Abstract
The Cbfa1/Runx2 transcription factor is essential for osteoblast differentiation. However, levels of Runx2 are often not well correlated with its transcriptional activity suggesting that this factor must be activated either by covalent modification or through interactions with other nuclear components. Runx2 is phosphorylated and activated by the mitogen-activated protein kinase (MAPK) pathway. This pathway is stimulated in at least two ways: by binding of type I collagen to β2β1 integrins on the osteoblast surface and by treatment of cells with the osteogenic growth factor, FGF2. Protein kinase A (PKA) also may phosphorylate/activate Runx2 under certain conditions. Runx2 activity also is enhanced by factors known to stimulate specific signal transduction pathways such as PTH/PTHrP (signals through PKA and PKC pathways) and BMPs (Signal through Smad proteins). Interactions with Runx2 are complex involving both binding of distinct components such as AP-1 factors and Smads to separate sites on DNA, direct interactions between Runx2 and AP-1/Smad factors and MAPK or PKA-dependent Runx2 phosphorylation. These findings suggest that Runx2 plays a central role in coordinating multiple signals involved in osteoblast differentiation.
Keywords: Bone, Osteoblast, Phosphorylation, Transcription
INTRODUCTION
Cbfa1/Runx2 is a bone-related transcription factor homologous to the Drosophila protein, Runt [1]. This protein is essential for the differentiation of osteoblasts from mesenchymal precursors and subsequent bone matrix mineralization in that Cbfa1−/− mice show a complete lack of functional osteoblasts [2]. Mutations in the Cbfa1 locus in humans cause cleidocranial dysplasia, an autosomal dominant disease characterized by the absence of clavicles, open fontanelles, supernumerary teeth, and short stature [3]. Runx2 can directly stimulate transcription of osteoblast-related genes such as those encoding osteocalcin (OCN), type I collagen, and collagenase 3 by binding to specific enhancer regions containing the core sequence, PuCCPuCA [4–6]. Beyond this, the molecular mechanism of Runx2 action is unknown.
Although Runx2 protein expression is restricted to mineralized tissues and their precursors, in many cases there is not a good correlation between actual Runx2 protein levels and the expression of osteoblast-related genes. Thus during development, Runx2 expression precedes osteoblast differentiation and osteocalcin expression by several days [4]. Similarly, presence of Runx2 protein in several osteoblast cell culture systems is not well correlated with expression of Runx2 targets. Both primary osteoblasts and the MC3T3-E1 osteoblast cell line do not exhibit major changes in levels of Runx2 protein during in vitro differentiation, even though expression levels of osteoblast marker genes like OCN, bone sialoprotein, and alkaline phosphatase are all dramatically increased [7]. Likewise, Runx2 is dramatically upregulated by both TGF-β and bone morphogenetic proteins (BMP)2 in C2C12 mesenchymal cells, but only BMP treatment can induce osteoblast-specific gene expression [8]. Taken together, these studies indicate that Runx2-dependent transcription is not simply regulated by levels of the Runx2 protein. Rather, they imply that this transcription factor is regulated either by post-translational mechanisms involving protein modification or by interactions with additional nuclear factors.
This paper presents four representative examples of Runx2 regulation using studies from the authors’ laboratory and the current literature. Specifically, we discuss Runx2 activation by binding of collagen to cell surface integrins, FGF2, PTH, and BMPs. These examples, while not covering all potential types of Runx2 regulation, will give the reader an appreciation of the breadth of signaling pathways that can control this important transcription factor.
ECM-DEPENDENT ACTIVATION
Differentiation of osteoblasts requires secretion of a type I collagen-containing extracellular matrix (ECM). Matrix synthesis is necessary for induction of osteoblast-related genes such as those encoding OCN, bone sialoprotein, alkaline phosphatase, and the parathyroid hormone/parathyroid hormone-related protein receptor and, ultimately, mineralization [9]. Recently, several groups showed that the ECM signals to the differentiating preosteoblast by binding to β1 subunit-containing integrins (β2β1 and, possibly, β1β1) [7, 10, 11]. Disruption of integrin signaling using either blocking antibodies or peptides that mimic the cell binding domain of collagen completely blocks ECM-dependent differentiation. This observation is highly significant for understanding osteoblast metabolism because it is through integrins that cells sense their ECM environment and respond to changes in mechanical loading of tissues [12].
We showed that Runx2 at least in part mediates the response of preosteoblasts to matrix signals. Specifically, ECM production by murine MC3T3-E1 osteoblast-like cells dramatically increases transcription of the OCN gene. This matrix response requires Runx2 and its cognate DNA-binding site in the OCN promoter, osteoblast-specific element 2 (OSE2) [13]. Interestingly, this Runx2-dependent increase in transcriptional activity is not accompanied by a significant change in Runx2 mRNA or protein although large increases are seen in the in vitro binding of Runx2 to OSE2 DNA as measured by gel retardation assays [7].
As one of the primary signal transduction pathways activated by integrins, the MEK/ERK branch of the mitogen-activated protein kinase (MAPK) pathway provides a plausible link between cell surface integrin activation and subsequent stimulation of Runx2-dependent transcription. We recently showed that the potent MAPK pathway inhibitor, U0126 (inhibits phosphorylation of ERK1/2 by MEK), rapidly and specifically inhibits both ERK phosphorylation and ECM-dependent induction of the OCN gene. Similarly, BMP actions in osteoblasts also require matrix signals and can be blocked by U0126 [14].
In separate studies, we showed that overexpression of a constitutively active MEK1 mutant induces both endogenous OCN mRNA and promoter activity and, further, that this response requires Runx2 and an intact OSE2 sequence. Finally, Runx2 phosphorylation was shown to be increased after transfection of cells with constitutively active MEK1, the kinase immediately before ERK1/2 in the MAPK cascade [15].
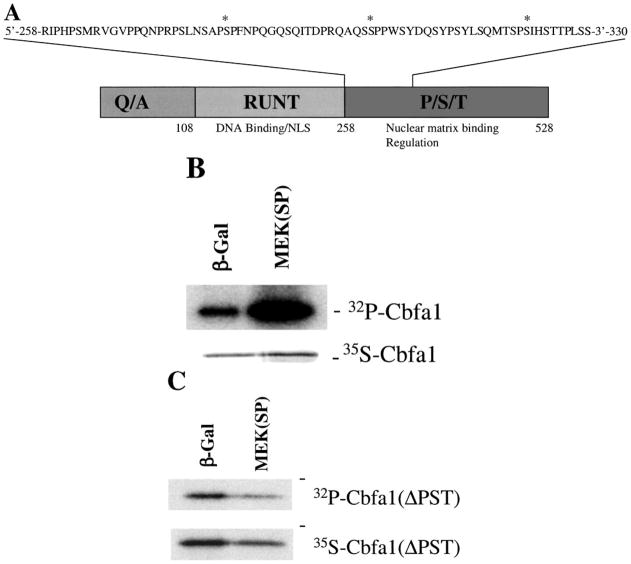
More recently, we focused our efforts on identifying regions of Runx2 that are necessary for MAPK regulation and phosphorylation. Figure 1A shows the overall domain structure of the Runx2 molecule that includes a glutamine/alanine-rich N-terminal region (Q/A), a central portion with the highest homology to the Drosophila Runt protein and containing the DNA-binding domain and nuclear localization sequences and the C-terminal proline/serine/threonine-rich (P/S/T) region [16]. Previous studies localized MAPK-responsive sequences to the P/S/T domain [15]. As shown in Figure 1B, this same region is also necessary for MEK-dependent increases in Runx2 phosphorylation since a truncated Runx2 molecule lacking this entire region did not show any MAPK-dependent changes in phosphorylation. Additional C-terminal deletions (not shown) further localized a minimal MAPK-responsive sequence between residues 286 and 330. This region contains three consensus MAPK phosphorylation sites, although we do not yet know if any of these can be directly phosphorylated by MAPK.
Figure 1.
Activation of the MAPK pathway stimulates phosphorylation within the P/S/T domain of Runx2. (A) Runx2 domains. Diagram shows the three major domains of Runx2: the glutamine/alanine-rich region (Q/A), the Runt domain that contains DNA binding and nuclear localization sequences (NLS), and the proline/serine/threonine (P/S/T) domains. Inset shows the amino acid sequence of a region of the P/S/T domain shown to be crucial for activation of transcription by the MAPK pathway. Consensus MAPK phosphorylation sites are indicated by asterisks. (B, C) Runx2 phosphorylation. Wild-type Runx2 (Panel B) or a P/S/T domain deletion mutant (ΔPST, Panel C) both containing a N-terminal HA tag were expressed in COS7 cells using a pcDNA5 expression vector. Cells were co-transfected with either pcDNA5 containing a β-galactosidase cDNA (β-gal) or a constitutively active MEK cDNA, MEK(SP). Cells were metabolically labeled with either 32P orthophosphate or 35S methionine/cysteine, and cell extracts immunoprecipitated with an anti-HA epitope monoclonal antibody. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography.
FGF2 REGULATION OF RUNX2
FGF2 is an important in vivo regulator of skeletal development and growth [17]. Intermittent FGF2 administation to rats stimulates bone formation and mineralization [18], whereas activating mutations in FGF receptors are associated with a series of craniosynostosis syndromes characterized by accellerated intramembranous bone formation in calvarial sutures [19, 20]. Of particular interest, activating mutations in FGFR1 upregulate Runx2 and enhance differentiation of calvarial osteoblasts [20]. FGF2 can also stimulate osteocalcin gene expression in MC3T3-E1 preosteoblast cells [21].
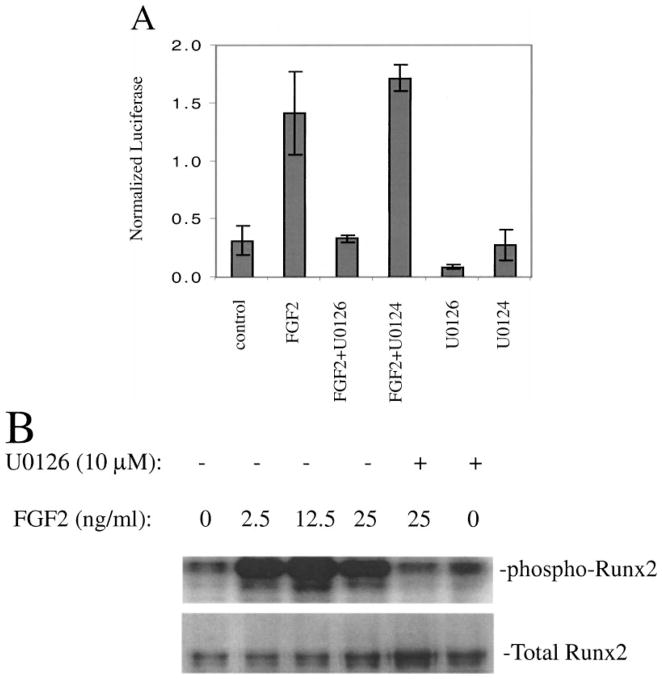
Since a major route for FGF receptor signaling involves activation of the MEK/ERK branch of the MAP kinase pathway [22], we initiated a series of studies to examine whether FGF2 induction of the osteocalcin gene required MAPK activity and Runx2 phosphorylation. Initial studies demonstrated that FGF2 could rapidly induce ERK phosphorylation and stimulate OCN mRNA in MC3T3-E1 cells (results not shown). FGF2 also stimulated activity of a 1.3 kb OCN promoter-luciferase reporter gene and this stimulation could be blocked by the MEK/ERK inhibitor, U0126, but not the inactive analogue, U0124 (Figure 2A). This stimulation was only seen in cells containing wild-type Runx2 (a P/S/T domain mutant was unresponsive) and also required an intact Runx2 binding site in the OCN promoter (results not shown). Metabolic labeling with [32P]-orthophosphate showed that the level of Runx2 phosphorylation in osteoblasts is increased by FGF2 treatment and that this response is also prevented by U0126 (Figure 2B).
Figure 2.
FGF2-dependent activation and phosphorylation of Runx2. (A) Activation of the OCN promoter. MC3T3-E1 preosteoblast cells stably transfected with a 1.3 kb fragment of the mouse osteocalcin gene 2 promoter driving a firefly luciferase reporter gene were treated with vehicle or 12.5 ng/mL FGF2 for 6 hr in the presence or absence U0126, a MEK inhibitor, or the inactive analogue, U0124. (B) Runx2 phosphorylation. MC3T3-E1 cells were metabolically labeled with 32P orthophosphate and treated with the indicated concentrations of FGF2 for 6 hr in the presence or absence of U0126 or U0124. Endogenous Runx2 was immunoprecipitated with a polyclonal antibody and analyzed by SDS-PAGE and autoradiography (upper panel). Total Runx2 in immunoprecipitates was measured by Western blotting (lower panel).
These studies show that actions of FGF2 on OCN gene expression require MAPK-dependent activation of Runx2 and that this activation is associated with increased phosphorylation of this transcription factor. Others have shown that FGF2 also can activate OCN transcription through an AP-1-like site that is immediately 5′ to the Runx2 binding site described above [21]. The nuclear factor/s binding this site are currently unknown, although their DNA-binding activity increases with FGF2 treatment. Of further interest, the FGF2 response is synergistically stimulated by the PKA pathway activator, forskolin, known to increase the activity of AP-1-related nuclear factors such as c-Fos and c-Jun. Taken together, these results raise the intriguing possibility that cooperative interactions take place between Runx2 and AP-1-like factors (see next section).
FGF2 has also been reported to stimulate the transcriptional activity of other osteoblast-related genes such as those encoding bone sialoprotein and interstitial collagenase (matrix metalloproteinase 1), although it is not known whether Runx2 is involved in either of these responses [23, 24]. Of interest, AP-1-like sites as well as MAP kinase activities were implicated in the regulation of both genes.
PTH ACTIVATES COLLAGENASE 3 GENE VIA RUNX2 AND AP-1 SITES
Actions of PTH on osteoblasts are complex, involving activation of both catabolic and anabolic pathways. The PTH receptor 1 activates both PKA and PKC-related signal transduction pathways. One early step in PTH-dependent bone remodeling is the PKA-dependent transcriptional activation of the collagenase 3 gene [25]. This collagenase specifically degrades fibrillar collagens.
Studies of the collagenase 3 promoter have been particularly informative for understanding how signals from different pathways can be integrated to regulate an osteoblast-related gene. Systematic analysis of the proximal collagenase 3 promoter by the laboratory of Dr. Partridge identified an 110 bp region as being essential for PTH responsiveness [6]. Attention was focused on two specific sequences within this region: a Runx2 binding site at −133 bp from the transcription start site and an AP-1 site at −48 bp. Mutations in either site that abrogated the binding of Runx2 or c-Fos/c-Jun, respectively, totally abolished PTH stimulation of promoter activity. The helical phasing between these two sites appears to be critical for the PTH response since insertion of additional bases interfered with promoter activation [26].
Hormonal responsiveness was enhanced by overexpression of both Runx2 and AP-1 binding proteins suggesting that cooperative interactions take place between these factors. More direct evidence for this concept comes from immunoprecipitation experiments that demonstrated direct interactions between Runx2 and c-Fos/c-Jun [26, 27]. This interaction requires the runt domain of Runx2 and the leucine zipper domain of the two AP-1 factors.
Interestingly, PTH stimulation of the collagenase gene, like the FGF2 or ECM stimulation of osteocalcin discussed above, was not accompanied by major changes in levels of the Runx2 protein [28]. Analysis of mutations in the P/S/T domain of Runx2 fused to the DNA-binding domain of the Gal4 yeast transcription factor localized a PTH responsive region to a PKA consensus phosphorylation site in the activation domain 3 (AD3) region of Runx2. This same site could be phosphorylated by purified PKA in vitro although it is not yet known if it is also phosphorylated in intact cells. These results suggest that PTH stimulates the collagenase 3 promoter by a PKA-dependent pathway that phosphorylates Runx2 and upregulates c-Fos and c-Jun via phosphorylation of CREB. This pathway is distinguished from the regulation of Runx2 by ECM or FGF2 which is mediated by the MEK/ERK MAP kinase pathway.
COOPERATIVE INTERACTIONS BETWEEN RUNX2 AND BMP/SMAD SIGNALING PATHWAYS
The bone morphogenetic proteins (BMPs) are the best described inducers of osteoblast and chondrocyte differentiation, as well as bone and cartilage formation in vivo. As is the case for other members of the TGF-β superfamily, BMP-initiated signals are transduced using specific Smad proteins. BMP signaling uses the receptor-regulated Smads 1, 5, and 8 (R-Smads) and the common partner protein, Smad4 [29]. BMP regulation of osteoblast gene expression is complex, involving direct interactions of R-Smad-Smad4 complexes with enhancer sequences on target genes (Smad binding elements or SBEs), binding of Smads to other nuclear factors as well as upregulation of separate transcription factors including Runx2 [4, 30].
BMP treatment or overexpression of Runx2 have both been reported to induce osteoblast-specific gene expression in mesenchymal cells [4, 31]. Since BMPs can upregulate Runx2, one possible explanation for how BMPs act in bone would be to assume that their effects are principally mediated by Runx2. Alternatively, BMPs may also activate other signaling pathways that, together with Runx2, stimulate gene expression. For example, consensus SBEs in the promoter regions of target genes could bind the R-Smad-Smad4 complex and this complex could cooperatively interact with Runx2 bound to another region of the promoter. In fact, there is some precedent for this concept: TGF-β stimulation of the IgCβ promoter is known to require interactions between Cbfa3, another Runt-related protein homologous to Runx2, and a protein complex containing Smad3/Smad4 bound to a separate SBE in this promoter [32].
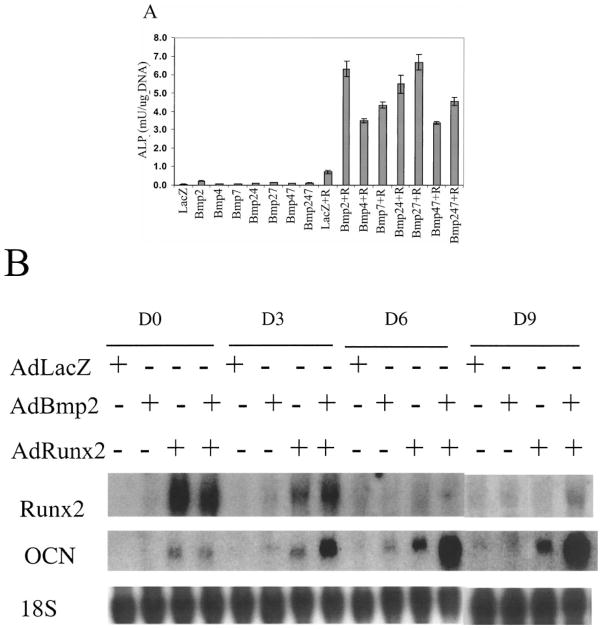
To specifically test for cooperativity between Runx2 and BMP signaling in osteoblast gene expression, we examined effects of BMP overexpression in the presence or absence of Runx2. For these studies, recombinant adenoviruses engineered to overexpress BMPs 2, 4, or 7 or Runx2 under the control of a CMV promoter [33] were used to transduce the pluripotent C3H10T1/2 mesenchymal cell line. Cells transduced with each virus were assayed for induction of osteoblast markers (alkaline phosphatase activity-ALP and OCN mRNA) as well as Runx2 mRNA. Preliminary experiments determined titers for each viral construct necessary to maximally induce osteoblast markers (results not shown). Subsequent experiments looked for cooperativity when optimal titers of viral constructs were combined. As shown in Figure 3A, individual BMPs or combinations of BMPs only modestly stimulated ALP activity. A somewhat higher level of induction was observed in cells transduced with the Runx2 virus alone. In contrast, co-transduction of cells with BMP and Runx2 viruses dramatically increased ALP activity relative to activities observed with either virus alone. Analysis of OCN mRNA levels showed a comparable degree of synergy between BMP2 and Runx2 (panel B). Consistent with previous reports on effects of BMPs and Runx2, cells transduced only with BMP2 virus showed a small stimulation in Runx2 and OCN mRNA at day 3 and 6 while transduction with the Runx2 virus induced a somewhat higher level of OCN mRNA at all times examined. However, combination of both viruses led to an approximately 10-fold increase in OCN mRNA levels. Interestingly, this synergy was seen even after Runx2 mRNA levels in transduced cells had declined nearly to control levels (see Figure 3B day 6 and 9 groups). In separate experiments, we saw a similar degree of synergy when control or Runx2 transduced cells were treated with saturating levels of recombinant BMP2 protein (results not shown).
Figure 3.
Synergistic stimulation of osteoblast gene expression by transduction of mesenchymal cells with adenoviruses overexpressing Runx2 and BMPs. (A) Alkaline phosphatase activity. C3H10T1/2 cells were transduced with optimal titers of adenoviruses containing a LacZ control cDNA (LacZ) or cDNAs encoding BMPs 2, 4, or 7 and/or Runx2 (R) as indicated. Cells were harvested after 6 days and assayed for alkaline phosphatase activity. (B) mRNA levels. Cells were transduced with AdLacZ, AdBmp2, or AdRunx2 adenovirus as indicated. After various times, total RNA was isolated for Northern blot analysis of Runx2, and OCN mRNAs as well as for 18S rRNA for blot normalization.
These studies strongly support the view that Runx2 and BMP signals cooperatively interact to stimulate osteoblast gene expression although they do not explore the basis for this cooperation. Possible explanations for the observed cooperativity include direct interactions between Runx2 and R-Smads, modulation of either R-Smad or Runx 2 transcriptional activity, or regulation of BMP receptor activity. Nevertheless, the observation that BMPs dramatically stimulate osteoblast gene expression even in cells already overexpressing Runx2 suggests that some aspects of BMP action cannot be explained simply by upregulation of Runx2.
SUMMARY AND CONCLUSIONS
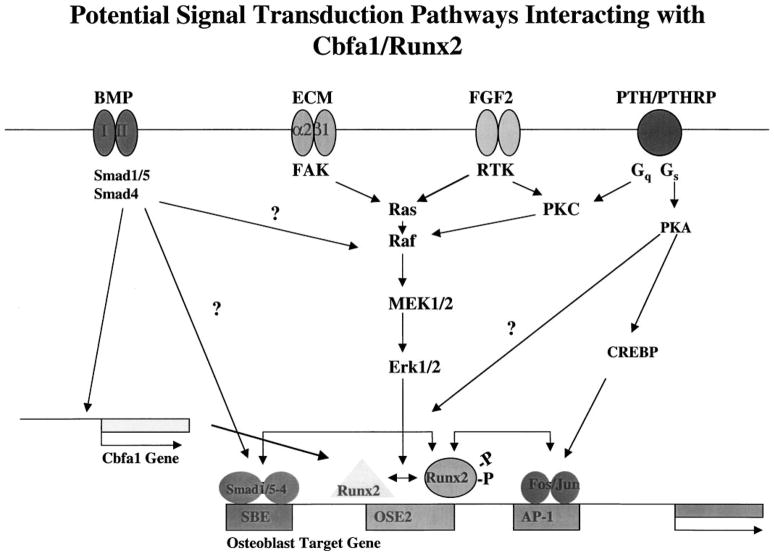
The four regulatory signals discussed in this article are shown in Figure 4 (i.e., those initiatied by BMPs, ECM, FGF2, or PTH/PTHrP). We believe that activation of Runx2 via phosphorylation is crucial for this factor to form an active complex with its DNA-binding site, OSE2, and stimulate transcription. Phosphorylation can be stimulated in three ways: (i) ECM binding to integrins on the cell surface activates focal adhesion kinase (FAK) and the MEK/ERK branch of the MAPK pathway; (ii) activation of receptor tyrosine kinase (RTK) activity in the FGF2 receptor also activates the MEK/ERK pathway; and (iii) the classic PKA pathway activated by the parathyroid hormone/parathyroid hormone-related peptide (PTH/PTHrP) receptor may also stimulate phosphorylation of Runx2 on sites distinct from those utilized by the MEK/ERK pathway. Alternatively, stimulation of the MAPK pathway via protein kinase C (PKC) is a potential route for cross-signaling from the PTH/PTHrP receptor via activation of Gq. The PKA pathway also upregulates AP-1-related factors like cFos and cJun by phosphorylation of cAMP response element binding protein (CREBP). AP-1 factors regulate gene expression by binding to AP-1 sites in osteoblast-related genes as well as by interacting with Runx2. Finally, the BMP/Smad pathway controls Runx2 gene expression possibly by directly upregulating the Cbfa1 gene. In addition, R-Smad-Smad4 heterodimers can directly interact with SBEs in regulatory regions of osteoblast-related genes as well as form complexes with Runx2.
Figure 4.
Overview of signal transduction pathways affecting Runx2 activity. Refer to text for explanation.
In our view, Runx2 can be considered a focal point for integration of a variety of signals affecting osteoblast activity. These signals include information about the extracellular matrix environment as detected through integrin-ECM interactions (i.e., Has an appropriate ECM been synthesized? Is the cell in contact with this ECM? Is the ECM experiencing mechanical loads?) and hormone/growth/differentiation factor levels in the extracellular milieu (i.e., Are the combined signals from endocrine/juxtacrine/autocrine factors telling the osteoblast or preosteoblast to grow or differentiate? Lay down new bone matrix or resorb existing matrix?) These disparate signals can affect Runx2 activity by altering transcription factor levels as is the case for BMP induction of Runx2, phosphorylation state (mediated by MAPK as well as possibly PKA-dependent pathways), or interactions between Runx2 and other transcription factors such as Smads and AP1-related factors.
Clearly, further study is required to assess the overall validity of this model as an explanation for how bone responds to diverse stimuli. Of particular importance will be the study of interactions between Runx2 and other potential partner proteins such as Smads or AP-1 factors and the degree to which such interactions are affected by the level of Runx2 phosphorylation.
Acknowledgments
This work was supported by National Institute of Dental and Craniofacial Research Grants DE13386, DE11732, and DE12211. Additional support was obtained from the Michigan Multipurpose Arthritis Center Grant AR20557 and the General Clinical Research Center Grant M01-RR00042.
References
- 1.Ducy P, Schinke T, Karsenty G. The osteoblast: A sophisticated fibroblast under central surveillance. Science. 2000;289(5484):1501–1504. doi: 10.1126/science.289.5484.1501. [DOI] [PubMed] [Google Scholar]
- 2.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89(5):765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 3.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 4.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 5.Kern B, Shen J, Starbuck M, Karsenty G. Cbfa1 contributes to the osteoblast-specific expression of type I collagen genes. J Biol Chem. 2000;5:5. doi: 10.1074/jbc.M006215200. [DOI] [PubMed] [Google Scholar]
- 6.Selvamurugan N, Chou WY, Pearman AT, Pulumati MR, Partridge NC. Parathyroid hormone regulates the rat collagenase-3 promoter in osteoblastic cells through the cooperative interaction of the activator protein-1 site and the runt domain binding sequence. J Biol Chem. 1998;273(17):10647–10657. doi: 10.1074/jbc.273.17.10647. [DOI] [PubMed] [Google Scholar]
- 7.Xiao G, Wang D, Benson MD, Karsenty G, Franceschi RT. Role of the alpha2-integrin in osteoblast-specific gene expression and activation of the Osf2 transcription factor. J Biol Chem. 1998;273(49):32988–32994. doi: 10.1074/jbc.273.49.32988. [DOI] [PubMed] [Google Scholar]
- 8.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20(23):8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franceschi RT. The developmental control of osteoblast-specific gene expression: Role of specific transcription factors and the extracellular matrix environment. Crit Rev Oral Biol Med. 1999;10(1):40–57. doi: 10.1177/10454411990100010201. [DOI] [PubMed] [Google Scholar]
- 10.Takeuchi Y, Suzawa M, Kikuchi T, Nishida E, Fujita T, Matsumoto T. Differentiation and transforming growth factor-beta receptor down-regulation by collagen-alpha2beta1 integrin interaction is mediated by focal adhesion kinase and its downstream signals in murine osteoblastic cells. J Biol Chem. 1997;272(46):29309–29316. doi: 10.1074/jbc.272.46.29309. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman D, Jin F, Leboy P, Hardy S, Damsky C. Impaired bone formation in transgenic mice resulting from altered integrin function in osteoblasts. Dev Biol. 2000;220(1):2–15. doi: 10.1006/dbio.2000.9633. [DOI] [PubMed] [Google Scholar]
- 12.Danen EH, Lafrenie RM, Miyamoto S, Yamada KM. Integrin signaling: Cytoskeletal complexes, MAP kinase activation, and regulation of gene expression. Cell Adhes Comm. 1998;6(2–3):217–224. doi: 10.3109/15419069809004477. [DOI] [PubMed] [Google Scholar]
- 13.Xiao G, Cui Y, Ducy P, Karsenty G, Franceschi RT. Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: Requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol Endocrinol. 1997;11(8):1103–1113. doi: 10.1210/mend.11.8.9955. [DOI] [PubMed] [Google Scholar]
- 14.Xiao G, Gopalakrishnan R, Jiang D, Reith E, Benson M, Franceschi RT. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J Bone Miner Res. 2002;17(1):101–110. doi: 10.1359/jbmr.2002.17.1.101. [DOI] [PubMed] [Google Scholar]
- 15.Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, Franceschi RT. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor. Cbfa1 J Biol Chem. 2000;275(6):4453–4459. doi: 10.1074/jbc.275.6.4453. [DOI] [PubMed] [Google Scholar]
- 16.Thirunavukkarasu K, Mahajan M, McLarren KW, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfbeta. Mol Cell Biol. 1998;18(7):4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12(11):1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 18.Mayahara H, Ito T, Nagai H, Miyajima H, Tsukuda R, Taketomi S, Mizoguchi J, Kato K. In vivo stimulation of endosteal bone formation by basic fibroblast growth factor in rats. Growth Factors. 1993;9(1):73–80. doi: 10.3109/08977199308991583. [DOI] [PubMed] [Google Scholar]
- 19.Kannan K, Givol D. FGF receptor mutations: Dimerization syndromes, cell growth suppression, and animal models. IUBMB Life. 2000;49(3):197–205. doi: 10.1080/713803609. [DOI] [PubMed] [Google Scholar]
- 20.Zhou YX, Xu X, Chen L, Li C, Brodie SG, Deng CX. A Pro250Arg substitution in mouse Fgfr1 causes increased expression of Cbfa1 and premature fusion of calvarial sutures. Hum Mol Genet. 2000;9(13):2001–2008. doi: 10.1093/hmg/9.13.2001. [DOI] [PubMed] [Google Scholar]
- 21.Boudreaux JM, Towler DA. Synergistic induction of osteocalcin gene expression: Identification of a bipartite element conferring fibroblast growth factor 2 and cyclic AMP responsiveness in the rat osteocalcin promoter. J Biol Chem. 1996;271(13):7508–7515. doi: 10.1074/jbc.271.13.7508. [DOI] [PubMed] [Google Scholar]
- 22.Nugent MA, Iozzo RV. Fibroblast growth factor-2. Int J Biochem Cell Biol. 2000;32(2):115–120. doi: 10.1016/s1357-2725(99)00123-5. [DOI] [PubMed] [Google Scholar]
- 23.Shimizu-Sasaki E, Yamazaki M, Furuyama S, Sugiya H, Sodek J, Ogata Y. Identification of a novel response element in the rat bone sialoprotein (BSP) gene promoter that mediates constitutive and fibroblast growth factor 2-induced expression of BSP. J Biol Chem. 2001;276(8):5459–5466. doi: 10.1074/jbc.M008971200. [DOI] [PubMed] [Google Scholar]
- 24.Newberry EP, Willis D, Latifi T, Boudreaux JM, Towler DA. Fibroblast growth factor receptor signaling activates the human interstitial collagenase promoter via the bipartite Ets-AP1 element. Mol Endocrinol. 1997;11(8):1129–1144. doi: 10.1210/mend.11.8.9958. [DOI] [PubMed] [Google Scholar]
- 25.Heath JK, Meikle MC, Atkinson SJ, Reynolds JJ. A factor synthesized by rabbit periosteal fibroblasts stimulates bone resorption and collagenase production by connective tissue cells in vitro. Biochim Biophys Acta. 1984;800(3):301–305. doi: 10.1016/0304-4165(84)90409-4. [DOI] [PubMed] [Google Scholar]
- 26.D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors, c-Fos and c- Jun, with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2001;18:18. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- 27.Hess J, Porte D, Munz C, Angel P. AP-1 and Cbfa/runt physically interact and regulate parathyroid hormone-dependent MMP13 expression in osteoblasts through a new osteoblast-specific element 2/AP-1 composite element. J Biol Chem. 2001;276(23):20029–20038. doi: 10.1074/jbc.M010601200. [DOI] [PubMed] [Google Scholar]
- 28.Selvamurugan N, Pulumati MR, Tyson DR, Partridge NC. Parathyroid hormone regulation of the rat collagenase-3 promoter by protein kinase A-dependent transactivation of core binding factor alpha1. J Biol Chem. 2000;275(7):5037–5042. doi: 10.1074/jbc.275.7.5037. [DOI] [PubMed] [Google Scholar]
- 29.Baker JC, Harland RM. From receptor to nucleus: The Smad pathway. Curr Opin Gen Dev. 1997;7(4):467–473. doi: 10.1016/s0959-437x(97)80072-x. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe M, Whitman M. The role of transcription factors involved in TGFbeta superfamily signaling during development. Cell Mol Biol (noisy-le-grand) 1999;45(5):537–543. [PubMed] [Google Scholar]
- 31.Xiao ZS, Hinson TK, Quarles LD. Cbfa1 isoform overexpression upregulates osteocalcin gene expression in non-osteoblastic and pre-osteoblastic cells. J Cell Biochem. 1999;74(4):596–605. [PubMed] [Google Scholar]
- 32.Hanai J, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, Stavnezer J, Kawabata M, Miyazono K, Ito Y. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem. 1999;274(44):31577–31582. doi: 10.1074/jbc.274.44.31577. [DOI] [PubMed] [Google Scholar]
- 33.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps ML. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71(3):1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]