2010-01-264432. [DOI] [PubMed] [Google Scholar]
- 43.White R, et al. Role of RIP140 in metabolic tissues: connections to disease. FEBS Lett. 2008;582:39–45. doi: 10.1016/j.febslet.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Williams SR, et al. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 46.Ito K, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. N Engl J Med. 2005;352:1967–1976. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 47.Ito K, et al. A molecular mechanism of action of theophylline: induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci USA. 2002;99:8921–8926. doi: 10.1073/pnas.132556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hew M, et al. Relative corticosteroid insensitivity of peripheral blood mononuclear cells in severe asthma. Am J Respir Crit Care Med. 2006;174:134–141. doi: 10.1164/rccm.200512-1930OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adcock IM, Ito K, Barnes PJ. Histone deacetylation: an important mechanism in inflammatory lung diseases. COPD. 2005;2:445–455. doi: 10.1080/15412550500346683. [DOI] [PubMed] [Google Scholar]
- 50.Wang S, et al. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci USA. 2009;106:151–156. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flågeng MH, et al. Nuclear receptor co-activators and HER-2/neu are upregulated in breast cancer patients during neo-adjuvant treatment with aromatase inhibitors. Br J Cancer. 2009;101:1253–1260. doi: 10.1038/sj.bjc.6605324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fluck MM, Schaffhausen BS. Lessons in signalling and tumourigenesis from polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2009;73:542–563. doi: 10.1128/MMBR.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, et al. Overexpression and gender-specific differences of SRC-3 (SRC-3/AIB1) immunoreactivity in human non-small cell lung cancer: an in vivo study. J Histochem Cytochem. 2010;58:1121–1127. doi: 10.1369/jhc.2010.956979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cai D, et al. Steroid receptor coactivator-3 expression in lung cancer and its role in the regulation of cancer cell survival and proliferation. Cancer Res. 2010;70:6477–6485. doi: 10.1158/0008-5472.CAN-10-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.John T, Liu G, Tsao MS. Overview of molecular testing in non-small-cell lung cancer: mutational analysis, gene copy number, protein expression and other biomarkers of EGFR for the prediction of response to tyrosine kinase inhibitors. Oncogene. 2009;28 (Suppl 1):S14–S23. doi: 10.1038/onc.2009.197. [DOI] [PubMed] [Google Scholar]
- 56.Adjei AA. K-ras as a target for lung cancer therapy. J Thorac Oncol. 2008;3 (Suppl 2):S160–S163. doi: 10.1097/JTO.0b013e318174dbf9. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto H, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmid K, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 59.Ye Y, Wang D, Su C, Rong T, Guo A. Combined detection of p53, p16, Rb, and EGFR mutations in lung cancer by suspension microarray. Genet Mol Res. 2009;8:1509–1518. doi: 10.4238/vol8-4gmr627. [DOI] [PubMed] [Google Scholar]
- 60.Sanford M, Scott LJ. Gefitinib: a review of its use in the treatment of locally advanced/ metastatic non-small-cell lung cancer. Drugs. 2009;69:2303–2328. doi: 10.2165/10489100-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 61.Carlson JJ. Erlotinib in non-small-cell lung cancer: a review of the clinical and economic evidence. Expert Rev Pharmacoecon Outcomes Res. 2009;9:409–416. doi: 10.1586/erp.09.49. [DOI] [PubMed] [Google Scholar]
- 62.Zhou HJ, et al. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 63.Wu RC, et al. Regulation of SRC-3 (pCIP/ACTR/ AIB-1/RAC-3/TRAM-1) Coactivator activity by IκB kinase. Mol Cell Biol. 2002;22:3549–3561. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yi P, et al. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell. 2008;29:465–476. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen T. Nuclear receptor drug discovery. Curr Opin Chem Biol. 2008;12:418–426. doi: 10.1016/j.cbpa.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 67.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein– protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 68.Goldsmith KC, Hogarty MD. Small-molecule BH3 mimetics to antagonize Bcl-2-homolog survival functions in cancer. Curr Opin Investig Drugs. 2009;10:559–571. [PubMed] [Google Scholar]
- 69.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Esposito E, Cuzzocrea S. TNF-α as a therapeutic target in inflammatory diseases, ischemia-reperfusion injury and trauma. Curr Med Chem. 2009;16:3152–3167. doi: 10.2174/092986709788803024. [DOI] [PubMed] [Google Scholar]
- 71.Marchioni F, Zheng Y. Targeting rho GTPases by peptidic structures. Curr Pharm Des. 2009;15:2481–2487. doi: 10.2174/138161209788682334. [DOI] [PubMed] [Google Scholar]
- 72.Kadow J, Wang HG, Lin PF. Small-molecule HIV-1 gp120 inhibitors to prevent HIV-1 entry: an emerging opportunity for drug development. Curr Opin Investig Drugs. 2006;7:721–726. [PubMed] [Google Scholar]
- 73.Rodriguez AL, Tamrazi A, Collins ML, Katzenellenbogen JA. Design, synthesis, and in vitro biological evaluation of small-molecule inhibitors of estrogen receptor α coactivator binding. J Med Chem. 2004;47:600–611. doi: 10.1021/jm030404c. [DOI] [PubMed] [Google Scholar]
- 74.Williams AB, Weiser PT, Hanson RN, Gunther JR, Katzenellenbogen JA. Synthesis of biphenyl proteomimetics as estrogen receptor α coactivator binding inhibitors. Org Lett. 2009;11:5370–5373. doi: 10.1021/ol901999f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mettu NB, et al. The nuclear receptor-coactivator interaction surface as a target for peptide antagonists of the peroxisome proliferator-activated receptors. Mol Endocrinol. 2007;21:2361–2377. doi: 10.1210/me.2007-0201. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, et al. Small-molecule inhibition of the steroid receptor coactivators, SRC-3 and SRC-1. Mol Endocrinol. 2011;25:2041–2053. doi: 10.1210/me.2011-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu J, et al. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu J, et al. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 79.Malovannaya A, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han SJ, Lonard DM, O’Malley BW. Multi-modulation of nuclear receptor coactivators through post-translational modifications. Trends Endocrinol Metab. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 82.Cooper JD, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang JJ, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hartmaier RJ, et al. Nuclear receptor coregulator SNP discovery and impact on breast cancer risk. BMC Cancer. 2009;9:438. doi: 10.1186/1471-2407-9-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]