Abstract
Forkhead box O (FOXO) transcription factors are key players in diverse cellular processes affecting tumorigenesis, stem cell maintenance and lifespan. To gain insight into the mechanisms of FOXO-regulated target gene expression, we studied genome-wide effects of FOXO3 activation. Profiling RNA polymerase II changes shows that FOXO3 regulates gene expression through transcription initiation. Correlative analysis of FOXO3 and RNA polymerase II ChIP-seq profiles demonstrates FOXO3 to act as a transcriptional activator. Furthermore, this analysis reveals a significant part of FOXO3 gene regulation proceeds through enhancer regions. FOXO3 binds to pre-existing enhancers and further activates these enhancers as shown by changes in histone acetylation and RNA polymerase II recruitment. In addition, FOXO3-mediated enhancer activation correlates with regulation of adjacent genes and pre-existence of chromatin loops between FOXO3 bound enhancers and target genes. Combined, our data elucidate how FOXOs regulate gene transcription and provide insight into mechanisms by which FOXOs can induce different gene expression programs depending on chromatin architecture.
Keywords: enhancer, FOXO, initiation, RNA pol II, transcription
Introduction
The Forkhead box O (FOXO) family of transcription factors consists of FOXO1, FOXO3, FOXO4 and FOXO6. In model organisms, FOXOs function as tumour suppressors (reviewed in Dansen and Burgering, 2008), and affect both lifespan (reviewed in Partridge and Bruning, 2008; Kenyon, 2010) and stem cell maintenance (Miyamoto et al, 2007; Paik et al, 2007; Tothova et al, 2007; Miyamoto et al, 2008; Yalcin et al, 2008; Paik et al, 2009; Renault et al, 2009; Zhang et al, 2011). An increasing number of FOXO target genes has been identified, responsible for modulating a variety of cellular functions including cell death, proliferation, metabolism and scavenging of reactive oxygen species (reviewed in van der Vos and Coffer, 2011). In general, two conserved pathways regulate FOXO activity. On the one hand, FOXOs are negatively regulated in response to insulin and growth factor signalling (Biggs et al, 1999; Brunet et al, 1999; Kops et al, 1999; Nakae et al, 1999), while positive regulation occurs in the presence of oxidative stress through JNK activity (Brunet et al, 2004; Essers et al, 2004; van den Berg and Burgering, 2011). Gene expression analysis of different cell types in Foxo1, 3, 4 triple knockout mice has shown that FOXO target gene activation is highly context dependent. Little overlap in FOXO-regulated genes between different primary cell types was found, suggesting that FOXOs regulate target gene expression in a cell type-specific manner (Paik et al, 2007). This context-dependent regulation has been attributed to both the output of specific signalling pathways, resulting in different combinations of post-translational modifications on FOXO (Calnan and Brunet, 2008) and to differential co-factor binding (van der Vos and Coffer, 2008, 2011).
Combining genome-wide FOXO-binding profiles with analysis of FOXO-induced changes in gene expression is crucial for understanding FOXO biology. Studies in Drosophila and Caenorhabditis elegans (C. elegans) linked genome-wide binding of FOXO orthologues to changes in gene expression in FOXO-deficient strains and have revealed a complex process in which binding was not the sole determinant of gene regulation (Teleman et al, 2008; Schuster et al, 2010; Alic et al, 2011). These studies are however limited, since changes in transcription are based on steady-state levels in mutants and likely include secondary and adaptive responses. Genome-wide binding studies of mammalian FOXOs using deep sequencing have been performed (Fan et al, 2010; Lin et al, 2010; Litvak et al, 2012), but are limited in the detection of direct consequences of FOXO activity.
In this study, we use a mammalian cell culture system with inducible FOXO3 to study direct consequences of FOXO3 activation. Changes in mRNA levels can be influenced by mRNA stability and might therefore not completely reflect direct effects of FOXO3 induction. We therefore determined changes in RNA polymerase II (RNAPII) occupancy as a more direct measure of transcription factor activity (Mokry et al, 2012). Combined with a FOXO3 genome-wide binding profile, we show FOXO3 acts as a transcriptional activator, regulating target gene expression through transcription initiation. We reveal a function for FOXO3 at distal locations outside of promoter regions and show the existence of loops between several FOXO3 bound distal regions and regulated genes. These data, together with the cell type specificity observed in other studies, suggest a model in which spatial organization of the genome is relevant for FOXO target gene regulation.
Results
Identification of FOXO3-induced transcriptional changes through genome-wide profiling of RNAPII occupancy
To study FOXO3-mediated changes in gene expression, we used DLD1 colon carcinoma cells containing a 4OH Tamoxifen (4OHT) inducible FOXO3A3-ER fusion (DLD1-F3 cells) (also called DL23 cells; Kops et al, 2002b).
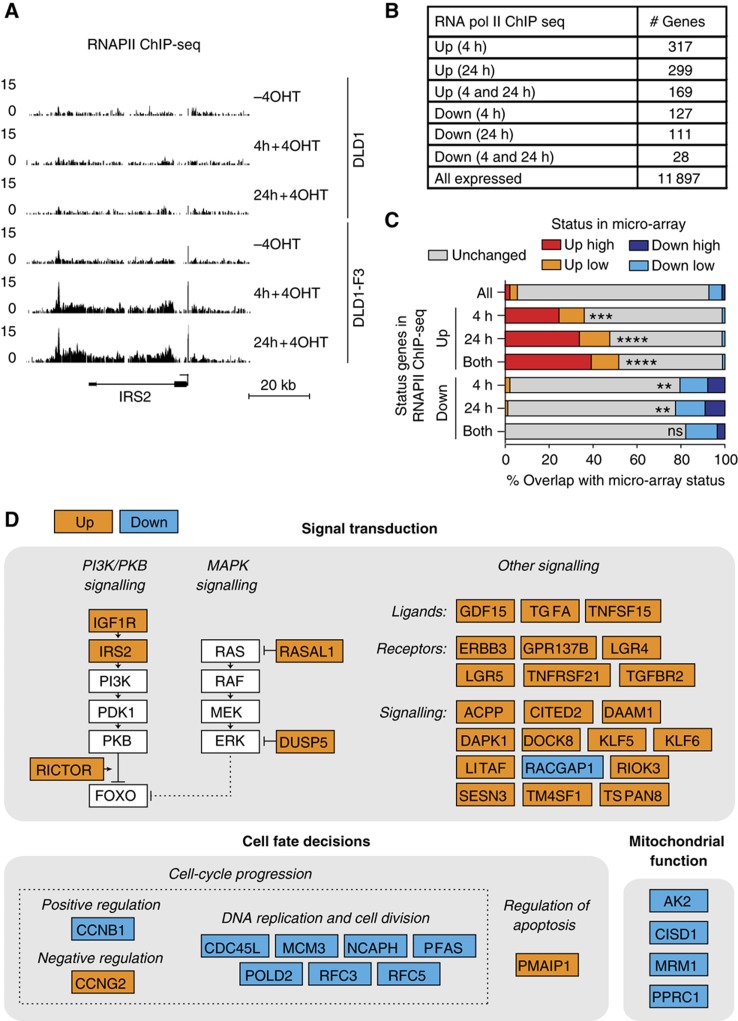
A constitutively active FOXO3A3 mutant, lacking three inhibitory PKB phosphorylation sites, is fused to the ligand responsive domain of the oestrogen receptor. This domain contains a point mutation and is therefore transcriptionally inactive and only responsive to 4OHT (Littlewood et al, 1995). DLD1-F3 cells contain comparable levels of inducible FOXO3A3 relative to endogenous FOXO3 and induction with 4OHT results in the characteristic FOXO effects on cell cycle (Kops et al, 2002b), making it a suitable system to study the global effects induced by the activation of a single transcription factor. Compared with physiological stimuli that activate endogenous FOXOs but also influence parallel pathways, this system allows us to study global effects induced by isolated activation of FOXO3. We first determined RNAPII occupancy by chromatin immunoprecipitation combined with deep-sequencing (ChIP-seq) in DLD1 and DLD1-F3 cells, providing a snapshot of global RNAPII distribution before or after 4 and 24 h upon addition of 4OHT. We calculated changes in RNAPII occupancy at the coding region of all annotated genes. We observed increased RNAPII occupancy specifically in DLD1-F3 cells after 4OHT treatment in numerous genes and identified many known FOXO3 targets as well as potential new FOXO3 target genes (Figure 1A and B; Supplementary Table 1; Supplementary Figure 1). Cross-reference with genes regulated in micro-array analysis (data set recently published in Ferber et al, 2012) revealed considerable overlap, with a higher degree of overlap for up- compared with downregulated genes (Figure 1C). This results in a list of target genes for which expression is altered through changes in transcription. Compared with FOXO micro-array studies, this excludes annotation of FOXO3 targets through transcription-independent effects.
Figure 1.
Genome-wide FOXO3-induced changes in RNAPII occupancy. RNAPII ChIP-seq was performed in DLD1-F3 cells containing 4OHT-inducible FOXO3 and DLD1 control cells. Profiles were generated in both cell lines without 4OHT and 4 and 24 h upon addition of 4OHT. (A) A representative example of RNAPII occupancy profiles. The genomic region surrounding the IRS2 gene locus is shown, for which RNAPII occupancy increases 2.8 (4 h) and 3.9-fold (24 h). Y-axis values represent tag coverage per base per 106 sequenced reads. (B) Numbers of genes in RNAPII occupancy analysis. Total number of genes with detectable RNAPII occupancy and changed RNAPII occupancy specifically in DLD1-F3 cells, 4 and/or 24 h upon addition of 4OHT. (C) Overlap of changes in RNAPII occupancy with changes in mRNA levels measured by micro-array gene expression analysis. Fold change in micro-array is distinguished in low (1.3–2-fold) and high (>2-fold) induction. Hypergeometric test for overlap for the subset of RNAPII changed genes versus all RNAPII occupied genes (ns=P>0.05, *P<0.05, **P<5 × 10−5, ***P<5 × 10−15, ****P<5 × 10−30). (D) Overview of FOXO3-regulated genes. Lead by processes identified in GO term analysis on changed genes, we manually categorized a subset of all genes changed in RNAPII occupancy upon FOXO3 activation, confirmed by changes in mRNA levels. Guided by literature, we subcategorized genes involved in signal transduction, cell fate decisions and mitochondrial function (details in main text).
To gain insight into the functional outcome of direct FOXO3-induced changes, we performed a Gene Ontology (GO) term analysis. The subset of upregulated targets reveals enrichment of genes involved in signalling (Supplementary Table 1). Manual inspection of the list of upregulated genes suggests that multiple signalling pathways are affected upon FOXO3 activation (Figure 1D). FOXO3 induces an auto-regulatory loop as increased expression of PI3K pathway components is observed (IGF1R and IRS2). This likely results in increased PKB/AKT activation in the presence of growth factors ultimately inhibiting FOXO (Kloet and Burgering, 2011). In addition, RICTOR expression increases, which is an mTOR complex 2 component necessary for efficient FOXO phosphorylation and cytoplasmic retention (Brown et al, 2011). Importantly, activation of the PI3K/PKB axis downstream of FOXO activation has been suggested for both mammalian cell systems and model organisms (Ide et al, 2004; Puig and Tjian, 2005; Hui et al, 2008), thus representing a conserved feature of FOXO biology. MAP kinase (MAPK) signalling is potentially inhibited upon FOXO3 activation, in line with the observations in other studies (van den Heuvel et al, 2005). MAPK activity negatively regulates FOXO3 stability (Yang et al, 2008) and in this manner FOXO3 could preserve its own expression. We also identified multiple ligands, receptors and other signalling components, involved in a variety of signal transduction pathways, including EGF, TGF, TNF, WNT and Rho signalling. This implies that FOXO3 activation results in an overall change in the wiring or activity of several signal transduction networks. PMAIP1 (NOXA), a previously identified FOXO target involved in the regulation of apoptosis (Valis et al, 2011), is also induced by FOXO3 activation. Enrichment in cell-cycle-related genes can be observed for downregulated targets (Figure 1D; Supplementary Table 1) as well as downregulation of genes with a role in mitochondrial biogenesis and function. This is in agreement with FOXO3-induced inhibition of cell-cycle progression (Kops et al, 2002b) and mitochondrial function (Ferber et al, 2012). In general, the observed transcriptional changes suggest sensitization of the PI3K/PKB axis, resulting in negative feedback on FOXO activation, inhibition of MAPK signalling and alteration of signalling networks, accompanied by inhibition of cell-cycle progression and mitochondrial function.
FOXO3 regulates target gene expression through RNAPII recruitment
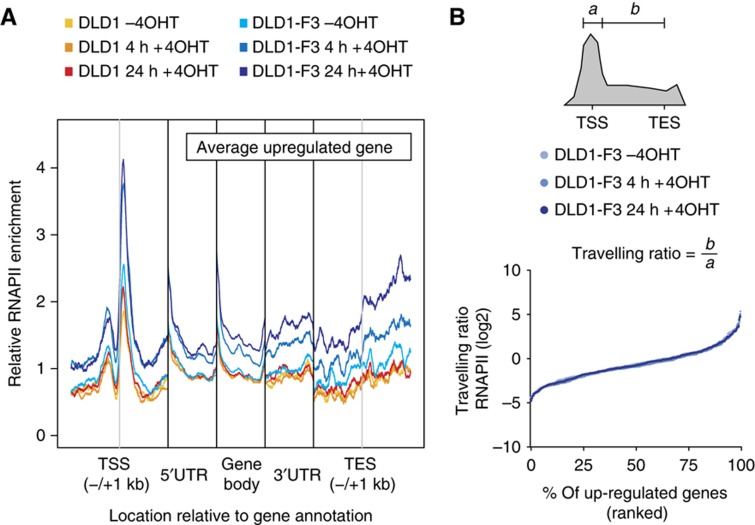
Transcription of eukaryotic genes is regulated at initial recruitment with additional regulation of pause release and elongation into the gene body (Nechaev and Adelman, 2011; Adelman and Lis, 2012). Using the RNAPII profiles, we sought to investigate whether FOXO3-mediated target gene regulation occurs through regulation of RNAPII recruitment or pause release. If regulation of target gene expression would occur through pause release, a shift in RNAPII occupancy from the TSS towards the gene body is expected. We performed meta-gene analysis of RNAPII occupancy over up- and downregulated genes independently categorized by micro-array analysis. Upon induction, an overall increase in RNAPII occupancy in DLD1-F3 profiles can be observed in the subset of upregulated genes (Figure 2A). In addition, we observed no changes in the relative ratio between RNAPII signal around the TSS and in the gene body (travelling ratio, Figure 2B). For transcription factors involved in the regulation of pause release, changes in this ratio have been observed upon alteration of transcription factor activity (Rahl et al, 2010). The preservation of overall distribution is consistent with FOXO3 activation regulating the recruitment of RNAPII, without affecting pause release and the efficiency of early elongation. The increase around the TSS in 4 and 24 h time points is similar, suggesting that full induction of RNAPII recruitment is achieved within 4 h of FOXO3 activation. The downregulated subset does not show any prominent change in the average level of RNAPII (Supplementary Figure 2). This could be due to the smaller overlap between micro-array and RNAPII experiments or different mechanisms of downregulation, which will be discussed later.
Figure 2.
FOXO3 induces target gene expression through recruitment of RNAPII. (A) Average RNAPII occupancy for upregulated genes independently classified by micro-array analysis. Genes were divided into 1 kb regions surrounding TSS and TES, the 5′UTR, 3′UTR and gene body. UTRs and gene body were normalized in size for every gene and average signal was determined for every position, excluding outliers (three highest and lowest values). Signal was normalized to 1 for the DLD1-F3 (-4OHT) profile for all regions separately. (B) Travelling ratio for upregulated genes classified by micro-array analysis. Ratios for RNAPII signals (a=0.3 kb up- and downstream of TSS, b=+0.3 kb from TSS to annotated TES) are ranked by increasing ratio for DLD1-F3 untreated, 4 and 24 h treated separately.
Genome-wide binding profile of human FOXO3 reveals a transcriptional activating role for FOXO3 binding
To further understand the mechanism of FOXO3-induced transcription initiation, we analysed genome-wide binding of FOXO3 by ChIP-seq. In total, we identified 9932 FOXO3-binding sites (peak coordinates in Supplementary Table 2), similar to the amount of FOXO1-binding sites observed by others (Fan et al, 2010; Lin et al, 2010). We confirmed 21 out of 21 FOXO3-binding sites with ChIP followed by qPCR (Supplementary Figure 3), showing reliability of the profile. We found increased FOXO3 binding in several previously described target gene promoters analysed by ChIP-PCR and the majority by reporter assay studies (among others: MXI1 (Delpuech et al, 2007), PIK3CA (Hui et al, 2008), PMAIP1 (Valis et al, 2011), SOD2 (Kops et al, 2002a), PRDX3 (Chiribau et al, 2008), CCNG2 and KLF6 (Terragni et al, 2008)) although not all regions fulfil the peak calling criteria.
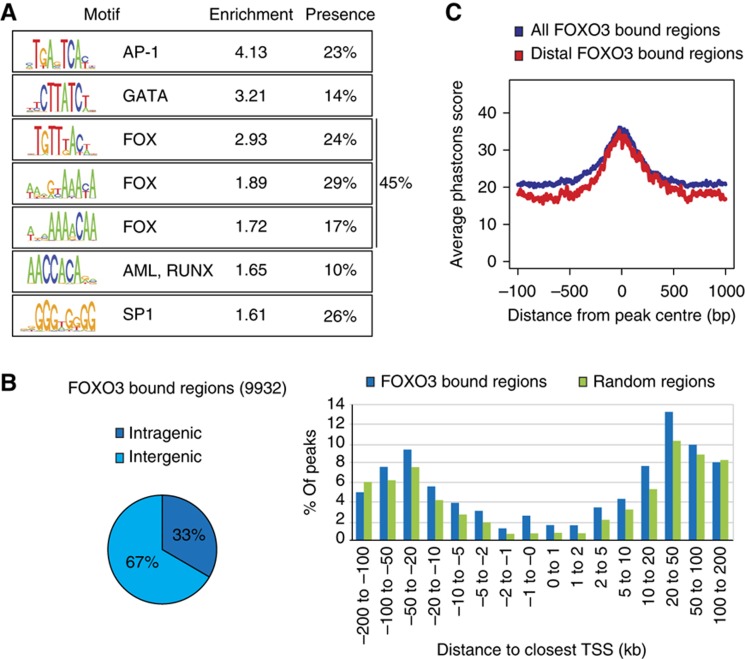
We performed de novo motif search to gain further insight into the sequence-requirements for FOXO3 DNA binding and identified eight sequence motifs enriched in FOXO3 bound regions compared with randomly selected regions. Identified motifs cluster close to the centres of FOXO3-binding sites, are evolutionary conserved and enriched in both strong and also weaker FOXO3-binding sites. Seven out of eight motifs were found to reliably associate with FOXO3 peaks (Figure 3A; Supplementary Figure 4). Three of these motifs fulfil the Forkhead motif criteria (Furuyama et al, 2000; Biggs et al, 2001), in line with direct DNA binding of FOXO3 to these regions. Lower stringency in mapping increased the total fraction of Forkhead motif containing peaks to 61%, showing a full or suboptimal Forkhead motif can be identified in the majority of peaks. In addition, another four identified motifs resemble known published motifs of AP-1 (Angel and Karin, 1991), GATA (Ko and Engel, 1993), RUNX (Meyers et al, 1993) and SP-1 (Kadonaga et al, 1987), suggesting a possible role for these factors in FOXO3 DNA-binding and target gene co-regulation.
Figure 3.
Genome-wide binding profile of human FOXO3. (A) ChIP-seq analysis was performed to identify FOXO3-binding regions in DLD1-F3 cells. De novo motif analysis revealed the presence of seven motifs enriched in the identified FOXO3-binding regions. Relative enrichment to control regions and percentage of peaks containing the motif are shown. (B) Distribution of peak location with respect to annotated genes. Intragenic denotes within transcribed regions. In addition, the distribution relative to the closest TSS, compared with random regions, is shown. (C) Evolutionary conservation of FOXO3 bound regions. Phastcons was used to produce base-by-base conservations scores of all FOXO3 bound regions and distal intergenic bound regions located >5 kb from the closest annotated transcript.
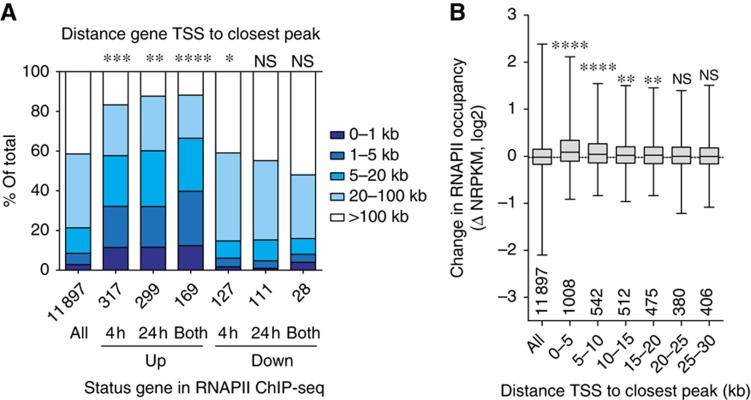
To further relate the location of peaks with respect to potential roles in gene regulation, we determined the distribution according to the closest TSS for all FOXO3 bound regions. The majority of identified binding sites is located outside of promoter regions in distal intergenic regions (>5 kb from any annotated transcript), with a substantial fraction of binding sites found within gene bodies (33%, Figure 3B). Interestingly, the subset of distal FOXO3-binding regions shows a high conservation around the peak centre (Figure 3C), indicating that these distal binding sites have biological relevance. To correlate changes in gene expression with FOXO3 binding, we determined the distance from the TSS to the closest FOXO3 peak for genes with changes in RNAPII occupancy (Figure 4A). On average, upregulated genes are closer to FOXO3 bound regions (up to 66% within 20 kb of the closest peak), compared with all genes (21%). Downregulated genes show a negative correlation (16%), but this is not significant due to the smaller differences and smaller number of genes. Similar conclusions can be drawn from changes in mRNA levels (Supplementary Figure 5). This suggests a pure activating role for FOXO3 DNA binding in recruitment of RNAPII to target genes, which is in agreement with binding data of FOXO orthologues in C. elegans and Drosophila (Schuster et al, 2010; Alic et al, 2011) and represents an evolutionary conserved feature of FOXO transcription factors.
Figure 4.
FOXO3 binding correlates with upregulation of RNAPII occupancy. (A) Comparison of gene status in RNAPII occupancy and distance between TSS and closest FOXO3 peak. The closest peak is determined by distance to the TSS only, irrespective of the location of other genes. The distribution of all RNAPII occupied genes is shown, as well as up- and downregulated (4 h, 24 h and both) genes (total number of genes in each category is indicated below x-axis). Hypergeometric test for number of genes with peak within 20 kb of subsets versus all (ns=P>0.05, *P<0.05, **P<5 × 10−5, ***P<5 × 10−15, ****P<5 × 10−30). (B) Fold change in RNAPII occupancy (4 h) for groups of genes binned by increasing distance of gene TSS to closest FOXO3 peak (group size is indicated above x-axis). The positive correlation is significant up to 20 kb (Mann–Whitney–Wilcoxon test, ns=P>0.05, *P<0.05, **P<0.005, ***P<0.0005, ****P<0.00005).
FOXO3 binds to and activates enhancers
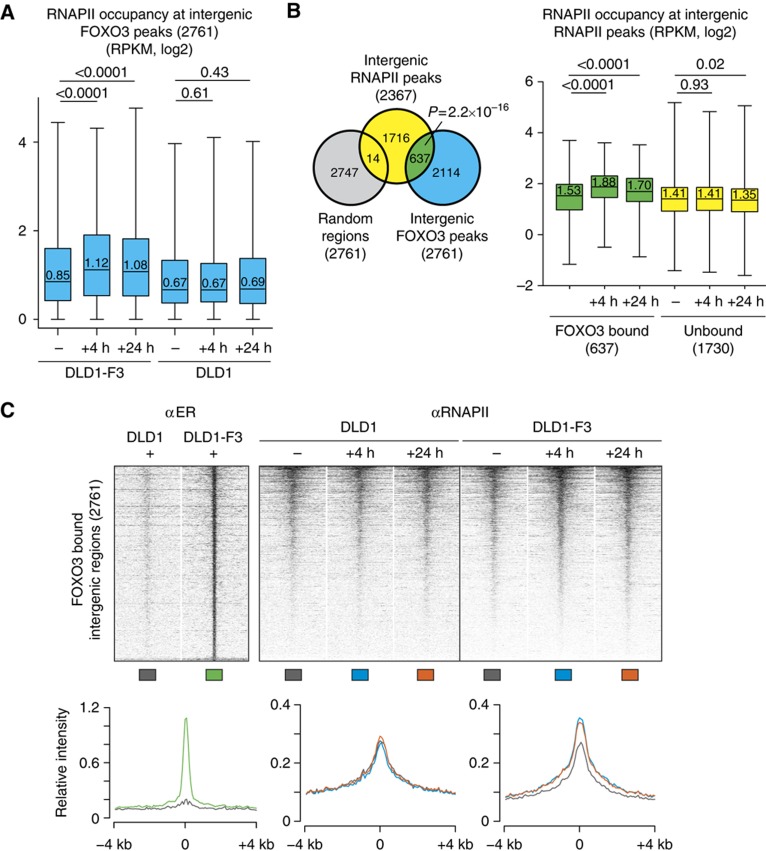
We reasoned that FOXO3 binding to distal regions might be relevant for transcriptional control, since we have observed binding of FOXO3 to many distal regions with increased conservation. Genes with the closest FOXO3 peak as distant as 15–20 kb still show a significant increase in RNAPII (Figure 4B), suggesting that FOXO3 can activate target gene expression from greater distances. Enhancers are DNA elements located distant from promoter regions that can bind transcription factors, transcriptional co-activators and chromatin regulators to affect transcription at promoters (Ong and Corces, 2011; Splinter and de Laat, 2011). Previous studies have reported the presence of RNAPII and transcription at enhancer regions (De Santa et al, 2010; Kim et al, 2010; Ernst et al, 2011). We therefore analysed RNAPII occupancy at distant FOXO3 bound regions and observed an increase in RNAPII levels upon FOXO3 activation (Figure 5A).
Figure 5.
FOXO3 recruits RNAPII to intergenic regions. (A) RNAPII levels at FOXO3 bound intergenic regions. Intergenic regions are located over 5 kb from any known transcript or known H3K4me3 peak. RNAPII signal is specifically increased in DLD1-F3 4 h and 24 h following 4OHT addition, with no change in DLD1 control cells (median within box, P-values from Mann–Whitney–Wilcoxon test, treated versus untreated). (B) RNAPII levels only increase in FOXO3 bound intergenic RNAPII peaks. Overlap between intergenic RNAPII peaks and intergenic FOXO3 peaks or matched random regions is determined. Overlap is higher as expected by chance with FOXO3 peaks compared with matched random regions (Pearson’s χ2-test with Yates’ continuity correction). Boxplot shows levels of RNAPII occupancy only increases in FOXO3 bound intergenic RNAPII peaks compared with unbound RNAPII peaks (category numbers indicated, median within box, P-values from Mann–Whitney–Wilcoxon test for treated versus untreated). (C) Heat map of RNAPII occupancy at all intergenic FOXO3 bound regions. Each row represents the 8-kb region surrounding the FOXO3 peak centre. Regions are sorted by descending RNAPII signal in DLD1-F3, 4 h. Signal intensities are normalized for sequencing depth. Maximum colour intensity corresponds to the 0.98th percentile of the normalized read density distribution. Graphs represent average RNAPII signal of the same 8 kb regions, line colours are indicated below heat maps.
To investigate the contribution of FOXO3-induced peaks to all intergenic RNAPII bound regions, we identified intergenic RNAPII peaks. A total of 2367 intergenic RNAPII peaks was found, similar to numbers from previous studies (De Santa et al, 2010). 637 peaks overlap with FOXO3 peaks (Figure 5B) accompanied by a specific increase in RNAPII occupancy in FOXO3 bound RNAPII peaks only, thereby excluding global changes in RNAPII ChIP efficiency upon FOXO3 activation. In an alternative approach, we created three classes of intergenic RNAPII peaks after FOXO3 induction based on occupancy status: increased, decreased and unchanged (Supplementary Figure 6A and B). The increased class shows the greatest overlap with FOXO3 peaks and, independently of the FOXO3 ChIP-seq, we identified the Forkhead motif by de novo motif search in these regions.
To further analyse the RNAPII binding profiles and changes, we generated heat maps covering 8 kb surrounding all FOXO3 bound distal intergenic regions and plots of the average signal intensity at these locations (Figure 5C). Prior to FOXO3 binding, the RNAPII intensity is high surrounding the FOXO3 peak centre, showing that RNAPII is already present in the majority of FOXO3 bound regions preceding induction. Upon FOXO3 activation, the largest RNAPII occupancy change can be observed directly at the FOXO3 peak centre and spreading up to several kb surrounding the FOXO3-binding site.
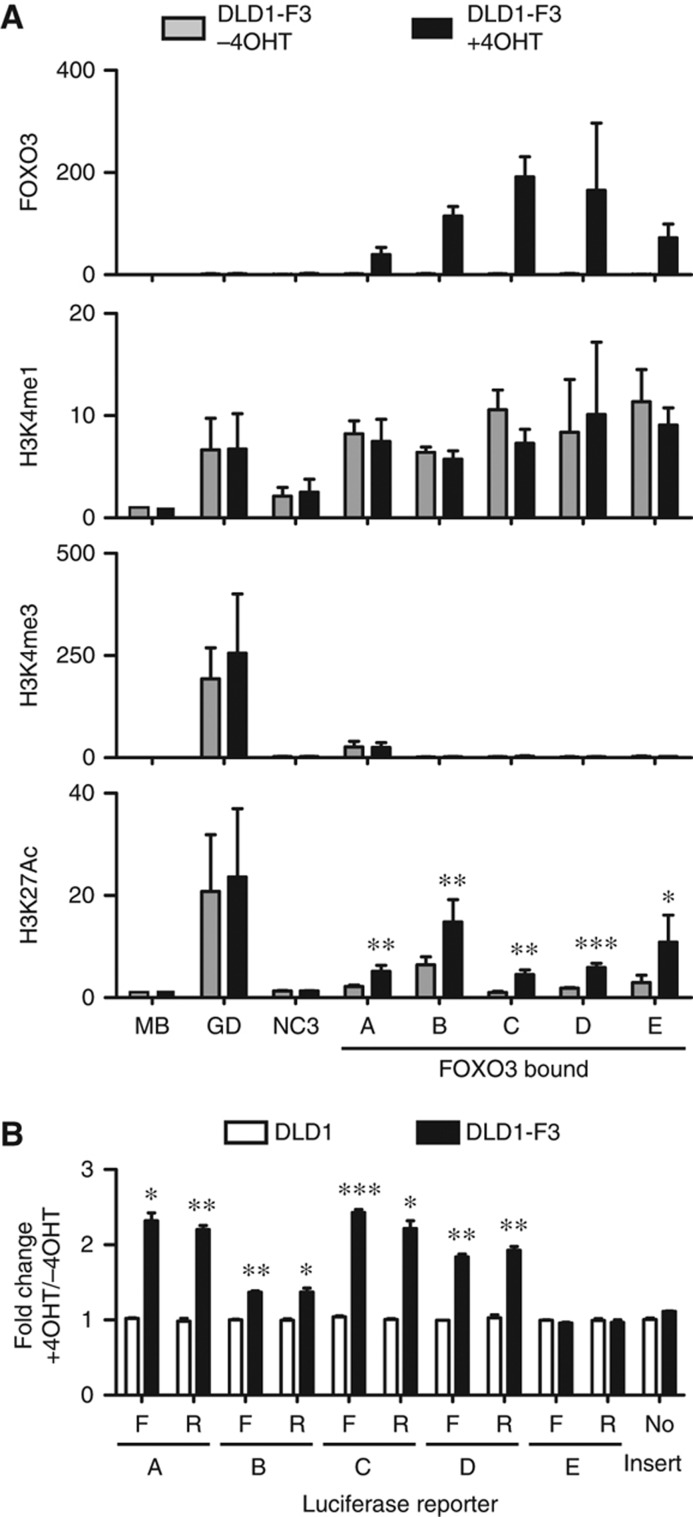
Combinations of modifications of the histone tails reflect the function and status of a locus (Zhou et al, 2011). The canonical chromatin signature of distant enhancers is the presence of Histone H3 Lysine 4 monomethylation (H3K4me1), while trimethylation (H3K4me3) is generally absent (Heintzman et al, 2007, 2009). Enhancers are also marked by the presence of H3K27 acetylation (Ac), associated with active enhancers (Creyghton et al, 2010). We performed ChIP-qPCR with antibodies recognizing specific histone modifications and show five FOXO3 bound regions are high in H3K4me1, while H3K4me3 is low, with an increase in H3K27Ac following FOXO3 activation (Figure 6A). We probed regions surrounding the FOXO3 peaks and could confirm the local enrichment of H3K4me1 and local increase of H3K27Ac (Supplementary Figure 6C). The presence of H3K27Ac prior to FOXO3 activation varies between tested regions. Similar variation can be observed for RNAPII occupancy (Figure 5C; Supplementary Figure 6D), likely reflecting differences in activity. In addition, four out of five sequences were responsive to FOXO3 induction in a luciferase-reporter assay (Figure 6B). This activation was orientation independent, a hallmark of enhancers (Banerji et al, 1981). Together, we show that distant FOXO3-binding sites are marked with a combination of histone modifications specific for enhancers and can obtain a more active signature following FOXO3 activation with increased histone acetylation and RNAPII occupancy.
Figure 6.
FOXO3 binds to and activates enhancers. (A) ChIPs on DLD1-F3 cells (not treated and 2 h following 4OHT treatment) with antibodies recognizing FOXO3, H3K4me1, H3K4me3 and H3K27Ac. Immunoprecipitated DNA was analysed by qPCR for five FOXO3 bound distal regions (closest gene-distance-location relative to TSS: A=DUSP5-25 kb-downstream, B=TGFBR2-128 kb-upstream, C=PDGFRA-5 kb-downstream (intron), D=HMGA-129 kb-downstream (intron), E=KC6-210 kb-upstream) and three unbound regions (GD=GAPDH promoter, MB=Myoglobin locus and NC3=negative control). GD is within the GAPDH promoter region as a positive control for histone ChIPs. Percentage of input was calculated and enrichment relative to MB control region is shown for three to five independent biological replicates (mean with s.e.m., P-value from paired t-test, treated versus untreated, *P<0.06, **P<0.05, ***P<0.005). (B) Luciferase assay on constructs containing a minimal promoter with upstream insertion of the same five FOXO3 bound regions. Orientation specificity was tested by analysing both orientations (F and R) of the same sequence. TK-renilla was co-transfected as internal control and luciferase/renilla values were determined. Fold change after 8 h 4OHT versus untreated were calculated for DLD1 and DLD1-F3 cells (mean of three independent biological replicates, s.e.m., P-value from paired t-test, DLD1-F3 versus DLD1, *P<0.05, **P<0.005, ***P<0.0005). Source data for this figure is available on the online supplementary information page.
FOXO3A3-ER faithfully mimics endogenous FOXO3
We made use of an inducible system to study direct effects of FOXO3 activation. Although considered to act neutral, we wished to exclude any influence from the ER-tag. Therefore, we generated DLD1-DBD cells containing a fusion of the ER moiety to only the FOXO3 DNA-binding domain (DBD) and Nuclear Localization Signal (NLS) (Supplementary Figure 7A). Upon activation with 4OHT, this fusion protein should bind to the same regions as full-length FOXO3, but as it lacks FOXO3 transactivation domains, this will allow us to observe any side effects induced by the ER-tag. We first confirmed binding of the DBD fusion to six regions previously identified to bind full-length FOXO3A3-ER (Supplementary Figure 7B). The binding of the ER-DBD did not induce H3K27 acetylation at these regions (Supplementary Figure 7C). To observe effects of the DBD fusion on RNAPII occupancy, we generated genome-wide profiles of ER-DBD binding and RNAPII changes upon 4OHT treatment in DLD1-DBD cells. We found substantial overlap between ChIP profiles (e.g., 18% of DBD peaks overlaps with called FOXO3A3-ER peaks) and in the vast majority of not overlapping peaks we found a clear signal which was below peak detection limit (Supplementary Figures 8 and 9A). The DBD fusion protein binds to intergenic FOXO3 bound regions, but we did not observe any changes in RNAPII occupancy at these regions or at intergenic DBD bound regions (Supplementary Figure 9). 4OHT treatment in DLD1-DBD cells also does not induce any similar changes as FOXO3A3-ER in gene expression, judged by both RNAPII occupancy (Supplementary Figure 10) and mRNA levels (Supplementary Figure 11). From this we conclude that the ER-tag acts neutral and that the FOXO3A3-ER fusion protein requires the FOXO3 transactivation domains to mediate the observed effects on histone acetylation and RNAPII occupancy.
We employed PI3K/PKB inhibition to induce and study endogenous FOXO3 activation. First, we confirmed binding of endogenous FOXO3 to the same six regions described above and PKB inhibition resulted in increased H3K27Ac at these locations, resembling the increase observed upon activation of exogenous FOXO3 (Supplementary Figure 7). Second, we also generated genome-wide binding profiles of endogenous FOXO3, in which we found considerable overlap between endogenous and exogenous ChIP profiles (Supplementary Figure 8), revealed by overlap in called peaks (34% of endogenous FOXO3 peaks overlaps with called FOXO3A3-ER peaks) and increased signal around peak centres from both data sets (Supplementary Figures 8B and 9A). In addition, to compare the above-described effects of exogenous FOXO3 activity with the effects of PKB inhibition, we generated RNAPII profiles in DLD1-F3 cells comparing addition of 4OHT and PKB inhibitor. At the distal intergenic endogenous and exogenous FOXO3 bound locations identified above, PKB inhibition mimics the increase in RNAPII occupancy upon activation by 4OHT (Supplementary Figure 9). Next, we calculated RNAPII occupancy changes at gene coding regions and compared these with above-described results (Figure 1). We identified the majority of genes (Supplementary Figure 10) to again be changed by 4OHT treatment: i.e., over 51% (0.5-fold cutoff) or 83% (0.2-fold cutoff) of genes previously identified to be upregulated by 4- and 24-h treatment with 4OHT were again identified to be upregulated. In addition, PKB inhibition induced similar changes with less fold induction, showing 17% (0.5-fold) and 60% (0.2-fold) of these same genes to be upregulated. Finally, we confirmed induction in mRNA levels for six genes upon activation of exogenous FOXO3 and PI3K/PKB inhibition (Supplementary Figure 11), repeatedly showing clear induction but slightly lower fold change compared with FOXO3A3-ER.
Our analysis shows that upon PI3K/PKB inhibition, endogenous FOXO3 binds to the same regions as FOXO3A3-ER. We also show concomitant similar changes on RNAPII profiles and histone acetylation. Generally, changes induced by PKB inhibition are somewhat smaller relative to activation of FOXO3A3-ER. Likely the FOXO3A3-ER system provides a more robust profile as FOXO3 activation through PKB inhibition still requires dephosphorylation of FOXO3, which likely results in a slower response. Alternatively, PKB inhibition might not induce the same level of constitutive activation like the A3 mutant does, since PKB inhibition will likely also affect signalling to other transcription factors that for example may enhance or reduce FOXO only effects. Thus, isolated activation of FOXO3 in our inducible system could also lack activation of other factors possibly cooperating with FOXO3 in target gene regulation. The great overlap in 4OHT and PKB inhibition induced changes (Supplementary Figure 10C), however, suggests that generally FOXO3 requires cooperation with activators already active under conditions studied.
FOXO3 bound enhancers communicate with target gene promoters through pre-existing DNA loops
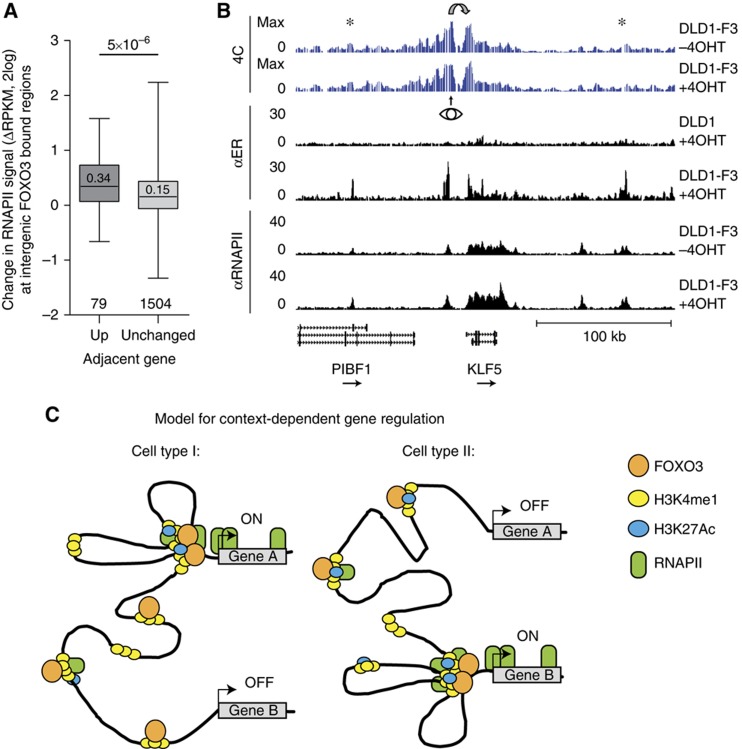
We initially employed the ER inducible system because we aimed to identify direct effects of FOXO3 activation. Unlike inhibition of the PI3K/PKB pathway, this system allows us to relate all observed changes specifically to activation of only FOXO3. We therefore sought to investigate a possible relation between the increase of RNAPII at intergenic FOXO3 bound regions and FOXO3-induced target gene regulation. Transcription of RNAPII occupied enhancer regions has been reported to precede the activation of adjacent protein-coding genes (De Santa et al, 2010). Indeed, we could observe a positive correlation between the RNAPII increase at FOXO3 bound regions and upregulation of the adjacent genes (Figure 7A; Supplementary Figure 12A), which could be explained by communication between FOXO3 bound regions and the neighbouring target gene. To examine whether FOXO3 bound regions can physically loop towards the TSS of a target gene, we performed high-resolution chromosome conformation capture analysis followed by sequencing (4C-seq) (Splinter et al, 2011). 4C can be used to generate a map of the spatial interactions of a selected locus. Based on visual inspection of regions around FOXO3-induced target genes, we selected candidate regions with increased RNAPII occupancy at both FOXO3 bound putative enhancer regions and target genes. Primers were designed for seven view points, consisting of four intergenic FOXO3 bound regions and three TSSs from induced genes (Supplementary Table 3). Figure 7B shows the region around the KLF5 locus, for which the RNAPII occupancy increases (1.9-fold after 4 h). 4C primers were designed around a FOXO3 bound region 18 kb upstream from the TSS for which the RNAPII occupancy also increases (2.1-fold after 4 h). Typical for a 4C profile, the frequency of captured genomic regions decreases as a function of the distance to the viewpoint. However, capture frequency greatly increases around the TSS of KLF5, indicating looping of the FOXO3 bound intergenic region towards the TSS region. The signal also increases slightly around other FOXO3 bound regions, hinting at the close proximity of multiple enhancers to the target gene TSS. The 4C profile is highly similar before and after FOXO3 induction, suggesting that FOXO3 binds to pre-existing loops and does not induce reorganization of the spatial environment of the locus. For three other regions, we find evidence of pre-existing loops between FOXO3 distant binding sites and target genes (Supplementary Figure 12). For the IRS2 gene, where we look from the TSS, we see looping to two upstream FOXO3-binding sites. It is important to note that the closest (and strongest) FOXO3 peak is not engaged in an interaction with the TSS of IRS2. Together, these results suggest pre-existing communication between several FOXO3 bound distal regions and regulated target genes.
Figure 7.
Communication between distal intergenic RNAPII and FOXO3 bound regions and adjacent target gene expression. (A) Status of the adjacent gene is predictive for RNAPII increase at intergenic FOXO3 bound regions. The status of the closest gene for every FOXO3 bound intergenic RNAPII peak was determined for all peaks within 5–100 kb to the TSS of the closest gene. Fold change in RNAPII occupancy (DLD1-F3 cells, 4 h versus untreated) for peaks with unchanged and upregulated adjacent gene are shown (category numbers indicated, median within box, P-value from Mann–Whitney–Wilcoxon test). (B) 4C signal and RNAPII and FOXO3 occupancy surrounding the KLF5 locus. An 18-kb upstream FOXO3 and RNAPII occupied putative enhancer was used as viewpoint (eye) for 4C analysis. Stars denote increased signal in other FOXO3 bound RNAPII occupied intergenic regions. (C) Model for FOXO3-induced target gene expression. Enhancers are marked by the presence of H3K4me1 and FOXO3 binding is accompanied by an increase of H3K27Ac and recruitment of RNAPII. Cell type specificity of enhancers and chromatin architecture could explain context dependency in gene activation. A model is shown in which pre-existing enhancers and promoter–enhancer loops are responsible for differential responses to FOXO activity; gene A is FOXO responsive in cell type I, while gene B is responsive only in cell type II.
Discussion
Using genome-wide approaches we have analysed how FOXO3 regulates target gene expression. To this end, we measured FOXO3 binding to genomic DNA and changes in RNAPII occupancy following FOXO3 activation. This analysis reveals several features of FOXO3-mediated gene regulation.
First, binding of FOXO3 to gene regulatory regions activates transcription and gene expression, whereas repression of gene expression following FOXO3 binding generally occurs through DNA-binding independent or indirect mechanisms. FOXO3-induced MYC-mediated gene repression possibly entails an important mechanism by which FOXO indirectly represses gene expression. We observe an increase in MXI1 expression, a previously reported FOXO3-induced negative regulator of MYC function (Delpuech et al, 2007). The MYC DNA-binding sequence is enriched in promoter regions of downregulated genes (CACGTG, (Blackwell et al, 1990), Supplementary Figure 13). This is in agreement with other studies that show an intricate and extensive inverse correlation between FOXO and MYC function (Kloet and Burgering, 2011). In addition, induction of cell-cycle inhibitor CCNG2 could be responsible for downregulation of cell-cycle genes. Other cell-cycle regulators previously reported to be regulated by FOXO (van der Vos and Coffer, 2011) were not changed above the thresholds used in this study, but possibly still contribute. Alternatively, the smaller overlap between RNAPII and mRNA levels in downregulated genes could reflect changes in mRNA stability rather than transcriptional changes. Although the activating role for FOXO3 DNA binding in gene expression is conserved and reported for C. elegans and Drosophila FOXO orthologues as well (Schuster et al, 2010; Alic et al, 2011), some examples of FOXO-mediated suppression of target gene expression have been reported. This includes repression of CCND1 (Schmidt et al, 2002), ID1 (Birkenkamp et al, 2007) and, during revision of this manuscript, FOXO3-mediated suppression of several antiviral genes in macrophages was reported (Litvak et al, 2012). Although our data set clearly shows FOXO3 DNA binding is correlated with transcriptional activation only, we do not wish to exclude the possibility that FOXO3 activation induces repression of individual genes.
Second, the changes observed in the RNAPII profile upon FOXO3 activation are consistent with FOXO3 regulating transcription initiation rather then pause release and elongation. This is in contrast to yeast forkheads, which have been reported to regulate RNAPII elongation (Morillon et al, 2003). We observe FOXO3 binding within the coding region of genes for roughly one-third of the peaks. In general, we can detect a slightly higher signal in the FOXO3 ChIP-seq throughout transcribed genes, but it is unclear whether this reflects genuine FOXO3 binding or a nonspecific increase in background signal. Binding of yeast forkheads and Drosophila FOXO within transcribed regions has been observed as well and has been proposed to be necessary for elongation (Morillon et al, 2003; Alic et al, 2011). Since our findings argue against a role for FOXO3 in pause release and elongation, the relevance of FOXO3 binding within genes is unclear.
Generally, pausing of RNAPII after initial recruitment seems specifically enriched at genes in signal-responsive pathways (Adelman and Lis, 2012) possibly allowing a quick response in gene expression upon stimulation. In light of this, we consider it surprising to only find evidence for the regulation of initiation by FOXO3 activation.
Third, the FOXO3 ChIP-seq resulted in the identification of nearly ten thousand FOXO3-binding sites. In addition to the Forkhead motif, we identified four motifs enriched in FOXO3 bound regions, resembling known published motifs for AP-1, GATA, RUNX and SP1. ChIPs on FOXO orthologues in Drosophila adults and C. elegans have identified AP-1 (C. elegans only) and GATA (both) binding sequences in a subset of FOXO bound regions as well, suggesting that the co-occurance is evolutionary conserved (Schuster et al, 2010; Alic et al, 2011). We could, however, not find any correlation between motif presence and RNAPII changes. From our data set it is therefore unclear to what extent these motifs, and the proteins that bind these sequences, influence FOXO3-induced target gene expression.
Pioneer transcription factors are transcription factors thought to be capable of initial binding to regulatory sequences, allowing binding of other factors by opening compacted chromatin, initiating the events that will ultimately result in transcriptional activation. Forkhead transcription factor FOXA1 has been studied extensively for its pioneering activity (reviewed in Zaret and Carroll, 2011). FOXO1 has been shown to similarly open compacted chromatin in vitro (Hatta and Cirillo, 2007; Hatta et al, 2009) suggesting that it may function as a pioneer factor as well. Reports on correlation of changes in histone modifications and FOXA1 activity are inconsistent, either reporting the presence of H3K4me1 upstream of, and importance for, FOXA1 binding and/or recruitment (Lupien et al, 2008) or induction of methylation after FOXA1 binding to regions lacking H3K4me1 (Serandour et al, 2011). Based on this it is unclear whether the higher level of H3K4me1 at several FOXO3-binding sites conflicts with the hypothesis of FOXO3 as a pioneer. The genome-wide RNAPII occupancy profiles revealed the local presence of RNAPII prior to FOXO3 binding in the majority of studied regions, suggesting that in general FOXO3-binding regions are already in a more open and potentially active state. This would argue against FOXO3 acting as a pioneering factor in these regions under the conditions used in this study, without excluding that FOXO3 acts as a pioneer in a subset of bound regions that lack RNAPII signal before FOXO3 binding. GATA has been described to function as a pioneering factor as well (reviewed in Zaret and Carroll, 2011), and RUNX1 was shown to be transiently required for chromatin unfolding for initial transcription (Hoogenkamp et al, 2009). The enrichment of GATA and RUNX binding motifs within FOXO3 bound regions might therefore indicate these to act as pioneering factors that thereby cooperate with and/or precede FOXO3 binding. Alternatively, FOXA1 may act as a pioneering factor for FOXO3 since FOXA1 binds the same Forkhead motif as FOXO3. However, for the same reason this possibility can therefore also not be addressed by our data through analysis of motif presence and would require detailed time course analysis of genome binding for multiple Forkhead members, including FOXA1 and FOXO3.
Fourth, we found a function for distal FOXO3 bound regions in target gene regulation. These regions are marked by the presence of canonical enhancer specific histone modifications and display a more active signature upon FOXO3 activation, including an increase in H3K27Ac and recruitment of RNAPII. Direct binding between p300 histone acetyltransferase and FOXOs has been observed in many studies (Calnan and Brunet, 2008). Since p300 has been linked to transcription initiation (Kraus et al, 1999) and binding to enhancers has been reported (Heintzman et al, 2007; Xi et al, 2007; Visel et al, 2009), recruitment of p300 by FOXO3 could be responsible for the observed increase in H3K27Ac at enhancers and RNAPII recruitment to enhancers and target genes.
Genome-wide studies for FOXO binding in Drosophila and C. elegans have not reported binding to distant sites, perhaps in part because these genomes are much smaller. Potentially, FOXO regulation of transcription from distant sites might be an acquired feature of mammalian FOXO biology not present in insects and nematodes. Other mammalian studies have identified FOXO binding in promoter regions and also at more distal locations, possibly at enhancers (Fan et al, 2010; Lin et al, 2010; Chen et al, 2011), but to what extent these contribute to regulation of gene expression was unclear. To our knowledge, no studies have reported looping of FOXO occupied regions to promoters of target genes. Over the past years, several proteins including GATA and CTCF, have been reported to be involved in looping in transcriptional regulation (Sexton et al, 2009). This could be an alternative explanation for the identification of the GATA motif in FOXO3 bound regions. In addition, the CTCF motif was found enriched in Drosophila FOXO bound regions (Alic et al, 2011). However, regulation and formation of enhancer–promoter looping is complex and establishing any involvement of these factors in FOXO target gene regulation requires thorough investigation. We observed that several FOXO3 bound regions might fold to one promoter. Regulation of target genes through several regulatory regions simultaneously could be necessary for tightly controlled gene expression, for example, through setting a threshold. This might therefore also explain why the number of FOXO3 bound regions greatly outnumbers the sum of regulated genes. Alternatively, many FOXO3 bound regions might not be functional in gene regulation in a given tissue or in general. In C. elegans a similar observation of DAF-16-binding sites outnumbering DAF-16-regulated genes has led to the suggestion that many sites are ‘parking sites’ occupied to ensure rapid DAF-16 transcriptional response when required (Schuster et al, 2010). Although we are not aware of reports showing chromatin looping being present and functionally relevant in C. elegans, our observation that several FOXO3-binding sites may co-localize in three dimensions clearly indicates an alternative possibility besides simple ‘parking’.
The pre-existing nature of the enhancer–promoter loops and the cell type specificity of enhancers and chromatin architecture (Ernst et al, 2011; Sanyal et al, 2012) suggest that cells are poised for specific behaviour. Variation in overall chromatin architecture between different systems may therefore at least in part explain the previously mentioned context-dependent nature of FOXO activation. Assessing chromatin architecture around differently responding target genes may prove necessary to determine the regulatory regions responsible for FOXO-induced changes in gene expression.
Our analysis has provided insight into the molecular mechanisms by which FOXO3 induces target gene regulation. We have revealed a function for FOXO3 outside of promoter regions. We have observed binding of FOXO3 to enhancers accompanied by the acquirement of a more active signature. Finally, our data suggest importance of pre-existing enhancers and chromatin architecture for FOXO3 activated transcription and context-dependent gene regulation (Figure 7C).
Materials and methods
Cell culture
DLD1, DLD1-F3 and DLD1-DBD cells were grown on RPMI-1640 medium with 10% FCS and standard supplements. 4-Hydroxy-tamoxifen (4OHT) from Sigma was dissolved in ethanol and added to cells at a final concentration of 1 μM. Cells were treated with PI3K inhibitor LY 294002 (Enzo Life Sciences) at a concentration of 10 μM and PKB inhibitor VIII (Santa Cruz) at a concentration of 10 μM. pcDNA3-ER-DBD-NLS was generated by amplification of DBD-NLS domains of FOXO3, which were amplified from FOXO3A3 plasmid using forward primer GCGCGCGGATCCGCTGGGGGCTCCGGGCAG and reverse primer GCGCGCCTCGAGTCAGGCTGCGCGGCCACGGCT, digested with BamHI and XhoI and ligated into pcDNA3-ER vector. DLD1-DBD cells were generated by transfection with fugene HD (Promega), selected with G418 (Santa Cruz, 500 ng/ml) and clonal lines were generated by limited dilution. Protein levels were determined by SDS–PAGE and western blot, with anti-ER (Santa Cruz, MC-20) and anti-tubulin (Calbiochem, CP06).
RNAPII chromatin immunoprecipitation and sequencing
Approximately 20 × 106 DLD1 and DLD1-F3 cells grown in the absence or presence of 4OHT for 4 and 24 h were used for ChIP-seq procedure. ChIP was performed according to previously described procedure (Hatzis et al, 2008; Mokry et al, 2010), details in Supplementary methods, using anti-RBP1 (PB-7C2) antibody (Euromedex). Chromatin was additionally sheared, end-repaired, sequencing adaptors were ligated and the library was amplified by LMPCR. After LMPCR, the library was purified and checked for the proper size range and for the absence of adaptor dimers on a 2% agarose gel and sequenced on SOLiD/AB sequencer to produce 50-bp long reads. Sequencing reads were mapped against the reference genome (hg19 assembly, NCBI build 37) using the BWA package (Li and Durbin, 2009). Nonuniquely placed reads were discarded. If more than five reads mapped to the same location and strand, only five reads were used for further analysis. For duplicate experiments, two times 20 × 106 DLD1-F3 cells were grown in the absence or presence of 4OHT or PKB inhibitor for 4 h. For profiling in DLD1-DBD cells, 20 × 106 cells were grown in the absence or presence of 4OHT for 4 h. Sequencing reads were again mapped against the reference genome and nonuniquely placed reads were discarded.
Quantification of RNAPII occupancy
To set gene expression from RNAPII ChIP-seq data, we counted the number of the sequencing tags aligned to annotated transcript coordinates (Mokry et al, 2012). To avoid transcripts with zero mapped tags to interfere with logarithmic transformation of read counts, one read per every 10 million sequencing tags was added to each transcript. Raw read counts were normalized to the transcript length (from TSS to TES) and sequencing depth and quantile normalized. All samples are presented as quantile normalized read counts per transcript per kb of transcript per million sequencing tags (NRPKM), log2 scale. Expressed gene is gene with an expression higher than −1.5 NRPKM in at least one condition within an experiment. Differentially transcribed genes were set as genes with at least 0.5 (log2) fold NRPKM change in DLD1-F3 4 and 24 h compared with untreated, while fold change in DLD1 cells is less than 0.2 (log2). Changes in RNAPII occupancy in DLD1 samples were used to estimate noise and determine cutoff values (Supplementary Figure 1). If differences in fold change between annotated transcripts of the same gene occur, the highest fold change was used for further analysis. To identify differentially transcribed genes in replicate experiments using 4 h of 4OHT or PKB inhibition, average NRPKM was determined prior to the calculation of fold changes. For comparison between data sets (Supplementary Figure 10), all genes below −1.5 NRPKM expression threshold in either one of the experiments or changed over 0.2-fold in DLD1 cells following 4OHT treatment were excluded from analysis.
GO term analysis
GO term analysis was performed using Database for Annotation, Visualization and Integrated Discovery (DAVID). Regulated genes were determined by a 0.5-fold (log2) change in RNAPII occupancy at 4 and/or 24 h in DLD1-F3 only, for which changes in mRNA levels are confirmed in micro-array gene expression analysis. All RNAPII occupied genes were used as a background list.
Determination of intergenic RNAP II peaks
Cisgenome software package (Ji et al, 2008) was used for the identification of RNAPII peaks from the ChIP-seq data. Regions within 5 kb of a known transcript (RefSeq) or H3K4me3 peak from a published data set in DLD1 cells (Tanimoto et al, 2010) are excluded. Regions larger than 3 kb in size were discarded as putative not-annotated genes. Only peaks identified in at least two out of four samples (DLD1 0 h, DLD1-F3 0 h, 4 h and 24 h) were included.
Chromatin immunoprecipitation of FOXO3 and histone modifications
DLD1, DLD1-F3 and DLD1-DBD cells were grown in the absence or presence of 4OHT or PKB inhibitor. For ER and FOXO3 ChIP-seq, 40 × 106 DLD1 and DLD1-F3 cells were used, while 10 × 106 DLD1-F3 cells were used for histone modifications. IPs were performed for 16 h with the following antibodies: 1 μg of H3K4me1, H3K4me3 and H3K27Ac and total H3 from Abcam (ab8895, ab8580 and ab4729, ab1791), 5 μg of αER or αFOXO3 (Santa Cruz, MC-20 and H144) and 5 μg of normal rabbit IgG (Santa Cruz). ChIPs were performed according to a previously described protocol from the Brunet lab (Renault et al, 2011), details in Supplementary methods. Sequencing library preparation, sequencing and mapping were performed for FOXO3 samples as for the RNAPII ChIP-seq. Multiple reads mapping to same location and strand have been collapsed to a single read and only uniquely placed reads were used for peak-calling. Cisgenome software package (Ji et al, 2008) was used for the identification of binding peaks from the ChIP-seq data and further analysis. Cisgenome 1.1 was used for calling initial DLD1-F3 aER data set: −c 0.2, −w 100, −s 25, −p 0.5, −g 200 –l 225, DLD1 aER sample was used as background, For all other data sets Cisgenome 2 with settings: −e 50, −maxgap 200, minlen 200; was used; for endogenous FOXO3 ChIPs IgG was used as background, for RNAPII human input DNA from (Mokry et al, 2010). Conservation was determined by preformatted PhastCon scores downloaded from Cisgenome. qPCR analysis was performed with IQ SYBR-Green mix (Bio-Rad), see primer sequences in Supplementary Table 3.
De novo motif discovery
Motif analysis was done using CisModule function (Zhou and Wong, 2004) incorporated in Cisgenome sofware, using the following parameters: motif number K=15, mean motif length Lambda=10, maximal motif length allowed=18, initial motif length=10, initial module size D=3.0, module length=100, order of background Markov chain=3, MCM iteration=500. For lower stringency in Forkhead motif mapping, r=50 instead of r=500 was used (−r is defined in the Cisgenome utility: motifmap_matrixscan_genome). A combination of Cisgenome functions, custom PERL and R scripts was used for additional data analysis. Similarity search with known transcription factor binding motifs was performed using TOMTOM motif comparison tool (Gupta et al, 2007).
Luciferase assay
DLD1 and DLD1-F3 cells were transfected using Fugene HD (Promega), with TK-renilla and pGL3-promoter vector without insert or with inserts generated by PCR and digestion with KpnI (primers in Supplementary Table 3) from genomic DLD1-F3 DNA. Lysis and subsequent determination of luciferase and renilla counts was performed 48 h after transfection using the Dual Luciferase Reporter Assay (Promega).
4C
4C analysis was performed with 20 × 106 DLD1-F3 cells cultured in the absence or presence of 4OHT for 4 h as described previously (Splinter et al, 2011), but with DpnII and Csp digestion. PCR was performed on 10 times 100 ng of 4C template for each primer pair and pooled prior to purification. Primers are listed in Supplementary Table 3.
Statistical analysis
Analysis were performed in R, Excel or Graph Pad. The details of tests used are given in figure captions.
Online access data sets
ChIP-seq and 4C raw data from this publication have been deposited at GEO with the accession numbers GSE35486 (ChIP-seq) and GSE36835 (4C).
Supplementary Material
Supplementary Figures S1–13, Supplementary Table Legends
FOXO3 induced transcriptional changes.
List of FOXO3A3-ER, endogenous FOXO3 and ER-DBD bound regions.
Sequences of all primers used for ChIP-qPCR, 4C, cloning of luciferase-reporters and qPCR on cDNA.
Acknowledgments
We thank S Holwerda for advice, H de Ruiter for technical support and HTM Timmers, DE Kloet and MC van den Berg for critical reading of the manuscript and discussion. We thank all members of the Burgering group for discussion and suggestions. We thank Ashley Webb (Brunet lab, Stanford University, USA) for sharing protocols. Research was in part funded by CBG (Centre for Biomedical Genetics), CGC (Cancer Genetics Centre) and CTMM (Centre for Translational Molecular Medicine).
Author contributions: AE, MM, BB, EW, AS, WL, EC designed the experiments. AE, MM, LS, PP, MT and RB performed the experiments. MM, AE and EW performed the data analysis. AE and BB wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adelman K, Lis JT (2012) Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alic N, Andrews TD, Giannakou ME, Papatheodorou I, Slack C, Hoddinott MP, Cocheme HM, Schuster EF, Thornton JM, Partridge L (2011) Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P, Karin M (1991) The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta 1072: 129–157 [DOI] [PubMed] [Google Scholar]
- Banerji J, Rusconi S, Schaffner W (1981) Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299–308 [DOI] [PubMed] [Google Scholar]
- Biggs WH 3rd, Cavenee WK, Arden KC (2001) Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mammalian Genome 12: 416–425 [DOI] [PubMed] [Google Scholar]
- Biggs WH 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC (1999) Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA 96: 7421–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp KU, Essafi A, van der Vos KE, da Costa M, Hui RC, Holstege F, Koenderman L, Lam EW, Coffer PJ (2007) FOXO3a induces differentiation of Bcr-Abl-transformed cells through transcriptional down-regulation of Id1. J Biol Chem 282: 2211–2220 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Kretzner L, Blackwood EM, Eisenman RN, Weintraub H (1990) Sequence-specific DNA binding by the c-Myc protein. Science 250: 1149–1151 [DOI] [PubMed] [Google Scholar]
- Brown J, Wang H, Suttles J, Graves DT, Martin M (2011) Mammalian target of rapamycin complex 2 (mTORC2) negatively regulates Toll-like receptor 4-mediated inflammatory response via FoxO1. J Biol Chem 286: 44295–44305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96: 857–868 [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Calnan DR, Brunet A (2008) The FoxO code. Oncogene 27: 2276–2288 [DOI] [PubMed] [Google Scholar]
- Chen Z, Xiao Y, Zhang J, Li J, Liu Y, Zhao Y, Ma C, Luo J, Qiu Y, Huang G, Korteweg C, Gu J (2011) Transcription factors E2A, FOXO1 and FOXP1 regulate recombination activating gene expression in cancer cells. PloS One 6: e20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiribau CB, Cheng L, Cucoranu IC, Yu YS, Clempus RE, Sorescu D (2008) FOXO3A regulates peroxiredoxin III expression in human cardiac fibroblasts. J Biol Chem 283: 8211–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB, Burgering BM (2008) Unravelling the tumor-suppressive functions of FOXO proteins. Tr Cell Biol 18: 421–429 [DOI] [PubMed] [Google Scholar]
- De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8: e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A (2007) Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol 27: 4917–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BM (2004) FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J 23: 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Morinaga H, Kim JJ, Bae E, Spann NJ, Heinz S, Glass CK, Olefsky JM (2010) FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J 29: 4223–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber EC, Peck B, Delpuech O, Bell GP, East P, Schulze A (2012) FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ 19: 968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Cirillo LA (2007) Chromatin opening and stable perturbation of core histone: DNA contacts by FoxO1. J Biol Chem 282: 35583–35593 [DOI] [PubMed] [Google Scholar]
- Hatta M, Liu F, Cirillo LA (2009) Acetylation curtails nucleosome binding, not stable nucleosome remodeling, by FoxO1. Biochem Biophys Res Commun 379: 1005–1008 [DOI] [PubMed] [Google Scholar]
- Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, Versteeg R, Cuppen E, van de Wetering M, Clevers H, Stunnenberg HG (2008) Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol 28: 2732–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Hoogenkamp M, Lichtinger M, Krysinska H, Lancrin C, Clarke D, Williamson A, Mazzarella L, Ingram R, Jorgensen H, Fisher A, Tenen DG, Kouskoff V, Lacaud G, Bonifer C (2009) Early chromatin unfolding by RUNX1: a molecular explanation for differential requirements during specification versus maintenance of the hematopoietic gene expression program. Blood 114: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui RC, Gomes AR, Constantinidou D, Costa JR, Karadedou CT, Fernandez de Mattos S, Wymann MP, Brosens JJ, Schulze A, Lam EW (2008) The forkhead transcription factor FOXO3a increases phosphoinositide-3 kinase/Akt activity in drug-resistant leukemic cells through induction of PIK3CA expression. Mol Cell Biol 28: 5886–5898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, Nakagawa Y, Takahashi A, Suzuki H, Sone H, Toyoshima H, Fukamizu A, Yamada N (2004) SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol 6: 351–357 [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT, Carner KR, Masiarz FR, Tjian R (1987) Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51: 1079–1090 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet DE, Burgering BM (2011) The PKB/FOXO switch in aging and cancer. Biochim Biophys Acta 1813: 1926–1937 [DOI] [PubMed] [Google Scholar]
- Ko LJ, Engel JD (1993) DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol 13: 4011–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM (2002a) Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419: 316–321 [DOI] [PubMed] [Google Scholar]
- Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM (1999) Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature 398: 630–634 [DOI] [PubMed] [Google Scholar]
- Kops GJ, Medema RH, Glassford J, Essers MA, Dijkers PF, Coffer PJ, Lam EW, Burgering BM (2002b) Control of cell cycle exit and entry by protein kinase B-regulated forkhead transcription factors. Mol Cell Biol 22: 2025–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Manning ET, Kadonaga JT (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol Cell Biol 19: 8123–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C (2010) A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI (1995) A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res 23: 1686–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, Rust AG, Bergthaler A, Aitchison JD, Aderem A (2012) A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature 490: 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers S, Downing JR, Hiebert SW (1993) Identification of AML-1 and the (8;21) translocation protein (AML-1/ETO) as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol Cell Biol 13: 6336–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, Chen C, Hosokawa K, Nakauchi H, Nakayama K, Nakayama KI, Harada M, Motoyama N, Suda T, Hirao A (2007) Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell 1: 101–112 [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Miyamoto T, Kato R, Yoshimura A, Motoyama N, Suda T (2008) FoxO3a regulates hematopoietic homeostasis through a negative feedback pathway in conditions of stress or aging. Blood 112: 4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry M, Hatzis P, de Bruijn E, Koster J, Versteeg R, Schuijers J, van de Wetering M, Guryev V, Clevers H, Cuppen E (2010) Efficient double fragmentation ChIP-seq provides nucleotide resolution protein-DNA binding profiles. PloS one 5: e15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokry M, Hatzis P, Schuijers J, Lansu N, Ruzius FP, Clevers H, Cuppen E (2012) Integrated genome-wide analysis of transcription factor occupancy, RNA polymerase II binding and steady-state RNA levels identify differentially regulated functional gene classes. Nucleic Acids Res 40: 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, O’Sullivan J, Azad A, Proudfoot N, Mellor J (2003) Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science 300: 492–495 [DOI] [PubMed] [Google Scholar]
- Nakae J, Park BC, Accili D (1999) Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem 274: 15982–15985 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K (2011) Pol II waiting in the starting gates: regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 1809: 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG (2011) Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, Hiller D, Jiang S, Protopopov A, Wong WH, Chin L, Ligon KL, DePinho RA (2009) FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell stem cell 5: 540–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Bruning JC (2008) Forkhead transcription factors and ageing. Oncogene 27: 2351–2363 [DOI] [PubMed] [Google Scholar]
- Puig O, Tjian R (2005) Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev 19: 2435–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA (2010) c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, Palmer TD, Butte AJ, Brunet A (2009) FoxO3 regulates neural stem cell homeostasis. Cell stem cell 5: 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault VM, Thekkat PU, Hoang KL, White JL, Brady CA, Kenzelmann Broz D, Venturelli OS, Johnson TM, Oskoui PR, Xuan Z, Santo EE, Zhang MQ, Vogel H, Attardi LD, Brunet A (2011) The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor. Oncogene 30: 3207–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long-range interaction landscape of gene promoters. Nature 489: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH (2002) Cell cycle inhibition by FoxO forkhead transcription factors involves downregulation of cyclin D. Mol Cell Biol 22: 7842–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster E, McElwee JJ, Tullet JM, Doonan R, Matthijssens F, Reece-Hoyes JS, Hope IA, Vanfleteren JR, Thornton JM, Gems D (2010) DamID in C. elegans reveals longevity-associated targets of DAF-16/FoxO. Mol Syst Biol 6: 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Metivier R, Salbert G, Eeckhoute J (2011) Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21: 555–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Bantignies F, Cavalli G (2009) Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol 20: 849–855 [DOI] [PubMed] [Google Scholar]
- Splinter E, de Laat W (2011) The complex transcription regulatory landscape of our genome: control in three dimensions. EMBO J 30: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, de Laat W (2011) The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25: 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Tsuchihara K, Kanai A, Arauchi T, Esumi H, Suzuki Y, Sugano S (2010) Genome-wide identification and annotation of HIF-1alpha binding sites in two cell lines using massively parallel sequencing. HUGO J 4: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Terragni J, Graham JR, Adams KW, Schaffer ME, Tullai JW, Cooper GM (2008) Phosphatidylinositol 3-kinase signaling in proliferating cells maintains an anti-apoptotic transcriptional program mediated by inhibition of FOXO and non-canonical activation of NFkappaB transcription factors. BMC Cell Biol 9: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho RA, Gilliland DG (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339 [DOI] [PubMed] [Google Scholar]
- Valis K, Prochazka L, Boura E, Chladova J, Obsil T, Rohlena J, Truksa J, Dong LF, Ralph SJ, Neuzil J (2011) Hippo/Mst1 stimulates transcription of the proapoptotic mediator NOXA in a FoxO1-dependent manner. Cancer Res 71: 946–954 [DOI] [PubMed] [Google Scholar]
- van den Berg MC, Burgering BM (2011) Integrating opposing signals toward Forkhead box O. Antioxid Redox Signal 14: 607–621 [DOI] [PubMed] [Google Scholar]
- van den Heuvel AP, Schulze A, Burgering BM (2005) Direct control of caveolin-1 expression by FOXO transcription factors. Biochem J 385: 795–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vos KE, Coffer PJ (2008) FOXO-binding partners: it takes two to tango. Oncogene 27: 2289–2299 [DOI] [PubMed] [Google Scholar]
- van der Vos KE, Coffer PJ (2011) The extending network of FOXO transcriptional target genes. Antioxid Redox Signal 14: 579–592 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RD, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE (2007) Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. Plos Genet 3: e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin S, Zhang X, Luciano JP, Mungamuri SK, Marinkovic D, Vercherat C, Sarkar A, Grisotto M, Taneja R, Ghaffari S (2008) Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J Biol Chem 283: 25692–25705 [DOI] [PubMed] [Google Scholar]
- Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, Huang H, Kuo HP, Lee DF, Li LY, Lien HC, Cheng X, Chang KJ, Hsiao CD, Tsai FJ, Tsai CH et al. (2008) ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 10: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yalcin S, Lee DF, Yeh TY, Lee SM, Su J, Mungamuri SK, Rimmele P, Kennedy M, Sellers R, Landthaler M, Tuschl T, Chi NW, Lemischka I, Keller G, Ghaffari S (2011) FOXO1 is an essential regulator of pluripotency in human embryonic stem cells. Nat Cell Biol 13: 1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wong WH (2004) CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc Natl Acad Sci USA 101: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–13, Supplementary Table Legends
FOXO3 induced transcriptional changes.
List of FOXO3A3-ER, endogenous FOXO3 and ER-DBD bound regions.
Sequences of all primers used for ChIP-qPCR, 4C, cloning of luciferase-reporters and qPCR on cDNA.