Abstract
The advent of fungal pathogens that are resistant to the classic repertoire of antifungal drugs has increased the need for new therapeutic agents. A prominent example of such novel compound is caspofungin, known to alter cell wall biogenesis by inhibiting β-1,3 D-glucan synthesis. Although much progress has been made in understanding the mechanism of action of caspofungin, little is known about its influence on the biophysical properties of the fungal cells. Here, we use atomic force microscopy to demonstrate that caspofungin induces major remodeling of the cell surface properties of Candida albicans. Caspofungin causes major morphological and structural alterations of the cells, which correlate with a decrease of the cell wall mechanical strength. Moreover, we find that the drug induces the massive exposure of the cell adhesion protein Als1 on the cell surface and leads to increased cell surface hydrophobicity, two features that trigger cell aggregation. This behaviour is not observed in yeast species lacking Als1, demonstrating the key role that the protein plays in determining the aggregation phenotype of C. albicans. The results show that AFM opens up new avenues for understanding the molecular bases of microbe-drug interactions and for developing new therapeutic agents.
The fungal pathogen Candida albicans shows strong cell adhesion properties that play essential roles in modulating pathogenesis and immune responses.1-5 In the therapeutic context, there is increasing evidence that cell adhesion and biofilm formation enable the pathogen to escape the effects of antifungal drugs, thereby contributing to the establishment of persistent fungal infections.3 Adhesion, aggregation and biofilm formation are mediated by cell adhesion molecules (adhesins), primarily the wall-anchored Als glycoproteins.6,7 Als proteins consist of an N-terminal Ig-like region which initiates cell adhesion, followed by a threonine-rich (T) region with an amyloid-forming sequence, a tandem repeat (TR) region that participates in cell-cell aggregation, and a stalk region projecting the molecule away from the cell surface. A key feature of Als adhesins is that their different protein domains synergize to bind an extremely broad range of ligands.7,8
The N-terminal Ig-like region binds to broad consensus “tϕ+” peptides (t, a residue common in turns; ϕ, a bulky hydrophobic residue; +, Lys or Arg), which constitute several percent of tripeptide sequences in the proteome.9 These Ig-like region interactions promote specific adhesion to host constituents, including epithelial cells, endothelial cells, and extracellular matrix proteins, and yeast-yeast aggregation. Because the tϕ+ motif is usually buried in native proteins, they become much better ligands after denaturation, consistent with the observation that C. albicans preferentially binds to damaged regions of tissues and denatured proteins. The hydrophobic TR domains of Als adhesins mediate non-specific adhesion to hydrophobic substrates, promote Als protein homotypic binding, and can be unravelled by mechanical force.10-12 The T domains have conserved amyloid-forming sequences that bind amyloid dyes like thioflavin T and Congo red and mediate aggregation through high avidity amyloid interactions.7,13 The strength of T-mediated cell adhesion results from the force-activated amyloid-like clustering of hundreds of proteins on the cell surface to form arrays of ordered multimeric binding sites.7,14 These amyloid clusters explain why Als proteins often show weak binding to specific ligands, yet mediate remarkably strong adherence.
Traditionally, fungal infections are treated by three groups of antifungal drugs: azoles which decrease synthesis of ergosterol, an essential plasma membrane sterol; polyenes which bind ergosterol and disturb cellular permeability; and fluorinated pyrimidines which inhibit nucleic acids and proteins synthesis.15,16 However, there is a constant need for new antifungal drugs owing to the emergence of resistant strains.17,18 An example of such novel drugs are echinocandins, like caspofungin, which target the synthesis of cell wall β-1,3 D-glucans.19,20 Treatment of C. albicans with caspofungin alters the cell morphology,21,22 lowers cell wall mechanical strength, increases sensitivity to osmotic pressure, and eventually leads to cell lysis and death.20,23 Caspofungin can also induce other cellular changes via stress response pathways, such as chitin synthesis up-regulation through high-osmolarity glycerol (HOG) pathway or invasive growth through mitogen-activated protein kinase (Mkc1) activation.24,25 Similar caspofungin-induced cellular responses have been demonstrated in the non-related species Saccharomyces cerevisiae, suggesting they are conserved among several yeast species.26,27 Recently, exposure of C. albicans to subinhibitory caspofungin concentrations was shown to induce cell aggregation through the expression of Als1,24 the major Als protein expressed on C. albicans yeast form cells, which is known to mediate cell aggregation and adherence.28 However, the molecular details underlying caspofungin-induced cell adhesion and cell wall remodeling remain poorly understood.
Atomic force microscopy (AFM) enables researchers to observe the supramolecular organization of cell surfaces, and to probe the localization, mechanical properties and adhesion of their individual molecules in relation to cellular function. Yet, the use of AFM to investigate drug-induced cell wall remodelling in C. albicans has thus far not been documented. Here, we track the cell surface molecular changes of single C. albicans cells exposed to caspofungin using different AFM modalities. We show that treatment of C. albicans with caspofungin causes cell swelling, as well as substantial cell wall structural and mechanical changes. We also demonstrate that the drug leads to the massive exposure of Als1 adhesins, accompanied by a major increase in cell surface hydrophobicity. These molecular changes trigger cell aggregation. Caspofungin-induced cell wall remodeling is not observed in C. glabrata and S. cerevisiae, two fungal species lacking Als adhesins. These nanoscale experiments offer exciting prospects in therapeutics, for understanding the action mode of antimicrobial drugs, and for screening new agents targeting the cell wall.
RESULTS AND DISCUSSION
Caspofungin dramatically enhances Als1-mediated adhesion
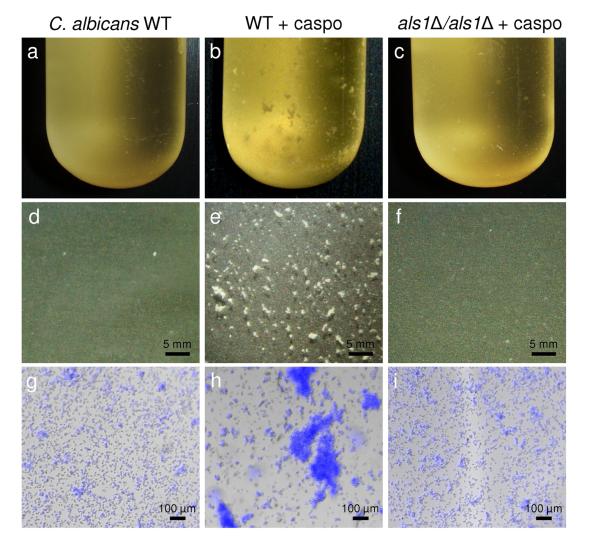
We first investigated the influence of subinhibitory concentrations of caspofungin on the ability of C. albicans to form cellular aggregates. Consistent with an earlier study,24 Fig. 1 shows that C. albicans cells grown for 2 h in the presence of 50 ng ml−1 caspofungin formed macroscopic aggregates, while cells grown in the absence of the drug did not. Cells from an als1Δ/als1Δ mutant strain deficient for Als1 expression did not form large aggregates, demonstrating that caspofungin-induced aggregation is mediated by Als1, the major adhesin expressed on C. albicans in the yeast form. Supporting further this view, caspofungin-induced aggregation was not observed in two fungal species lacking Als adhesins, i.e. C. glabrata and S. cerevisiae.
Fig. 1. Influence of caspofungin on Als1-mediated aggregation.
(a-c) Stereomicrographs, (d-f) optical microscopy, and (g-i) overlaid optical and fluorescence microscopy images of C. albicans WT cells grown in the absence (a, d, g) or in the presence (b, e, h) of 50 ng ml−1 caspofungin (caspo), and of als1Δ/als1Δ mutant cells grown in the presence of 50 ng ml−1 caspofungin (c, f, i). Cells were stained with Calcofluor White for fluorescence imaging.
Caspofungin alters the morphology and structure of C. albicans
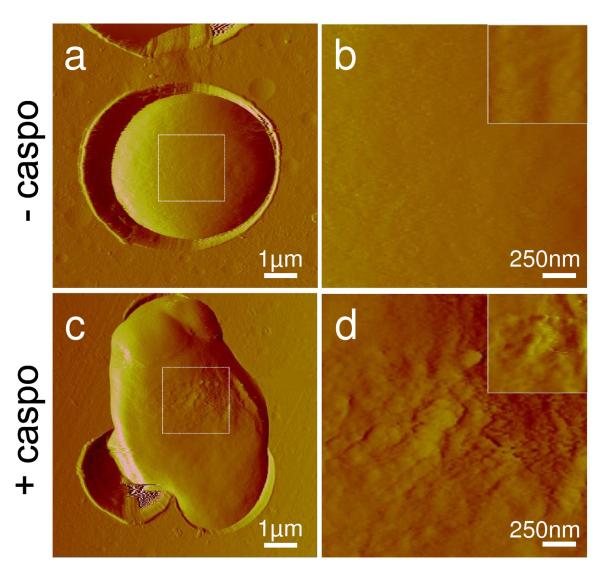
We used AFM imaging in buffer to observe structural changes on the surface of yeast cells exposed to caspofungin. A representative low-resolution deflection image of a single C. albicans cell immobilized in a porous membrane is shown in Fig. 2a. Using small imaging forces (~100 pN), images were obtained repeatedly without detaching the cell or altering significantly the surface morphology. High resolution images (Fig. 2b) revealed a smooth and homogeneous surface, consistent with the presence of an outer layer of mannoproteins and with earlier electron microscopy data.21,22
Fig. 2. AFM images of native and caspofungin-treated C. albicans cells.
Low (a, c) and high (b, d) resolution deflection images recorded in buffer for a native C. albicans cell (a, b), and a C. albicans cell treated for 2 h with 50 ng ml−1 caspofungin (c, d). The insets show the height images corresponding to the deflection images in (b, d). Similar data were obtained in multiple experiments using different tips and cell cultures.
Fig. 2c and 2d show low and high resolution images of a cell grown for 2 h in the presence of 50 ng ml−1 caspofungin. As can be seen, the drug changed the cell shape and increased the average cell size, as observed earlier by scanning electron microscopy (SEM) and transmission electron microscopy (TEM).21,22 As the cell wall target of caspofungin, β-1,3 D-glucan, plays a critical role in maintaining cell shape, mechanical rigidity and resistance to osmotic pressure,15,29 we interpret such cell swelling as evidence for the formation of caspofungin-induced osmotically fragile cells resulting from compromised cell wall synthesis and abnormal division. These changes are reminiscent of the behavior of Staphylococcus aureus exposed to lysostaphin, an enzyme that specifically cleaves the bacterial cell wall peptidoglycan, thereby lysing the bacteria.30 Moreover, substantial alteration of the C. albicans cell surface structure was clearly observed, the surface roughness (calculated on 500 nm × 500 nm height images) increasing from ~1 nm, for untreated cells, to ~5 nm, for treated cells. These AFM observations are qualitatively consistent with electron microscopy investigations22 showing progressive disintegration of the cell wall after exposure to caspofungin. We note that similar morphological and structural changes were observed in the yeasts C. glabrata and S. cerevisiae (Supplementary Fig. 1).
Caspofungin decreases cell wall mechanical strength
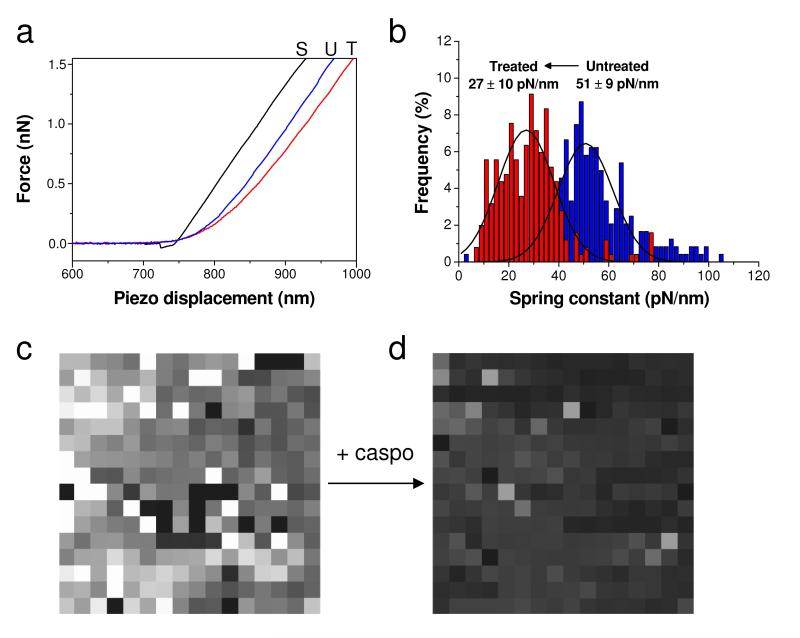
As caspofungin alters the cell wall mechanical strength, we then asked whether the observed structural changes were correlated with differences in cell wall mechanics, using AFM force spectroscopy. Fig. 3a shows typical force vs piezo displacement curves obtained for solid substrata, for native C. albicans cells, and for cells treated with caspofungin for 2 h. Consistent with earlier studies,30,31 the curves recorded on the cells showed two regimes, i.e. a nonlinear domain at low loading forces, followed by a linear one at high loading forces. From the shape of these curves, it can already be seen that the cell wall was substantially softer than the substratum, and that caspofungin caused a softening of the cell wall.
Fig. 3. Mechanical properties of native and caspofungin-treated C. albicans cells.
(a) Representative force vs displacement curves recorded on the polymer substratum (black curve, S), on an untreated cell (blue curve, U), and on a cell treated with caspofungin at 50 ng ml−1 for 2 h (red curve, T). (b) Stiffness histogram and (c, d) stiffness maps (1 μm × 1 μm; z-range = 75 and 100 pN/nm, for c and d respectively) documenting the homogenous decrease in cell spring constant upon treatment with the drug. Similar data were obtained in multiple experiments using different tips and cell cultures.
To quantify the changes in cell wall nanomechanics, the cell spring constant was extracted from multiple force curves (n = 256) recorded across the cell surface.32,33 The spring constant of untreated cells was found to be 51 ± 9 pN/nm, a value roughly in the range of that reported for S. cerevisiae,34 considering that it strongly depends on the cell wall organization, ionic strength, and tip stiffness and geometry. Notably, incubation with caspofungin for 2 h decreased the cell spring constant to 27 ± 10 pN/nm (n = 256). This change may be attributed to a decrease of the inner turgor pressure of the cell,32,33 in agreement with the observed cell swelling (Fig. 2) and with the formation of osmotically fragile cells. As shown in Fig. 3c and 3d, stiffness maps recorded on different areas of the native and treated cell surfaces showed rather homogeneous contrasts, indicating that the drug-induced softening affected the entire cells.
Probing single Als1 proteins on C. albicans cells
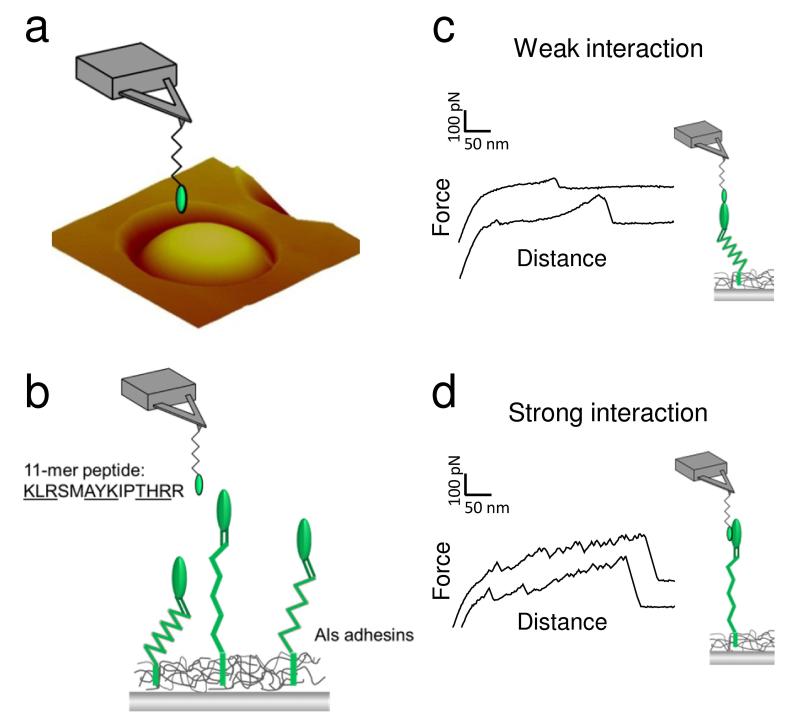
Next, we probed individual cell surface Als1 proteins to address the following pertinent questions: what are the surface densities and surface expression patterns, what are their biophysical properties (adhesion, mechanics, extension), and how does caspofungin change these characteristics? To probe single Als1 proteins, the AFM tip was functionalized with a short adhesion peptide (KLRSMAYKIPTHRR) containing three “tϕ+” structural motifs (underlined) that bind to the N-terminal Ig-like region of Als proteins (Fig. 4a and 4b).9 Peptides were attached through their NH2 groups using a well-established protocol that favours the detection of single - or only few – molecules.35 This technique results in predominant coupling to the amino end of peptides, because they have a lower pK than the Lys epsilon NH2, and the amino groups in Arg are generally not reactive. Force-distance curves recorded across the surface of C. albicans cells using AFM tips bearing the adhesion peptide showed two types of adhesion patterns, i.e. low adhesion force profiles displaying single small adhesion forces at short distances, reflecting weak, non-specific interactions between the peptide probe and the Als Ig-like region (Fig. 4c), and large adhesion force curves showing sawtooth patterns with multiple large force peaks and long ruptures distances, that we attribute to strong interaction of the structural adhesion motifs of the peptide with the Als Ig-like region (Fig. 4d).36
Fig. 4. Single-molecule detection of Als1 proteins.
(a, b) To probe single Als1 proteins, C. albicans yeast cells were immobilized in a porous polymer membrane (a) and imaged in buffer using AFM tips terminated with a short adhesion peptide (KLRSMAYKIPTHRR) containing three amino acid structural motifs (underlined sequences) that bind to the N-terminal Ig-like region of Als1 (b). (c, d) Force-distance curves recorded between the peptide-tip and the yeast surface featured two types of adhesion signatures, i.e. weak adhesion events with short rupture distances (c) and strong adhesion events with extended sawtooth patterns (d). While the weak adhesion forces reflect non-specific interaction with Als1 and other surface molecules, the strong adhesion forces are due to the specific detection and unfolding of single Als1 (see text for details). Similar data were obtained in several independent experiments using different tip preparations and cell cultures.
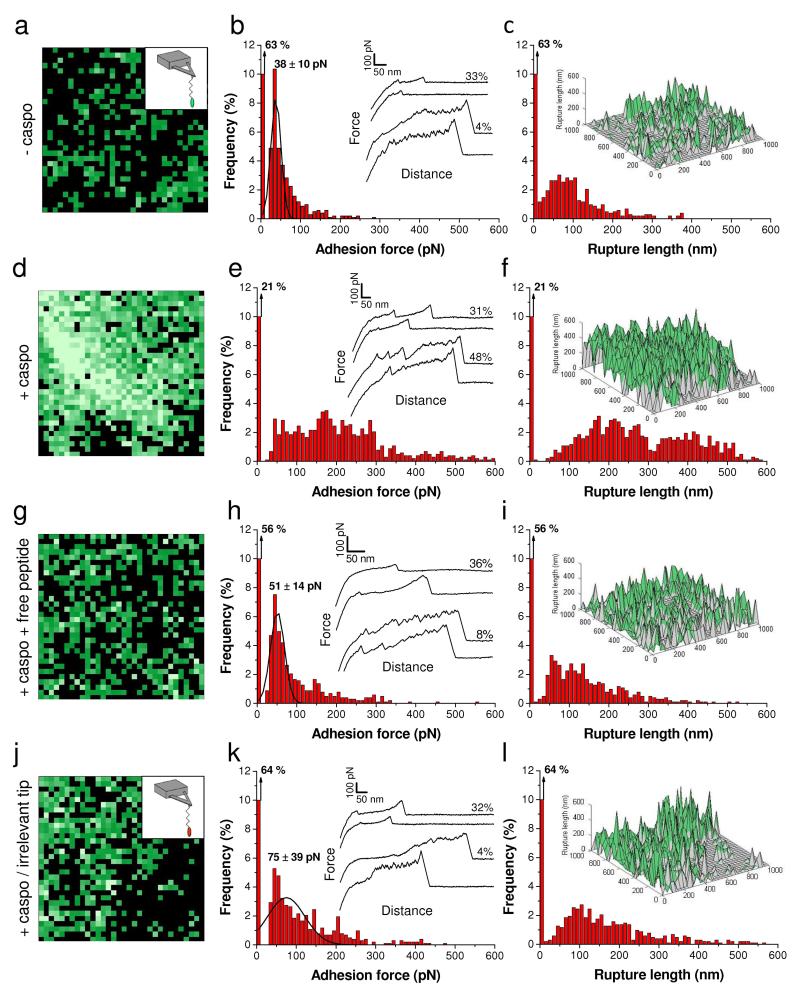
Fig. 5a-5c shows the adhesion map, adhesion histogram with representative force curves and rupture length histogram obtained on a WT C. albicans cell with a peptide-tip. A substantial fraction (37 %) of the curves showed adhesion signatures, the vast majority (33 %) of interactions being weak (38 ± 10 pN) and short (10-100 nm) (Fig. 5b, upper curves). These weak interactions are non-specific as they were not competed by free adhesion peptides and were observed even when using an irrelevant peptide probe lacking the “tϕ+” adhesion motifs (Fig. 5g-5l, see below for details). In contrast, the remaining 4 % of adhesion curves showed strong interactions, with multiple large force peaks (331 ± 38 pN; Fig. 5b, lower curves) and long ruptures (200-400 nm; Fig. 5c). These strong interactions are characteristic of specific, multipoint binding to the Als Ig-like regions, with sawtooth sequential unfoldings of the T and TR regions of the protein.10 Adhesion maps indicated that most of the interacting regions formed clusters of 100-200 nm, corresponding to a minimum surface density of 200 sites/μm2 (Fig. 5a).
Fig. 5. Caspofungin increases the expression of Als1 in C. albicans wild-type cells.
(a, d, g) Adhesion force maps (1 μm × 1 μm; black and green pixels correspond to adhesion forces smaller and larger than 50 pN, respectively; brighter green color means larger adhesion forces) recorded in buffer between an AFM tip bearing a short adhesion peptide (KLRSMAYKIPTHRR) and the surface of a native C. albicans cell (a), a C. albicans cell treated with 50 ng ml−1 caspofungin (d), and a C. albicans cell treated with 50 ng ml−1 caspofungin, harvested and further blocked by injection of free adhesion peptides (0.1 mg ml−1) (g). (j) Adhesion force map (1 μm × 1 μm, color scale: 350 pN) recorded in buffer between a tip bearing an irrelevant peptide (ESTTTTLNISSE) and a native C. albicans cell. (b, e, h, k) Corresponding adhesion force histograms (n = 1024) together with representative force curves. (c, f, i, l) Histograms of rupture distances (n = 1024), and 3-D reconstructed polymer maps of x, y position vs rupture length (false colors, adhesion forces in green). Similar data were obtained in multiple experiments using different tips and cell cultures.
Single-molecule AFM demonstrates that caspofungin increases Als1 exposure
We then used peptide-terminated tips to probe the distribution, adhesion and extension of Als1 on cells treated for 2 h with caspofungin (Fig. 5d-5f). Treated cells showed a much higher detection frequency (79 % vs 37 % on cells not exposed to caspofungin), corresponding to a minimum protein surface density of 600 proteins/μm2. Notably, the increased detection level corresponded to a 12-fold increase in strong/long unfolding interactions, thus reflecting the massive binding and unfolding of Als1 proteins. These differences in distribution, adhesion and extension are clearly visible in the three-dimensional (3-D) maps constructed by combining adhesion forces and rupture distances measured on every position (Fig. 5f vs 5c). Accordingly, the above single-molecule analyses, which correlate with the structural and elasticity changes, indicate that the amount of Als1 proteins exposed on the cell surface dramatically increased upon treatment with caspofungin, thereby leading to major changes in cell surface biophysical properties (adhesion, elasticity, conformation and extension of the surface proteins).
To assess the specificity of the detection events, we carried out several control experiments, i.e. blocking with free adhesion peptides, use of irrelevant peptide probes, and use of a yeast strain deficient for Als1 expression. Fig. 5g-5i shows that blocking the cell surface with free adhesion peptides led to a major reduction of the frequency of strong/long unfolding events (from 48 % to 8 %), the fraction of weak/short events remaining essentially unchanged. As can be seen in Fig. 5j-5l, the same behavior was observed when the AFM tip was functionalized with a random peptide (ESTTTTLNISSE) lacking the “tϕ+” consensus adhesion peptide. This demonstrates that, unlike the weak/short binding interactions, the strong/long unfolding interactions are specific to Als1 and rely on the “tϕ+” structural motif.
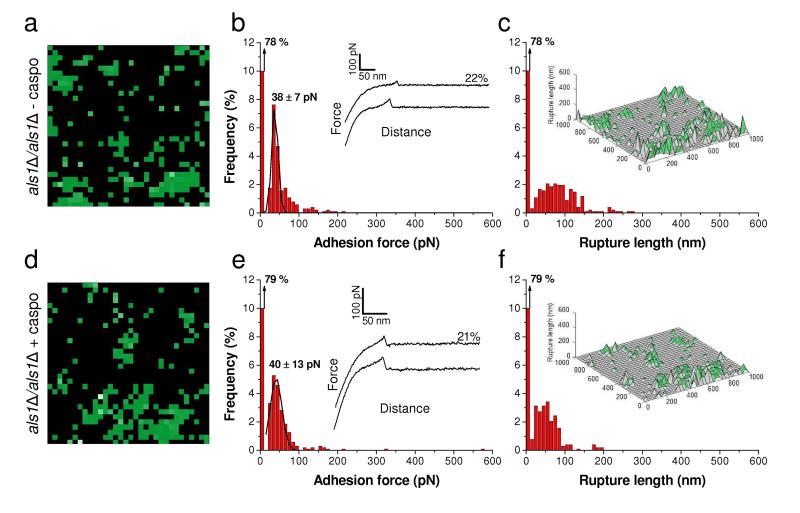
However, because our adhesion peptide should also bind Als5 and perhaps other Als adhesins,9 we cannot exclusively attribute binding to Als1. We therefore analyzed an als1Δ/als1Δ mutant strain to clarify this issue (Fig. 6). In the absence of caspofungin treatment, the Als1-deleted cells (Fig. 6a-6c) displayed only weak/short binding events, at a lower density than on WT cells (22 % vs 37 %). This suggests that nearly half of the non-specific interactions on the WT were associated with Als1, the remaining fraction being due to other surface molecules. Notably, treatment of the Als1-deleted cells with caspofungin did not induce any change in the frequency and nature of the binding events, demonstrating that the dramatic changes observed on WT cells are associated with Als1. Similarly, we found the same behavior (no strong unfolding events, moderate frequency of weak binding events, no effect of the drug) for the two yeasts C. glabrata and S. cerevisiae (Supplementary Fig. 2). The above single-molecule data were strongly correlated with the macroscale phenotypic behavior of the cells, as neither the als1Δals1Δ mutant, nor C. glabrata, nor S. cerevisiae aggregated upon exposure to caspofungin.
Fig. 6. Control experiment using an als1Δ/als1Δ mutant strain.
(a, d) Adhesion force maps (1 μm × 1 μm, color scale: 350 pN) recorded in buffer with a peptide-tip on als1Δ/als1Δ C. albicans cells grown without (a) or with (d) caspofungin (50 ng ml-1). (b, e) Corresponding adhesion force histograms (n = 1024) together with representative force curves. (c, f) Histograms of rupture distances (n = 1024), and 3-D reconstructed polymer maps (false colors, adhesion forces in green). Similar data were obtained in several independent experiments using different tip preparations and cell cultures.
Caspofungin increases cell surface hydrophobicity through Als1 expression
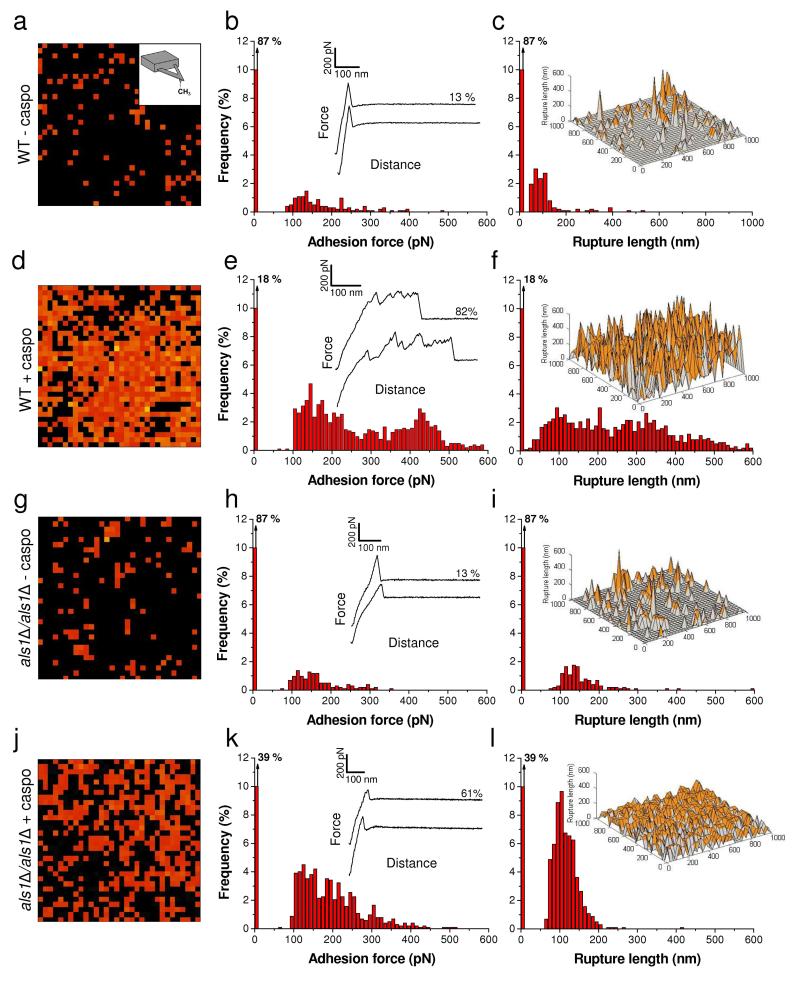
As hydrophobic interactions represent an important driving force for C. albicans adhesion, we then asked whether these could also be involved in caspofungin-induced aggregation. Therefore, we used chemical force microscopy (CFM) with hydrophobic (CH ) tips37,38 3 to quantify the nanoscale hydrophobic character of C. albicans cells prior and after treatment with the drug (Fig. 7). Force curves recorded on WT yeast cells showed few, randomly distributed adhesion forces (13 %) with moderate magnitude (50-300 pN) and short rupture lengths (50-100 nm) (Fig. 7a-7c), documenting a rather hydrophilic surface.38 By contrast, a large fraction of the curves (82 %) obtained on WT cells treated for 2 h with caspofungin showed large adhesion forces (300-1000 pN) with extended rupture lengths (400-1000 nm) (Fig. 7d-7f), which are characteristic of a more hydrophobic surface.38 We suggest that these signatures reflect the simultaneous unfolding of several Als1 proteins. This interpretation is supported by our finding that als1Δ/als1Δ cells – treated or not with the drug - showed much weaker (100-400 pN) and shorter (100 nm) force signatures than the treated WT cells (Fig. 7g-7l). These data lead us to conclude that overexpression of Als1 proteins increases the cell surface hydrophobicity, an effect that is likely to promote cell aggregation. As a matter of fact, the hydrophobic TR domains of Als proteins are thought to contribute to cell adhesion by promoting hydrophobic interactions.7,11,12 Hence, the large amounts of TR domains on caspofungin-treated cells, and their force-induced unfolding, will lead to extended conformations in which hydrophobic groups are freshly exposed and favor hydrophobic interactions with other cells.
Fig. 7. Caspofungin increases cell surface hydrophobicity.
(a, d, g, j) Adhesion force maps (1 μm × 1 μm, color scale: 1000 pN) recorded in buffer on C. albicans WT cells (a, d) or als1Δ/als1Δ mutant cells (g, j), grown without (a, g) or with (d, j) caspofungin (50 ng ml−1) using hydrophobic tips. (b, e, h, k) Corresponding adhesion force histograms (n = 1024) together with representative force curves. (c, f, I, l) Histograms of rupture distances (n = 1024), and 3-D reconstructed hydrophobicity maps (false colors, adhesion forces in orange). Similar data were obtained in several independent experiments using different tip preparations and cell cultures.
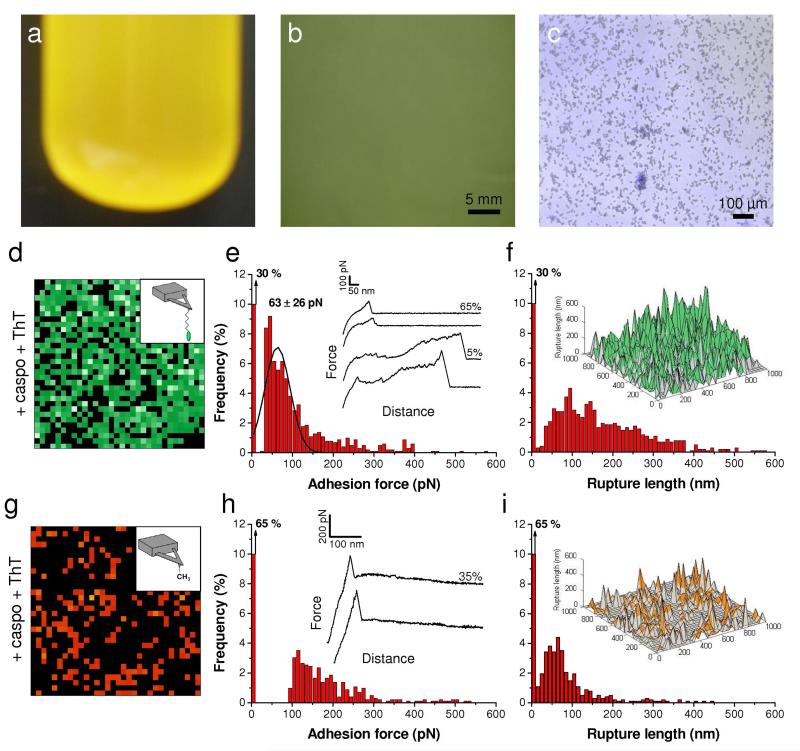
Amyloid interactions are essential for cell aggregation, Als1 unfolding and surface hydrophobicity
Als1-mediated adhesion and aggregation is dependent on amyloid-like interactions that cluster the adhesin molecules on the wall surface to form adhesion nanodomains.7,13,14,39,40 These interactions can be inhibited with anti-amyloid dyes such as Congo red or thioflavin T. Therefore, we explored the influence of the anti-amyloid agent thioflavin T (Fig. 8). C. albicans cells grown in the presence of caspofungin and thioflavin T (100 μM) for 2 h did not form large aggregates (Fig. 8a-8c). Remarkably, single-molecule imaging of caspofungin-treated cells in the presence of thioflavin T detected cell surface binding, but most strong/long unfolding events were abolished (Fig. 8d-8f). This suggests that Als1 proteins were still exposed in large amount but that their conformational properties were altered. Supporting this view, CFM with hydrophobic tips revealed much weaker (50-300 pN) and shorter (<200 nm) adhesion signatures than on cells treated with caspofungin alone. Hence, thioflavin T is able to disrupt aggregation, Als1 biophysical properties and surface hydrophobicity. We suggest that the loss of amyloid formation by Als1 leads to more disordered protein conformational properties, particularly to a lack of TR unfolding and a loss of hydrophobicity, and in turn to weaker interactions between the Als1 Ig-like region and peptide ligands (on the tip or on opposing cells).
Fig. 8. Cell aggregation, Als1 unfolding and cell surface hydrophobicity require amyloid interactions.
(a, b, c) Micrograph (a), optical microscopy (b) and overlaid optical and fluorescence microscopy (c) images of C. albicans WT cells grown in the presence of 50 ng ml−1 caspofungin and 100 μM thioflavin T (ThT) documenting the inhibition of cell aggregation by ThT. (d, g) Adhesion force maps (1 μm × 1 μm, color scale: 350 pN) recorded in buffer with an adhesion peptide-tip (d) or a hydrophobic-tip (g) on C. albicans WT cells grown in the presence of caspofungin (50 ng ml−1), harvested and further treated with 100 μM ThT. (e, h) Corresponding adhesion force histograms (n = 1024) together with representative force curves. (f, i) Histograms of rupture distances (n = 1024), and 3-D reconstructed polymer maps (false colors, adhesion forces in green for peptide-tip and orange for hydrophobic-tip).
Biological implications and concluding remarks
A hot topic in current antifungal therapy is to unravel the mechanisms of action of new agents (e.g. caspofungin) exhibiting novel targets (e.g. cell wall glycans). Using AFM techniques, we have shown that treatment of C. albicans with caspofungin leads to major changes in the structural, mechanical and biophysical properties of the cell surface, which, in turn, trigger cell aggregation. Our main findings are as follows: i) AFM topographic imaging reveals that caspofungin induces major morphological and ultrastructural alterations of the cell surface, consistent with the notion that the drug targets cell wall β-1,3 D-glucans; ii) AFM force spectroscopy demonstrates that these structural changes correlate with a substantial decrease of the cell wall mechanical strength, in agreement with the role of β-1,3 D-glucan in maintaining cell shape, mechanical rigidity and resistance to osmotic pressure; iii) single-molecule imaging and manipulation show that the drug induces the massive exposure of Als1 adhesins on the cell surface, which specifically bind to “tϕ+” adhesion structural motifs (on the peptide probe or on opposing cells); iv) Als1 overexpression dramatically increases cell surface hydrophobicity; v) the above cell surface molecular changes represent the driving force for cell aggregation; vi) these features are not observed in yeast species lacking Als1.
In summary, our single-cell and single-molecule experiments demonstrate major remodelling of the C. albicans cell wall in response to antifungals (nanostructure, cell wall elasticity, increased expression of Als1 proteins, hydrophobicity, and cell aggregation). We believe these findings are of great biological significance as they may represent a general mechanism of drug resistance. A unique feature of microbial biofilms is their resistance to a variety of antimicrobial agents, including antibiotics, antiseptics and industrial biocides.1,3,41 In fungi, biofilm drug resistance is believed to reflect multiple mechanisms, including phenotypic changes resulting from a decreased growth rate or nutrient limitation, surface-induced expression of resistance genes, and the presence of a subpopulation of phenotypic variants (“persisters”) that are tolerant to a range of treatments, and restricted penetration of drugs through the biofilm matrix.1,3 Moreover, cell aggregates are also known to be resistant to antifungal drugs. In S. cerevisiae, for instance, cell aggregation mediated by specific cell-surface proteins encoded by the FLO genes (also referred to as “flocculation”) has been shown to protect the cells from multiple stresses, including antimicrobials.42 As Als1 is known to have critical roles in C. albicans adhesion, aggregation and biofilm formation,6,7,43 the Als1 surface remodeling demonstrated here, and the resulting aggregation phenotype, may provide a general strategy used by the pathogen to become resistant against antimicrobial agents. As suggested by Gregory et al., caspofungin-induced cell wall damages are likely to trigger the activation of genes that operate to provide protection from further damage and to coordinate cell wall remodeling during antifungal stress.24
Methods
Microorganisms and cultures conditions
C. albicans SC5314, C. albicans als1Δ/als1Δ,43 C. glabrata ATCC 90030 and S. cerevisiae S288C (MUCL 38902) were cultivated in YPD medium (1 % yeast extract, 2 % Bacto-peptone, 2 % D-glucose) at 30°C and with shaking at 200 rpm. For caspofungin treatment, yeast cells in the early logarithmic growth phase were inoculated for 2 h with caspofungin (kindly provided by Jürgen Heinisch, Universität Osnabrück, Germany) to a final concentration of 50 ng ml−1. In some experiments, thioflavin T (Sigma-Aldrich, Vienna, Austria) was also added to a final concentration of 100 μM, either during growth together with caspofungin (for aggregation assays) or after cell harvest (for AFM experiments). Prior to analysis, yeast cells were always harvested by centrifugation, washed 3 times with sodium acetate buffer and resuspended in 10 ml buffer to a concentration of ~1×106 cells ml−1.
Aggregation assays
The aggregation phenotype was directly observed before and after caspofungin treatment. Washed cells were observed at low magnification by optical microscopy (Zeiss Stemi DV4 Stereo Microscope, Oberkochen, Germany) or at high magnification by optical and fluorescence microscopy (Zeiss Axio Observer Z1 equipped with a Hamamatsu camera C10600, Oberkochen, Germany). In the latter case, Calcofluor White M2R (Sigma-Aldrich, Vienna, Austria) was added at a final concentration of 5 μg ml−1.
Atomic force microscopy
AFM measurements were performed at room temperature (20°C) in sodium acetate buffer, using Nanoscope V Multimode (topographic imaging, single-molecule imaging and chemical force microscopy) and Bioscope Catalyst (stiffness measurements) AFMs from Bruker corporation (Santa Barbara, CA). We used oxide sharpened microfabricated Si3N4 cantilevers with a nominal spring constant of ~0.01 N m−1 (Microlevers, Bruker corporation). The spring constants of the cantilevers were measured using the thermal noise method (Picoforce, Bruker corporation). Yeast cells were immobilized by mechanical trapping into porous polycarbonate membranes (Millipore, Billerica, MA) with a pore size similar to the cell size.44 After filtering a cell suspension, the filter was gently rinsed with the buffer, carefully cut (1 cm × 1 cm), attached to a steel sample puck using a small piece of double face adhesive tape, and the mounted sample was transferred into the AFM liquid cell while avoiding dewetting.
For single-molecule imaging, AFM tips were first functionalized with 14 amino acids peptides, (KLRSMAYKIPTHRR, Eurogentec) containing three adhesion motifs (underlined) described as ligands for Als1,9 by using PEG-benzaldehyde linkers.35 Cantilevers were washed with chloroform and ethanol, placed in an UV-ozone-cleaner for 30 min, immersed overnight into an ethanolamine solution (3.3 g ethanolamine into 6 ml of DMSO), then washed 3 times with DMSO and 2 times with ethanol, and dried with N2. The ethanolamine-coated cantilevers were immersed for two hours in a solution prepared by mixing 1 mg Acetal-PEG-NHS dissolved in 0.5 ml of chloroform with 10 μl triethylamine, then washed with chloroform and dried with N2. Cantilevers were further immersed for 5 min in a 1 % citric acid solution, washed in MilliQ water, and then covered with a 200 μl droplet of PBS solution containing the peptides (0.2 mg ml−1) to which 2 μl of a 1 M NaCNBH3 solution were added. After 50 min, cantilevers were incubated with 5 μl of a 1 M ethanolamine solution in order to passivate unreacted aldehyde groups, and then washed with and stored in buffer. A single cell was first localized using a bare tip, after which the tip was changed with a peptide-functionalized tip. Adhesion maps were obtained by recording 32 × 32 force-distance curves on areas of 1 μm2, calculating the adhesion force for each force curve and displaying the value as a color pixel using the AFM Nanoscope Analysis software 8.15r3 (Bruker corporation, Santa Barbara, CA). For some experiments, buffer solutions containing free peptides (0.1 mg ml−1) or thioflavin T (100 μM) were injected into the AFM chamber. Control experiments were also performed by functionalizing the tips with the irrelevant peptide ESTTTTLNISSE. Unless specified otherwise, all force curves were recorded with a maximum applied force of 250 pN, using a constant approach and retraction speed of 1000 nm s−1.
For quantifying cell surface hydrophobicity by means of chemical force microscopy,38 hydrophobic tips were prepared by immersing gold-coated cantilevers (OMCL-TR4, Olympus Ltd., Tokyo, Japan; nominal spring constant ~0.02 N m−) for 12 h in 1 mM solutions of HS(CH2)11CH3 in ethanol and then rinsed with ethanol. Force curves were recorded with a maximum applied force of 500 pN. Adhesion force values were extracted from each force curve using the AFM Nanoscope Analysis software 8.15r3 (Bruker corporation, Santa Barbara, CA).
Mechanical properties were measured by recording force curves on the cell surface using a maximum applied force of 2 nN. Cell spring constants were extracted from force curves using the AFM Nanoscope Analysis software v1.40r2 (Bruker corporation, Santa Barbara, CA).
Supplementary Material
Acknowledgements
Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS), the Université catholique de Louvain (Fonds Spéciaux de Recherche), the Région Wallonne, the Federal Office for Scientific, Technical and Cultural Affairs (Interuniversity Poles of Attraction Programme), and the Research Department of the Communauté française de Belgique (Concerted Research Action). Work at Brooklyn College was supported by NIH grants SC1 GM083756 and R01 GM098616. We thank J. Heinisch for providing us with caspofungin. Y.F.D. and D.A. are Senior Research Associate and Postdoctoral Researcher of the FRS-FNRS.
Footnotes
Author contributions. S.E.K.C., A.B., D.A., D.N.J., P.N.L., and Y.F.D. designed the research, analysed the data and wrote the paper; S.E.K.C. and A.B. performed the research.
SUPPLEMENTARY INFORMATION AVAILABLE This material is available free of charge via the Internet at http://pubs.rsc.org.
References
- 1.Douglas LJ. Trends Microbiol. 2003;11:30. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 2.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. Microbiol. Mol. Biol. Rev. 2007;71:282. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finkel JS, Mitchell AP. Nat. Rev. Microbiol. 2011;9:109. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. Nat. Rev. Microbiol. 2012;10:112. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verstrepen KJ, Klis FM. Mol. Microbiol. 2006;60:5. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- 6.Hoyer LL. Trends Microbiol. 2001;9:176. doi: 10.1016/s0966-842x(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 7.Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrene YF. Trends Microbiol. 2012;20:59. doi: 10.1016/j.tim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JE., Jr. J. Biol. Chem. 2004;279:30480. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- 9.Klotz SA, Gaur NK, Lake DF, Chan V, Rauceo J, Lipke PN. Inf. Immun. 2004;72:2029. doi: 10.1128/IAI.72.4.2029-2034.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alsteens D, Dupres V, Klotz SA, Gaur NK, Lipke PN, Dufrene YF. ACS Nano. 2009;3:1677. doi: 10.1021/nn900078p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank AT, Ramsook CB, Otoo HN, Tan C, Soybelman G, Rauceo JM, Gaur NK, Klotz SA, Lipke PN. Eukaryot. Cell. 2010;9:405. doi: 10.1128/EC.00235-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rauceo JM, De Armond R, Otoo H, Kahn PC, Klotz SA, Gaur NK, Lipke PN. Eukaryot. Cell. 2006;5:1664. doi: 10.1128/EC.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrene YF, Lipke PN. PLoS One. 2011;6:e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. Proc. Natl. Acad. Sci. 2010;107:20744. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odds FC, Brown AJ, Gow NA. Trends Microbiol. 2003;11:272. doi: 10.1016/s0966-842x(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 16.Pfaller MA. Am. J. Med. 2012;125:S3. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Gow NA, Knox Y, Munro CA, Thompson WD. Med. Mycol. 2003;41:331. doi: 10.1080/13693780310001600859. [DOI] [PubMed] [Google Scholar]
- 18.Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Jones RN. J. Clin. Microbiol. 2011;49:3800. doi: 10.1128/JCM.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douglas CM, D’Ippolito JA, Shei GJ, Meinz M, Onishi J, Marrinan JA, Li W, Abruzzo GK, Flattery A, Bartizal K, Mitchell A, Kurtz MB. Antimicrob. Agents Chemother. 1997;41:2471. doi: 10.1128/aac.41.11.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Letscher-Bru V, Herbrecht R. J. Antimicrob. Chemother. 2003;51:513. doi: 10.1093/jac/dkg117. [DOI] [PubMed] [Google Scholar]
- 21.Bizerra FC, Melo AS, Katchburian E, Freymuller E, Straus AH, Takahashi HK, Colombo AL. Antimicrob. Agents Chemother. 2011;55:302. doi: 10.1128/AAC.00633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunyach C, Drakulovski P, Bertout S, Jouvert S, Reynes J, Mallie M. Mycoses. 2011;54:e62. doi: 10.1111/j.1439-0507.2009.01834.x. [DOI] [PubMed] [Google Scholar]
- 23.Chaffin WL, Lopez-Ribot JL, Casanova M, Gozalbo D, Martinez JP. Microbiol. Mol. Biol. Rev. 1998;62:130. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregori C, Glaser W, Frohner IE, Reinoso-Martin C, Rupp S, Schuller C, Kuchler K. Eukaryot. Cell. 2011;10:1694. doi: 10.1128/EC.05187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. PLoS Pathog. 2008;4:e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markovich S, Yekutiel A, Shalit I, Shadkchan Y, Osherov N. Antimicrob. Agents Chemother. 2004;48:3871. doi: 10.1128/AAC.48.10.3871-3876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinoso-Martin C, Schuller C, Schuetzer-Muehlbauer M, Kuchler K. Eukaryot. Cell. 2003;2:1200. doi: 10.1128/EC.2.6.1200-1210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE., Jr Mol. Microbiol. 2002;44:61. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- 29.Lesage G, Bussey H. Microbiol. Mol. Biol. Rev. 2006;70:317. doi: 10.1128/MMBR.00038-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francius G, Domenech O, Mingeot-Leclercq MP, Dufrêne YF. J. Bacteriol. 2008;190:7904. doi: 10.1128/JB.01116-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaboriaud F, Parcha BS, Gee ML, Holden JA, Strugnell RA. Colloids Surf. B Biointerfaces. 2008;62:206. doi: 10.1016/j.colsurfb.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Arnoldi M, Fritz M, Bauerlein E, Radmacher M, Sackmann E, Boulbitch A. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 2000;62:1034. doi: 10.1103/physreve.62.1034. [DOI] [PubMed] [Google Scholar]
- 33.Gaboriaud F, Bailet S, Dague E, Jorand F. J. Bacteriol. 2005;187:3864. doi: 10.1128/JB.187.11.3864-3868.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karreman RJ, Dague E, Gaboriaud F, Quiles F, Duval JF, Lindsey GG. Biochim. Biophys. Acta. 2007;1774:131. doi: 10.1016/j.bbapap.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Ebner A, Wildling L, Kamruzzahan ASM, Rankl C, Wruss J, Hahn CD, Hölzl M, Zhu R, Kienberger F, Blaas D, Hinterdorfer P, Gruber HJ. Bioconjug. Chem. 2007;18:1176. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- 36.Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. Proc. Natl. Acad. Sci. 2011;108:15775. doi: 10.1073/pnas.1103496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frisbie CD, Rozsnyai LF, Noy A, Wrighton MS, Lieber CM. Science. 1994;265:207. doi: 10.1126/science.265.5181.2071. [DOI] [PubMed] [Google Scholar]
- 38.Dague E, Alsteens D, Latge JP, Verbelen C, Raze D, Baulard AR, Dufrene YF. Nano Lett. 2007;7:3026. doi: 10.1021/nl071476k. [DOI] [PubMed] [Google Scholar]
- 39.Ramsook CB, Tan C, Garcia MC, Fung R, Soybelman G, Henry R, Litewka A, O’Meally S, Otoo HN, Khalaf RA, Dranginis AM, Gaur NK, Klotz SA, Rauceo JM, Jue CK, Lipke PN. Eukaryot. Cell. 2010;9:393. doi: 10.1128/EC.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nobbs AH, Vickerman MM, Jenkinson HF. Eukaryot. Cell. 2010;9:1622. doi: 10.1128/EC.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costerton JW, Stewart PS, Greenberg EP. Science. 1999;284:1318. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 42.Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latge JP, Fink GR, Foster KR, Verstrepen KJ. Cell. 2008;135:726. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. Curr. Biol. 2008;18:1017. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasas S, Ikai A. Biophys J. 1995;68:1678. doi: 10.1016/S0006-3495(95)80344-9. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.