Abstract
Landmark events occur in a coordinated manner during pre-implantation development of the mammalian embryo, yet the regulatory network that orchestrates these events remains largely unknown. Here, we present the first systematic investigation of the network in pre-implantation mouse embryos using morpholino-mediated gene knockdowns of key embryonic stem cell (ESC) factors followed by detailed transcriptome analysis of pooled embryos, single embryos, and individual blastomeres. We delineated the regulons of Oct4, Sall4, and Nanog and identified a set of metabolism- and transport-related genes that were controlled by these transcription factors in embryos but not in ESCs. Strikingly, the knockdown embryos arrested at a range of developmental stages. We provided evidence that the DNA methyltransferase Dnmt3b has a role in determining the extent to which a knockdown embryo can develop. We further showed that the feed-forward loop comprising Dnmt3b, the pluripotency factors, and the miR-290-295 cluster exemplifies a network motif that buffers embryos against gene expression noise. Our findings indicate that Oct4, Sall4, and Nanog form a robust and integrated network to govern mammalian pre-implantation development.
Keywords: pluripotency factors, pre-implantation development, transcriptional networks
Introduction
Many critical cellular events occur within a few days during the pre-implantation development of a mammalian embryo. First, the paternal and maternal genomes are reprogrammed to a totipotent state. Second, the embryonic genome is activated after a period of reliance on maternal factors. Third, polarity is established when the inner and outer cells of a multicell embryo are biased toward different cell fates. Finally, two important cell fate decisions occur before implantation. In the first cell fate decision, the outside cells develop into extraembryonic trophectoderm, while cells that are inside the embryo retain pluripotency and form the inner cell mass (ICM). In the second cell fate decision, some of the ICM cells further differentiate into the primitive endoderm (Zernicka-Goetz et al, 2009). Given their complexities, all of these early cellular events have to be carefully orchestrated by a gene regulatory network to ensure the proper development of the embryo.
The gene regulatory network of embryonic stem cells (ESCs) has been extensively investigated in recent years. ESCs, which are derived from the ICM of mammalian embryos, are pluripotent like cells in the ICM but they can also self-renew indefinitely in culture. The transcription factor Oct4, also known as Pou5f1, has been shown to have a key role in the maintenance of an undifferentiated state in both human and mouse ESCs. Chromatin immunoprecipitation (chIP) assays in ESCs reveal that Oct4 binds to more than a thousand loci in the genome, suggesting that it may regulate many genes as a guardian of pluripotency (Boyer et al, 2005; Loh et al, 2006; Chen et al, 2008a). In addition, other key transcription factors that control the self-renewing and pluripotent properties of ESCs include Sall4 and Nanog. Previous studies indicate that Oct4, Sall4, and Nanog share a close functional relationship. Co-immunoprecipitation experiments show that they interact with one another in vivo (Wu et al, 2006; Liang et al, 2008; Yang et al, 2008). Furthermore, the three transcription factors co-occupy the promoters of many target genes (Boyer et al, 2005; Wu et al, 2006; Yang et al, 2008), indicating that they form an integrated network in ESCs.
Compared with ESCs, much less is known about the gene regulatory network in early mammalian embryos largely due to technical challenges. In particular, limited number of embryos or cells has so far precluded extensive studies of any regulatory network operating during pre-implantation development. For example, chIP experiments to map transcription factor binding sites in pre-implantation embryos are currently unfeasible. Fortunately, advances in gene expression profiling are now allowing studies into small number of cells or even single cells. Several laboratories used microarrays to profile the transcriptome during pre-implantation development of mouse embryos (Hamatani et al, 2004; Wang et al, 2004; Zeng et al, 2004; Xie et al, 2010). Expression analysis of 48 genes in individual cells from the one-cell zygote to the blastocyst identified markers of outer and inner cells (Guo et al, 2010). In addition, next-generation sequencing technology was used to investigate the pre-implantation transcriptome in single mouse blastomeres (Tang et al, 2011). While these previous studies were important for understanding early mammalian development, they relied primarily on transcriptome profiling of wild-type embryos and did not perform perturbation experiments to dissect interactions between the different genes.
Here, we show that the key pluripotency factors in ESCs also form critical nodes in the regulatory network governing pre-implantation development of the mammalian embryo. We used gene-specific knockdowns followed by genome-wide microarray analysis as well as extensive single-embryo and single-cell expression profiling to deconstruct the Oct4-Sall4-Nanog transcriptional network in pre-implantation embryos. Our study yielded novel and unexpected insights into the regulatory mechanisms that control the earliest stages of mammalian development.
Results
Oct4, Sall4, and Nanog are essential for pre-implantation development of the mouse embryo
Since a network of transcription factors has been shown to regulate pluripotency in ESCs (Zhou et al, 2007; Chen et al, 2008a), we hypothesize that some of these factors may also have an integral role in controlling developmental progression of the pre-implantation embryo. We previously found that Oct4 may play an important role at the maternal-zygotic transition (Foygel et al, 2008). Here, we sought to undertake a more comprehensive study of the functions of key ESC factors in pre-implantation embryos. We first measured the abundance of several pluripotency factors in individual one-cell zygotes using a microfluidic real-time PCR system known as Biomark (Fluidigm). While the Krüppel-like factor Klf5 was absent, we detected the transcripts of Oct4, Sall4, and Nanog in multiple one-cell embryos (Supplementary Figure S1). Hence, we chose to focus on the three transcription factors in this study.
To determine if Oct4, Sall4, or Nanog played an important role during pre-implantation development of the mammalian embryo, we depleted each of the proteins by injecting translation-blocking morpholinos (MOs) (Supplementary Figure S2), which targeted both maternal and embryonic transcripts, into one-cell fertilized zygotes and then cultured the injected embryos together with control embryos in serum supplemented Human Tubal Fluid (SAGE In-Vitro Fertilization Inc.). For each MO injection, we confirmed that the protein was absent by immunocytochemistry (Figure 1A). After 4 days of in vitro culture, 97.7% of uninjected embryos and 85.1% of control MO-injected embryos developed at least until the morula stage, but surprisingly, <25% of embryos that were depleted of any of the three pluripotency factors compacted. In addition, 40–60% of embryos injected with a MO targeting Oct4, Sall4, or Nanog arrested by the 4-cell stage after 4 days of in vitro culture, compared with <10% for the uninjected or control MO-injected embryos (P<0.05, Student’s t-test). To verify that our findings were not due to off-target effects, we performed rescue experiments for each pluripotency factor (Figure 1B and C). The percentage of embryos that arrested by the 4-cell stage decreased by 25.3% when we co-injected Oct4 MO with Oct4 mRNA, while the percentage decrease for Sall4 or Nanog was 18.3 or 24.7% respectively (P<0.05, Student’s t-test). These and other additional rescue data (Supplementary Results and Supplementary Figures S3 and S4) indicated that the MOs were specific for their intended targets and that our observed phenotypes were a direct consequence of knocking down the pluripotency factors. Taken together, our experiments indicate that Oct4, Sall4, and Nanog are essential for pre-implantation development. Interestingly, these results contradicted published knockout mouse models, where embryos that were homozygous null for Oct4, Sall4, or Nanog appeared to develop in vivo at least until the blastocyst stage, although they were still embryonic lethal (Nichols et al, 1998; Mitsui et al, 2003; Sakaki-Yumoto et al, 2006). A likely reason for the apparent discrepancy is that maternal transcripts are not eliminated in knockout mouse models (see Supplementary information for more discussion).
Figure 1.
MO-mediated knockdown of Oct4, Sall4, or Nanog in pre-implantation mouse embryos was effective and specific and resulted in developmental arrest. (A) Confocal microscopy images of embryos stained with α-Oct4 (left panel), α-Sall4 (middle panel), or α-Nanog (right panel) antibodies. The embryos were stained for the protein-of-interest 2 days after microinjection. Oct4 and Nanog proteins were completely undetectable by immunostaining in 100% of the embryos injected with the corresponding MO, while the uninjected embryos and the control MO-injected embryos were strongly stained. In addition, while a small amount of Sall4 protein was detected in embryos injected with a Sall4 MO, the protein level in these embryos was significantly less than that in all the control embryos. (Scale bar=45 μm). (B) Representative images of pre-implantation embryos under various experimental conditions. For every set of rescue experiments, there were four conditions: (i) no injection, (ii) injection with a control MO, (iii) injection with the experimental MO alone, and (iv) co-injection with the experimental MO and the corresponding mRNA that was in vitro transcribed. (C) Percentages of embryos that arrested by the 4-cell stage during 4 days of in vitro culture. Left panel (green bars): compared with embryos injected with the Oct4 MO only, we observed that co-injecting 0.6 mM Oct4 MO with 0.20–39 ng/μl Oct4 mRNA resulted in a smaller percentage of embryos arresting by the 4-cell stage (9 independent experiments) (P<0.05, Student’s t-test). Middle panel (blue bars): compared with embryos injected with the Sall4 MO alone, we found that co-injecting 0.6 mM Sall4 MO with 7–146 ng/μl Sall4 mRNA lowered the rate of developmental arrest by the 4-cell stage (8 independent experiments) (P<0.05, Student’s t-test). Right panel (red bars): compared with embryos injected with the Nanog MO alone, we discovered that co-injecting 0.6 mM Nanog MO with 5 ng/μl Nanog mRNA also resulted in partial rescue of the phenotype (7 independent experiments) (P<0.05, Student’s t-test).
Global transcriptome analysis identifies genes dependent on Oct4, Sall4, and Nanog
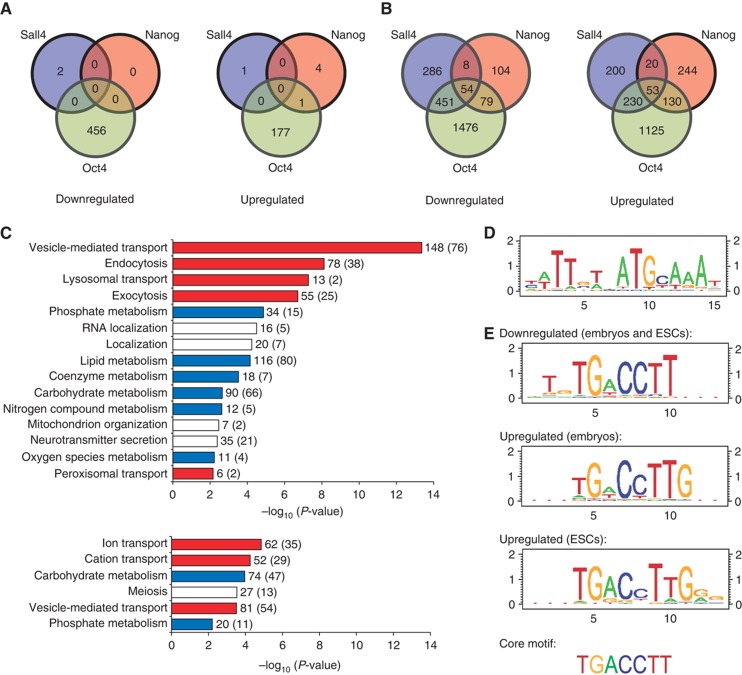
To understand the molecular mechanisms that might cause the embryos to arrest during knockdown of a pluripotency factor, we investigated transcriptome changes in pooled embryos using Affymetrix microarrays at ∼20 and 44 h after MO injection (Supplementary Files S1–S5). At 20 h, uninjected control embryos are at the 2-cell stage, while at 44 h, almost all the uninjected embryos are at the multicell stage. Comparison of the earlier timepoint with the later timepoint revealed that depletion of a pluripotency factor reduced the extent of zygotic genome activation (see Supplementary information and Supplementary Figure S5). We also found that MO-mediated knockdown of Oct4 had the most prominent effect, compared with knockdowns of Sall4 and Nanog. At the 20-h timepoint, a total of 634 genes were differentially expressed in Oct4 MO-injected embryos, while <10 genes were differentially expressed in Sall4 MO- and Nanog MO-injected embryos (false discovery rate (FDR)<0.1) (Figure 2A) (see Materials and methods for details). However, at 44 h post injection, a clear effect on global expression levels was observed for all three transcription factors (Figure 2B). Notably, 505 out of 799 genes that were downregulated during Sall4 knockdown were also downregulated during Oct4 knockdown and 133 out of 245 genes that were downregulated during Nanog knockdown were also downregulated during Oct4 knockdown, indicating that the three transcription factors shared a close functional relationship during pre-implantation embryonic development (P<10−100, chi-square test).
Figure 2.
Microarray analysis of pooled embryos identified the protein-coding regulons of Oct4, Sall4, and Nanog. (A) Venn diagram showing the number of genes that were differentially expressed 20 h after MO injection to knock down one of the three pluripotency factors. At this earlier timepoint, only knockdown of Oct4 exerted a strong effect on global gene expression levels. (B) Venn diagram showing the number of genes that was differentially expressed 44 h after MO injection. At this later timepoint, knockdown of any of the three transcription factors caused hundreds of genes to be significantly downregulated or upregulated (FDR<0.1). (C) GO analysis of genes that were differentially expressed during knockdown of Oct4, Sall4, or Nanog in pre-implantation embryos but showed no evidence of being controlled by the pluripotency factors in ESCs. These genes may be novel direct targets of the three transcription factors in the embryos or they may also be indirectly regulated. Upper panel: GO analysis of the embryo-specific genes that were downregulated. Lower panel: GO analysis of the embryo-specific genes that were upregulated. All the biological processes that are unique to the embryo (P<0.01) are plotted. The actual numbers of genes for each process (with the expected numbers given in brackets) are shown at the end of the bars. Many of the functional categories are related to either transport (in red) or metabolism (in blue). (D) A 15-bp motif within Oct4-bound regulatory regions of genes that were also differentially expressed during knockdown of Oct4 in pre-implantation embryos or ESCs. (E) A 7-bp core motif was found to be enriched in Nanog-bound regulatory regions of genes that were also differentially expressed during knockdown of Nanog in pre-implantation embryos or ESCs. The core motif was extended at the 5′ end for genes that were downregulated during Nanog knockdown and it was extended at the 3′ end for genes that were upregulated during Nanog knockdown.
We asked which of the differentially expressed genes are directly controlled by Oct4, Sall4, or Nanog. Since chIP experiments could not be readily performed on pre-implantation embryos, we compared our microarray results with publicly available chIP data sets in ESCs instead (Boyer et al, 2005; Loh et al, 2006; Lim et al, 2008; Yang et al, 2008; Chen et al, 2008a; Ouyang et al, 2009). 20.4% of the downregulated genes and 18.8% of the upregulated genes have promoters or enhancers that are bound by the corresponding transcription factor (Supplementary Figure S6). Gene Ontology (GO) analysis using the Panther classification system (Thomas et al, 2003) further revealed that the downregulated genes are involved in the establishment or maintenance of chromatin architecture, while the upregulated genes are involved in stress response and differentiation or developmental processes. Many of the direct targets are also involved in the cell cycle or are components of key signaling pathways (Supplementary Figure S7; Supplementary File S6).
Since ESCs do not exist in a natural developmental context, the pluripotency factors may control additional genes in pre-implantation embryos but not in ESCs. To determine the embryo-specific genes, we subtracted away from our microarray results genes that were present in the ESC chIP data sets as well as genes that responded to Oct4, Sall4, or Nanog depletion in ESCs (Supplementary Figure S6). GO analysis revealed that many of these embryo-specific genes were associated with metabolism- and transport-related functions (Figure 2C; Supplementary File S7), which reflected the unique embryonic conditions that are absent from ESCs.
To identify cis-acting elements that mediate the direct targeting of Oct4, Sall4, or Nanog to the genome in pre-implantation embryos, we searched the bound loci for known motifs and also performed de novo motif discovery using CisGenome (Ji et al, 2008). The Oct4-Sox2 consensus sequence was present in the promoter or enhancer regions of 83 downregulated genes (out of 284 genes) and 35 upregulated genes (out of 153 genes) (Figure 2D; Supplementary File S8). In addition, we obtained with high confidence a core motif, TGACCTT, that was enriched in the Nanog-bound promoter or enhancer regions of 25 downregulated genes (out of 40 genes) and 31 upregulated genes (out of 46 genes) (Supplementary File S9). Notably, the motif for downregulated genes is extended at the 5′ end by two basepairs, while the motif for upregulated genes is extended at the 3′ end by one to three basepairs (Figure 2E). A de novo search for cis-regulatory elements using a different program, SCOPE, also returned the same core motif (Carlson et al, 2007; Chakravarty et al, 2007; Supplementary Figure S8). Taken together, our results suggest that different cofactors may mediate Nanog’s function as a transcriptional activator or repressor. Furthermore, the core motif is similar to previously identified motifs for the retinoic acid receptor Rara as well as the estrogen-related receptors Esrra and Esrrb (Supplementary Figure S9), which suggests that Nanog may co-regulate many genes together with Rara and Esrra/Esrrb.
MicroRNAs form an integral component of the Oct4-Sall4-Nanog regulatory network in pre-implantation embryos
MicroRNAs (miRNAs) are an important class of post-transcriptional regulators that have diverse and critical roles during animal development and regeneration (Yang and Wu, 2007; Stefani and Slack, 2008). It is likely that they also have major roles in pre-implantation developmental processes. We therefore quantified miRNA expression from the 1-cell to the blastocyst stage in uninjected mouse embryos and detected 297 distinct miRNAs in at least one of these stages (Supplementary File S10). K-means clustering further revealed that many of the miRNAs were temporally regulated over development (Figure 3A).
Figure 3.
Oct4, Sall4, and Nanog control the expression of numerous developmentally regulated miRNAs in the pre-implantation embryo. (A) K-means clustered expression profiles for 235 miRNAs that were clearly expressed in uninjected (normal) pre-implantation embryos. Genes are in rows, with temporal progression from left to right. Red indicates high expression levels, while green indicates low expression levels. The majority of these miRNAs show a general increase in expression levels (cluster I and cluster V) from the 1-cell to the blastocyst stage. (B) Venn diagram showing the number of miRNAs that were differentially expressed during knockdown of Oct4, Sall4, or Nanog (P<0.05). MiRNAs in the miR-290-295 cluster were differentially expressed under all three knockdown conditions. (C) An example of an integrated network controlled by Oct4, Sall4, and Nanog in pre-implantation embryos. The protein-coding genes are depicted in various colors to signify different functional categories. Dark brown: pluripotency transcription factors; light blue: epigenetic regulators; light brown: signaling molecules; pink: differentiation factors; purple: embryo-specific factors. The listed miRNAs are within the top 20 most abundant miRNAs in the embryos. X→Y means ‘X regulates Y.’ For the arrows emanating from the center, a solid arrow indicates confirmed direct regulation, while a dotted arrow indicates either indirect regulation or potential direct regulation.
We hypothesize that some of the highly expressed miRNAs are integral components of the Oct4-Sall4-Nanog gene regulatory network in the pre-implantation embryo. To determine which miRNAs are regulated by Oct4, Sall4, or Nanog, we knocked down each transcription factor separately and measured miRNA expression levels 46 h after microinjection when the knockdown phenotypes were readily observable. More miRNAs were downregulated than upregulated (Figure 3B; Supplementary File S11), indicating that Oct4, Sall4, and Nanog served primarily as positive regulators of miRNA expression in pre-implantation embryos. In addition, most of the downregulated miRNAs were significant (P<0.05) in at least two different knockdown conditions, suggesting that the expression of many miRNAs was under combinatorial control by the transcription factors (Figure 3B). In total, 120 distinct miRNAs were differentially expressed. 40 of them (33.3%) have promoters or enhancers that are occupied by Oct4, Sall4, or Nanog in ESCs (Lim et al, 2008; Marson et al, 2008). The remaining 80 miRNA genes may be regulated indirectly by the three transcription factors or they may represent novel direct targets of Oct4, Sall4, or Nanog in the pre-implantation embryo.
Next, we sought to understand how the pluripotency factors and their protein-coding and non-coding regulons are interconnected. 61.1% of the protein-coding genes (2646 out of 4329 genes) that were differentially expressed during knockdown of Oct4, Sall4, or Nanog were predicted by the miRanda algorithm ( www.microrna.org) (Betel et al, 2008) to be targeted by at least one miRNA that was also regulated directly or indirectly by the three transcription factors (Supplementary File S12). Similar results were obtained when we restricted our target predictions to protein-coding genes and miRNAs that were present in the ESC chIP data sets (62.7%; 622 out of 992 genes) (Supplementary File S13). In comparison, when we randomly selected 10 000 distinct sets of protein-coding genes and miRNAs, we found that only 52.8±0.7% of the protein-coding genes were predicted to be miRNA targets. Hence, our analysis indicates that Oct4, Sall4, and Nanog and their protein-coding and non-coding regulons are more interconnected than random (P<10−6, Z-test). To increase confidence in the miRNA-target predictions, we further determined which of the miRNAs outputted by miRanda were also predicted by at least one other algorithm (see Materials and methods and Supplementary Files S14 and S15). An example of a subnetwork is illustrated in Figure 3C.
Knockdown of Oct4, Sall4, or Nanog causes gene expression changes even in embryos that appear to develop normally
As our microarray data were generated from pools of embryos and the ensemble averages may have masked interesting embryo-to-embryo variability or important expression dynamics at the single embryo level, we sought to monitor gene expression levels in individual embryos using BioMark (Fluidigm). We focused our single embryo analysis on 48 protein-coding genes, 43 of which were found from our microarray experiments to be differentially expressed during Oct4, Sall4, or Nanog knockdown. The 43 genes were chosen because they perform essential functions in ESCs, are key epigenetic regulators or signaling molecules, have important roles during differentiation events, or may have embryo-specific functions. The five remaining genes had unchanged expression levels and served as negative controls.
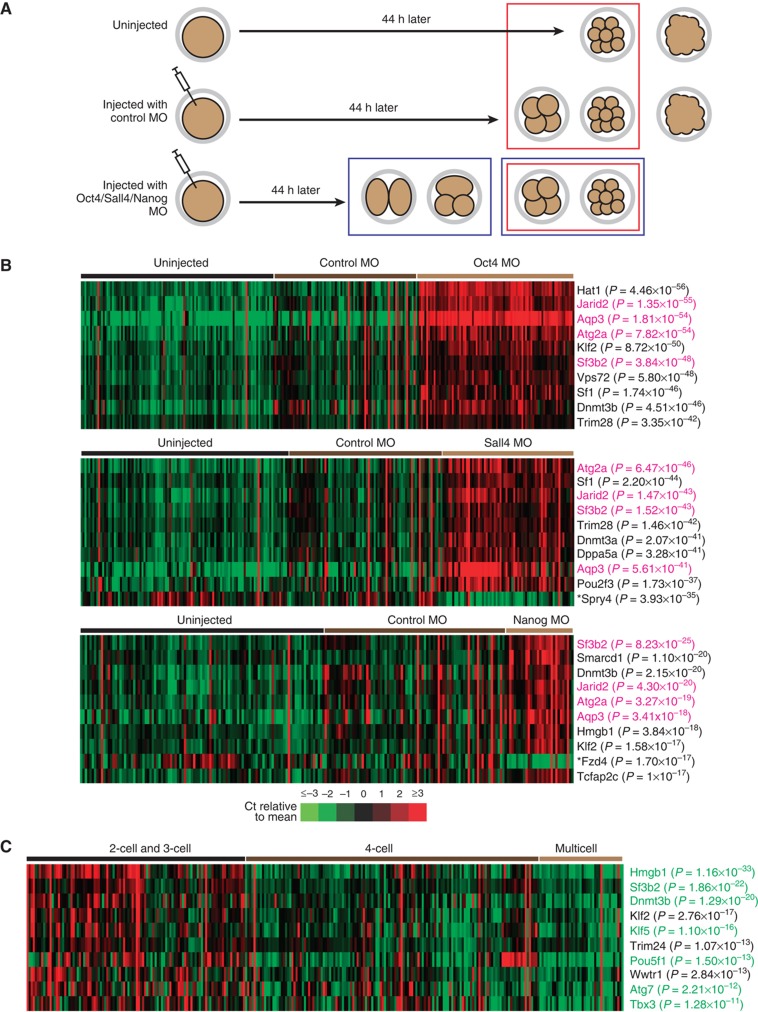
In our first set of analyses, we compared between the various experimental conditions, namely (1) no injection, (2) injection with a control MO, and (3) injection with a MO targeting Oct4, Sall4, or Nanog. For each condition, we collected between 80 and 120 embryos at 44 h after MO injection. When all the embryos were considered in the analysis, we found that our BioMark and microarray results agreed reasonably well with each other (Supplementary Information, Supplementary Figure S10, and Supplementary File S16). However, since depletion of a pluripotency factor caused embryos to arrest at different developmental stages (Supplementary Figure S11), some of the transcript changes observed between experimental conditions could be due to expression changes between the various stages. Hence, we next compared 4-cell and multicell control embryos with knockdown embryos that were also at the 4-cell and multicell stages (red boxes in Figure 4A). Intriguingly, we found that many genes were still significantly downregulated or upregulated (Figure 4B). In particular, at least four genes, namely Jarid2, Aqp3, Atg2a, and Sf3b2, were differentially expressed in all three knockdown conditions. Hence, although some of the knockdown embryos appeared to be morphologically normal, numerous molecular changes had already occurred in them.
Figure 4.
Gene expression analysis of individual embryos reveals hidden molecular changes and transcript differences between early- and late-arrested embryos. (A) A schematic diagram showing the developmental progression of uninjected and injected embryos. Embryos were collected at 44 h post-injection for expression analysis. Two main analyses were performed. First, we compared the expression of control embryos that were at the 4-cell and multicell stages with the expression of knockdown embryos that were also at the 4-cell and multicell stages (red boxes). Second, we compared the expression of knockdown embryos that were at the 2-cell and 3-cell stages (early-arrested) with the expression of knockdown embryos that were at the 4-cell and multicell stages (late-arrested) (blue boxes). (B) The first comparison. Although all the embryos in this analysis were morphologically similar (4-cell and multicell stages), many genes were still differentially expressed in the knockdown embryos. The individual Ct values for the top 10 ranking genes of each knockdown condition are plotted as heatmaps. P-values are given in brackets. Top panel: Oct4 knockdown; middle panel: Sall4 knockdown; bottom panel: Nanog knockdown. Most of the genes shown are downregulated upon knockdown of a pluripotency factor. The genes that are upregulated are marked by an asterisk (*). In addition, genes that are highlighted in pink are common among all the three knockdown conditions and they participate in critical functions such as epigenetic gene silencing (Peng et al, 2009; Li et al, 2010; Pasini et al, 2010), autophagy (Tsukamoto et al, 2008), and alternative splicing (Tang et al, 2010). (C) The second comparison. The individual Ct values for the top 10 ranking genes are plotted as a heatmap, with the P-values given in brackets. The expression levels of the genes highlighted in green are also significantly different between the 4-cell stage and the multicell stage.
Expression of Dnmt3b distinguishes early-arrested embryos from late-arrested embryos during Oct4, Sall4, or Nanog knockdown
Strikingly, we observed that, while uninjected and control-injected embryos were able to develop in a stereotypical manner, embryos that lacked any of the three pluripotency factors arrested over a range of developmental stages instead (Supplementary Figure S11). At 44 h after MO injection, the knockdown embryos were distributed mainly over the 2-cell, 3-cell, 4-cell, and more advanced multicell stages. Hence, in our second set of analyses, we asked if our BioMark experiments could identify genes that would differentiate early-arrested embryos (2-cell and 3-cell embryos) from late-arrested embryos (4-cell and multicell embryos) during knockdown of a pluripotency factor (blue boxes in Figure 4A).
Interestingly, the expression of a handful of genes correlates with the extent to which a knockdown embryo develops. Figure 4C shows a heatmap of the top 10 genes that are differentially expressed between early-arrested embryos and late-arrested embryos. Most of them encode DNA-binding proteins, such as the transcription factor Tbx3, which shares many common targets with Oct4 and Nanog (Han et al, 2010). We further compared 4-cell knockdown embryos with multicell knockdown embryos (Table I) and found that 7 of the 10 genes that were differentially expressed between early-arrested embryos and late-arrested embryos were also more highly expressed in multicell embryos than in 4-cell embryos. Hence, the expression of these seven genes (highlighted in green in Figure 4C and Table I) is low in 2-cell and 3-cell knockdown embryos, high in 4-cell knockdown embryos, and even higher in multicell knockdown embryos.
Table 1. The top 10 genes that were differentially expressed between 4-cell knockdown embryos and multicell knockdown embryos.

|
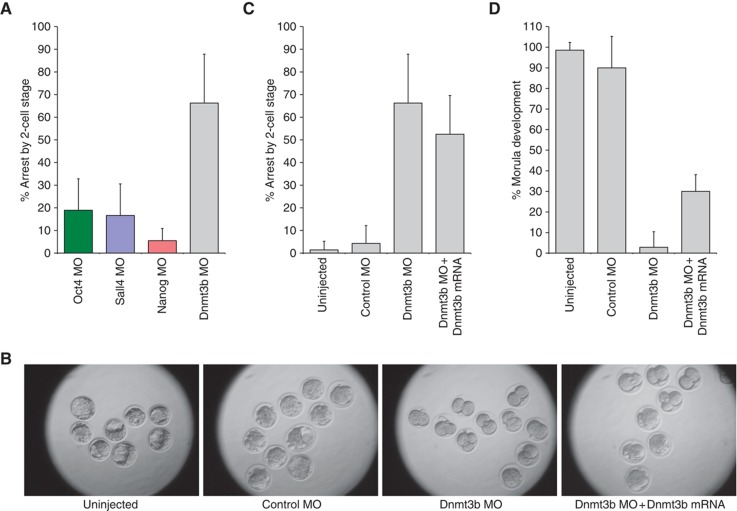
Since promoter DNA methylation contributes to ESC gene regulation and methylation-deficient ESCs are restricted in their developmental potential (Fouse et al, 2008), we decided to investigate the DNA methyltransferase Dnmt3b in greater depth. Knockdown of Dnmt3b using a translation-blocking MO revealed a more severe phenotype than that for any of the three pluripotency factors. Specifically, 66.3±21.6% of embryos injected with the Dnmt3b MO arrested by the 2-cell stage, even though the percentages for Oct4, Sall4, and Nanog were only between 5.5 and 19.0% (P<0.001, Student’s t-test) (Figure 5A). Rescue experiments with in vitro transcribed Dnmt3b mRNA confirmed the specificity of the Dnmt3b MO (Figure 5B–D). These results suggest that Dnmt3b is essential for pre-implantation development and that it may have a role in determining the extent to which an embryo depleted of Oct4, Sall4, or Nanog can develop. In addition, although Dnmt3b−/− embryos had been reported to develop normally before E9.5 (Okano et al, 1999b; Ueda et al, 2006), it is likely that maternal Dnmt3b, which we detected in 1-cell mouse zygotes (Supplementary Figure S1), may be driving the earliest developmental stages in the knockout mouse model.
Figure 5.
The DNA methyltransferase Dnmt3b is essential for pre-implantation development. (A) Percentages of embryos that arrested by the 2-cell stage for four different knockdown conditions. Unexpectedly, we observed that the majority of embryos injected with a MO targeting Dnmt3b arrested around the maternal-zygotic transition (P<0.001, Student’s t-test, when comparing Dnmt3b with any of the pluripotency factors). (B) Representative microscopy images of pre-implantation mouse embryos under various experimental conditions. (C) Quantification of observed phenotypes after 4 days of in vitro culture. Compared with embryos that were injected with 0.6 mM Dnmt3b MO only, co-injection of 0.6 mM Dnmt3b MO with 1–10 ng/μl Dnmt3b mRNA reduced the fraction of embryos that arrested by the 2-cell stage (7 independent experiments) (P<0.05, Student’s t-test). (D) Percentages of embryos that developed until at least the morula stage after 4 days of culture. Compared with embryos that were injected with Dnmt3b MO only, co-injection of Dnmt3b MO with in vitro transcribed Dnmt3b mRNA increased the percentage of embryos that reached the morula stage (P<0.05, Student’s t-test).
A coherent feed-forward loop controls the expression of Dnmt3b
In ESCs, chIP experiments showed that the pluripotency factors can control the transcription of Dnmt3b directly. Interestingly, the factors can also regulate Dnmt3b expression indirectly via the ESC-specific miRNAs in the miR-290-295 cluster. Specifically, Oct4 and Nanog directly control the expression of these miRNAs, which shows a general upward trend from the 1-cell to the blastocyst stage (Supplementary Figure S12). The miRNAs then target the 3′UTR of the retinoblastoma-like 2 (Rbl2) mRNA, whose protein product functions as a transcriptional repressor of Dnmt3b (Benetti et al, 2008; Sinkkonen et al, 2008). In the pre-implantation embryo, we found from our microarray analysis that although Rbl2 did not pass our cutoff criterion for significant genes, its expression is still upregulated 1.6- to 2.9-fold during knockdown of Oct4, Sall4, or Nanog. Our microarray analysis also revealed that Dnmt3b expression was downregulated by 3- to 19-fold when Oct4, Sall4, or Nanog was depleted (FDR<0.1; Supplementary Files S3 and S5). Hence, a coherent feed-forward loop (FFL) consisting of the pluripotency factors, the miR-290-295 cluster, Rbl2, and Dnmt3b regulates DNA methylation in the pre-specified embryo and in ESCs (Figure 8B).
To obtain a quantitative understanding of the feed-forward architecture, we constructed a model of non-linear ordinary differential equations (ODEs) based on Michaelis-Menten kinetics with cooperativity (Figure 6A) and performed the simulation using a Runge-Kutta algorithm. We chose the parameters so that the model would reproduce known experimental observations. In uninjected (wild-type) embryos, the pluripotency factors are present at high levels and activate the expression of the miR-290-295 cluster. The miRNAs then downregulate the production of Rbl2. When the level of Rbl2 is low, Oct4, Sall4, and Nanog can then fully activate the expression of Dnmt3b (Figure 6B, left panel). However, the converse is true during knockdown of the pluripotency factors (Figure 6B, right panel). Furthermore, in Dicer-null ESCs, both the transcript and protein levels of Dnmt3b were significantly lower than that in wild-type ESCs (Benetti et al, 2008; Sinkkonen et al, 2008). In agreement with these observations, when we set the expression of the miRNAs to zero in our model, we found that the peak Dnmt3b expression level dropped by 1.7-fold for our chosen set of parameters (Supplementary File S17).
Figure 6.
A coherent feed-forward motif sharpens the Dnmt3b transcriptional output into a bistable switch. (A) A model of non-linear ODEs describing the behavior of the FFL. In the model, both the rate of transcription and the miRNAs’ silencing of Rbl2 are modeled on Michaelis-Menten kinetics with cooperativity. In the equations, B represents basal promoter activity, γ stands for the rate of degradation, K stands for the Michaelis constant, and H is the Hill coefficient. A list of parameter values that reproduces known experimental observations is provided in Supplementary File S17. (B) Simulations of the system showing how the concentrations of the miRNAs (blue), Dnmt3b (red), and Rbl2 (green) change over time. Left panel: high levels of the pluripotency factors; right panel: low levels of the pluripotency factors. (C) A plot showing how the concentration of Dnmt3b varies as a function of the concentration of the pluripotency factors. Both the original FFL (blue curve) and a simplified two-component system comprising only the pluripotency factors and Dnmt3b (red curve) were simulated. Only the FFL exhibits a threshold or switch-like behavior. (D) A plot showing how the Dnmt3b transcriptional output depends on the relative regulatory strengths of different transcription factors. A larger βa/βb ratio indicates that Dnmt3b transcription is more reliant on the pluripotency factors, while a smaller βa/βb ratio indicates that the expression of Dnmt3b is more dependent on the Rbl2 transcriptional repressor. (E) RT-qPCR results for single Dicer knockout (Dcr KO) ESCs, Dcr KO ESCs transfected with a control miRNA mimic, Dcr KO ESCs transfected with a miR-295 mimic, and wild-type ESCs. Upper panel: A boxplot for the Ct values of Dnmt3b. Lower panel: A boxplot for the Ct values of the pluripotency factors. The bottom and top of every orange box represent the 25th and 75th percentiles and the band inside each box is the median. The dotted lines extend to the furthest datapoint that is within 1.5 times the interquartile range from the box, while the black circles indicate the outliers. The Ct values of Dnmt3b for the Dicer-null ESCs are significantly larger than those for the miR-295 transfected cells or the wild-type ESCs (P<0.005, Wilcoxon rank sum test) but are comparable to the Ct values for the control-transfected cells (P>0.01, Wilcoxon rank sum test). In contrast, the Ct values of the pluripotency factors are similar between the Dicer-null ESCs and the other three cell populations (P>0.01, Wilcoxon rank sum test).
We next investigated the advantages that the FFL confers on the pluripotency factors’ regulation of DNA methylation. Superficially, the pathway through the miRNAs and Rbl2 appears to be unnecessary, as the pluripotency factors can simply activate the expression of Dnmt3b. To identify differences between this straightforward design and the FFL, we also simulated the two-component model consisting of the pluripotency factors and Dnmt3b only (Supplementary File S18). When we varied the concentration of the pluripotency factors from low (0) to high (1) and examined the output (normalized Dnmt3b expression level) of both models, we found that the simple two-component model produced a more graded output, while the FFL gave a more switch-like response (Figure 6C). Hence, the miRNA-Rbl2 pathway served to sharpen the system output so that it has a more switch-like behavior.
Since the performance of a non-linear dynamical system is dependent on the precise values of the parameters, we wondered how our results may change with different parameter choices. We first found that the feed-forward network motif is robust with respect to the synthesis rate of the Rbl2 transcriptional repressor (see Supplementary information and Supplementary Figure S13). Next, we studied the behavior of the dynamical system when we varied the synthesis rate of Dnmt3b. The expression of Dnmt3b is under combinatorial regulation by the pluripotency factors and Rbl2. In our mathematical description of the FFL, we have modeled the influences of each transcription factor as both mutually independent (second and third terms on the right-hand side of the third ODE) and cooperative (fourth term of the third ODE). However, the relative contributions of each regulator to the transcription of Dnmt3b are unknown. Thus, we varied the ratio of the two parameters, βa and βb, and observed how the concentration of Dnmt3b responds to changes in the level of the pluripotency factors. We found that when βa/βb is approximately bigger than 5, the hypersensitive response is lost and the FFL essentially behaves like the simple two component system. However, when the ratio becomes smaller, the switch-like behavior becomes more pronounced (Figure 6D; Supplementary Figure S14). In contrast, the steepest slope of the response curve is unaffected by the parameter βc (see Supplementary information and Supplementary Figure S15). Hence, our analysis showed that in order to achieve a hypersensitive response, the regulation of Dnmt3b transcription cannot be dominated by the pluripotency factors, so that Rbl2 can exert its effect.
A prediction from our model is that at high levels of pluripotency factors, for example in pre-specified embryos and in ESCs, the concentration of Dnmt3b becomes more susceptible to intrinsic gene expression noise when the FFL is broken. To test the prediction, we determined the cell-to-cell variability of Dnmt3b transcript level for wild-type ESCs and for Dicer−/− ESCs (Calabrese et al, 2007). Specifically, we performed quantitative real-time PCR (RT-qPCR) experiments on four different sets of cells: (1) Dicer−/− ESCs, (2) Dicer−/− ESCs transfected with a control miRNA mimic that has minimal sequence identity with miRNAs in humans and mice (Dharmacon), (3) Dicer−/− ESCs transfected with a miR-295 mimic (Dharmacon), and (4) wild-type ESCs. To verify that the transfection of miR-295 was successful, we measured the levels of its target genes Rbl2, p21, and Casp2 (Benetti et al, 2008; Sinkkonen et al, 2008; Wang et al, 2008; Zheng et al, 2011) and confirmed that they were all downregulated (Supplementary Figure S16). We also transfected each of the four cell populations with a plasmid encoding green fluorescent protein (GFP), so that we could sort for single cells by flow cytometry, and then determined the transcript levels of Dnmt3b, Oct4, Sall4, and Nanog in individual ESCs using Biomark (Fluidigm). Strikingly, Dnmt3b expression exhibited the widest spread of Ct values in Dicer-null ESCs. The interquartile range for Dicer-null ESCs and for cells transfected with the control miRNA was 8.94 and 3.33 respectively, while the range for Dicer-null ESCs transfected with miR-295 and for wild-type ESCs was 2.87 and 2.79 respectively (Figure 6E, upper panel). The standard deviation of Dnmt3b expression level was also larger in Dicer-null ESCs and control-transfected cells than in miR-295-transfected cells and wild-type ESCs (Supplementary Figure S17A). Furthermore, these results are not due to the expression of the pluripotency factors exhibiting different degrees of variability between the four sets of cells (Figure 6E, lower panel, and Supplementary Figure S17B). When corrected for the standard deviation of the pluripotency factors, the expression of Dnmt3b was still more variable in Dicer-null cells and control-transfected cells than in miR-295-transfected cells and wild-type ESCs (Supplementary Figure S17C). Hence, our results indicate that the FFL serves to buffer Dnmt3b expression against intrinsic variability in the concentrations of the pluripotency factors.
Single-cell analysis reveals an increase in gene expression noise during knockdown of Oct4 or Sall4
Our simulations suggest an intriguing explanation for why during knockdown of a pluripotency factor, some embryos might express a higher level of Dnmt3b and progress further in development, while other embryos might express a lower level of Dnmt3b and arrest earlier in development. In uninjected embryos, the pluripotency factors are present at high levels and fluctuations in their concentrations due to intrinsic noise do not affect the level of Dnmt3b significantly (right side of blue graph in Figure 6C). However, when a pluripotency factor has been knocked down to a low level, the gradient of the response curve increases dramatically and the system exists in the switch regime instead (left side of blue graph in Figure 6C). In this case, even small fluctuations in the concentrations of the pluripotency factors can affect the level of Dnmt3b appreciably.
To test the hypothesis that the expression of Dnmt3b and possibly many other genes become noisier during knockdown of a pluripotency factor, we performed single-cell real-time PCR experiments using BioMark (Fluidigm). In these experiments, we focused on Oct4 and Sall4, since knockdown of Nanog gave a milder phenotype that is consistent with the expression of Nanog being significantly lower than that of Oct4 or Sall4 in the earliest developmental stages. For each experimental condition (no injection, injection with control MO, injection with Oct4 MO, and injection with Sall4 MO), we collected embryos that were morphologically at the 4-cell stage and manually separated them into individual cells, so that we could profile gene expression in each blastomere. All the cells were harvested at 39 h post-injection, as uninjected embryos were at the 4-cell stage at this timepoint.
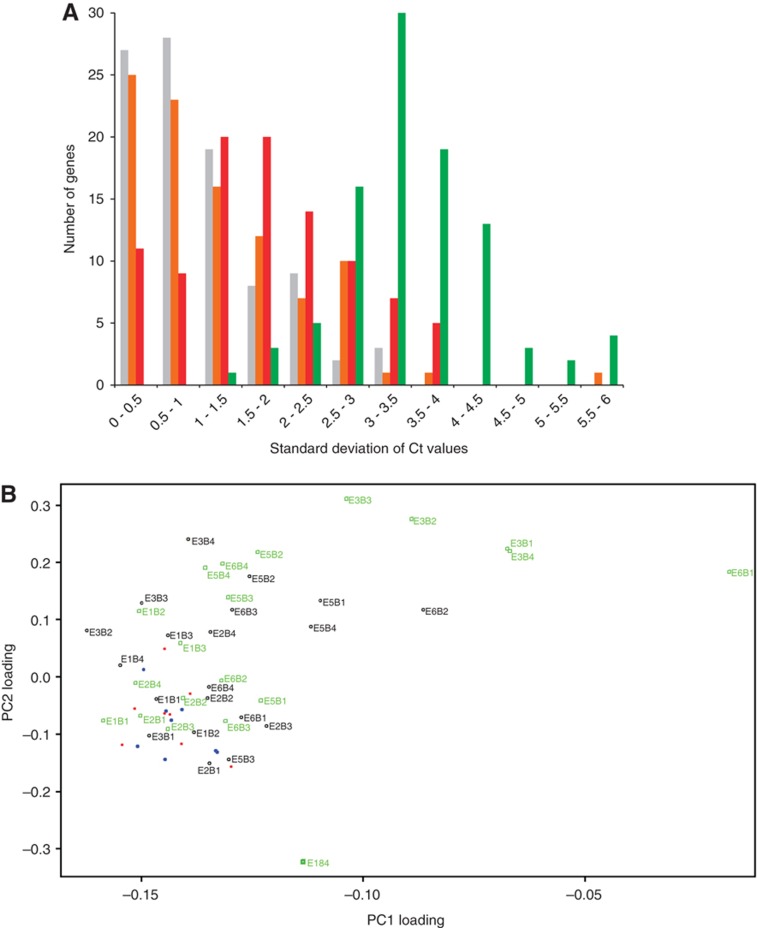
We monitored the expression of 84 functionally diverse genes, including Dnmt3b (Supplementary File S19). 68 of these genes were selected because they were differentially expressed during knockdown of one of the pluripotency factors, while the remaining genes were chosen because they were related family members or performed similar functions. Hierarchical clustering of our single-cell data revealed that all the blastomeres from uninjected embryos grouped together in a single tight cluster. However, the cells from Oct4 or Sall4 MO-injected embryos did not appear to form any coherent cluster. We further calculated the standard deviations of Ct values for every gene within each experimental condition and then plotted a histogram for these standard deviations (Figure 7A). Notably, the distributions for Oct4- and Sall4-depleted blastomeres were shifted to the right, indicating that gene expression was more variable in the knockdown cells compared with the control cells. The average standard deviation for uninjected cells was 1.00, while the average standard deviations for Oct4-depleted and Sall4-depleted cells were 1.74 and 3.47 respectively and were significantly higher than that for uninjected cells (P<10−7 and P<10−15 respectively, Wilcoxon rank sum test). In contrast, cells from control MO-injected embryos showed an average standard deviation of only 1.21 (P>0.1, Wilcoxon rank sum test). Hence, knockdown of Oct4 or Sall4 resulted in noisier gene expression, not just for Dnmt3b but also for the majority of the genes that we profiled.
Figure 7.
Single-cell analysis reveals that gene expression becomes noisier during knockdown of a pluripotency factor. (A) A plot showing how the standard deviations of gene expression are distributed for each experimental condition. Gray bars: uninjected embryos, orange bars: embryos injected with a control MO, red bars: embryos injected with a MO targeting Oct4, and green bars: embryos injected with a MO targeting Sall4. Most genes showed an increased variability in expression when Oct4 or Sall4 was depleted (P<10−7 and P<10−15 respectively, Wilcoxon rank sum test). We also noted that the gene expression variability of our uninjected and control-injected cells is similar to the variability observed in a previously published single-cell profiling data set for wild-type blastomeres (Tang et al, 2011) (see Supplementary information). (B) Principal component projections of individual blastomeres showing how similar or different the cells are to one another. Blastomeres that were derived from uninjected 4-cell embryos (solid blue circles) or from embryos injected with a control MO (solid red circles) clustered close together. Blastomeres that were derived from embryos injected with a MO targeting Oct4 (unfilled black circles) or Sall4 (unfilled green squares) were more spread out. Each knockdown cell is also labeled as ‘E_B_’, where ‘E’ stands for embryo and ‘B’ stands for blastomere. From the plot, we observed that embryo-to-embryo variability is noticeably larger than the variability between cells from the same embryo. For example, for the Sall4 knockdown condition, E2B1, E2B2, E2B3, and E2B4 are positioned relatively close to one another but they are all far away from any blastomere from E3 (also see Supplementary information).
To confirm the results, we performed principal component analysis (PCA) on our single-cell data. The first principal component accounted for 70% of the gene expression variability between all the cells, while the second principal component accounted for 7% of the variability. A projection of the cells’ expression vectors onto these two principal components showed that the uninjected blastomeres and the control MO-injected cells clustered together, while the knockdown cells were clearly more spread out (Figure 7B). In addition, we observed that among all the knockdown blastomeres, cells from different embryos were generally further apart from one another, while cells from the same embryo were closer to one another, indicating that the gene expression from embryo to embryo is noisier than the gene expression between cells of the same embryo. Nevertheless, increases in both inter-embryo variability and intra-embryo variability contributed to noisier gene expression during depletion of a pluripotency factor.
The increase in gene expression noise is not a result of variable knockdown efficiency among the different embryos, as determined by quantification of protein levels in our immunostaining images of control and knockdown embryos (Figure 1A; Table II; also see Supplementary information and Supplementary Figure S18). We further co-injected each MO with the corresponding mRNA and showed that the increase in gene expression noise could be rescued (see Supplementary information and Supplementary Figure S19). Collectively, our data demonstrate that consistent depletion of a pluripotency factor in preimplantation mouse embryos leads to an increased variability in the expression of numerous genes, including Dnmt3b.
Table 2. Variability in protein level (standard deviation/mean) based on quantification of pixel intensities.
| Antibody | Uninjected | Control injected | Knockdown | Background |
|---|---|---|---|---|
| α-Oct4 | 0.172 | 0.123 | 0.166 | 0.167 |
| α-Sall4 | 0.170 | 0.284 | 0.292 | 0.082 |
Discussion
How multiple cellular events are coordinated in the correct spatiotemporal order to ensure proper development of the 1-cell fertilized zygote into a pluripotent blastocyst that can give rise to all cell types in the body is a largely unsolved problem in mammalian developmental biology. While gene regulatory networks have been extensively studied in ESCs in the past decade, the limiting amount of material available for experiments has hampered the application of chIP to map transcription factor binding sites in pre-implantation embryos. Furthermore, the technical challenge of microinjecting the small and fragile embryos poses a barrier to extensive gene knockdown studies.
Here, we leveraged on recent advances in gene expression profiling methods to deconstruct the pluripotency gene regulatory network that controls pre-implantation development in the mouse embryo. We knocked down Oct4, Sall4, and Nanog by microinjecting MOs into the embryos and profiled the gene expression of pooled embryos, single embryos, and individual blastomeres to identify the regulon of the pluripotency factors. Similar to their roles in ESCs, Oct4, Sall4, and Nanog control the expression of numerous genes involved in the cell cycle, establishment or maintenance of chromatin architecture, signaling, and differentiation processes in the pre-implantation embryo. We also discovered that many of the genes that were differentially expressed in the embryo during knockdown of Oct4, Sall4, or Nanog were not regulated directly or indirectly by the three pluripotency factors in ESCs. GO analysis of these embryo-specific genes revealed that they largely performed metabolism- or transport-related functions. Interestingly, a recent study found that the expression of metabolism genes undergoes major changes during the derivation of ESCs from the ICM, which may be a result of the cells accommodating a dramatic change in their environment (Tang et al, 2010). Hence, since the embryonic environment is distinct from the in vitro culture conditions of ESCs, Oct4, Sall4, and Nanog may have an extended role in the pre-implantation embryo by regulating the unique metabolic requirements and cellular conditions of the embryo.
There are other pluripotency factors that may perform important roles in pre-implantation embryos, in particular Sox2, which is known to co-regulate many genes with Oct4 and Nanog in ESCs. However, we found that maternal Sox2 protein is difficult to deplete. In embryos injected with a MO targeting the translation start site of Sox2, we could still detect by immunocytochemistry a substantial amount of Sox2 protein 24 h after injection. Unsurprisingly, these embryos did not show an obvious phenotype until the morula stage. Future advances in chIP technologies will allow us to study the Sox2 regulatory network in the earliest developmental stages.
Strikingly, we observed that embryos injected with Oct4, Sall4, or Nanog MOs arrested over a range of developmental stages. Uninjected wild-type mouse embryos showed a relatively synchronous rate of pre-implantation development with over 90% developing to the blastocyst stage by Day 4 of in vitro culture. However, the majority of embryos injected with Oct4, Sall4, or Nanog MOs arrested at 2-cell, 3-cell, 4-cell, and multicell stages. This loss in developmental control is not due merely to the act of injecting and possibly injuring the embryos because, even though embryos injected with a control MO developed slightly slower than uninjected embryos, over 80% of them still progressed to at least the morula stage by Day 4.
Why did some knockdown embryos arrest early, while others arrest at a later stage? From our single-cell analysis, we discovered that depletion of a pluripotency factor caused an increase in gene expression noise. The increase is contributed predominantly by larger variations in gene expression between different embryos and also partly by a larger spread of expression levels between individual cells of the same embryo. As a result, many genes are expressed at high levels in some embryos but at low levels in other embryos. A subset of these genes might be driver genes that perform key functions and determine the developmental potential of an embryo. Hence, embryos that express higher levels of the driver genes will progress further in development, while embryos that express lower levels of the driver genes will arrest earlier in development (Figure 8A).
Figure 8.
Schematics of an exemplary regulatory circuit governing developmental progression in the pre-implantation embryo and the consequences of its perturbations. (A) A graphical model outlining the sequence of events that occurs during knockdown of Oct4, Sall4, or Nanog. Dnmt3b is a representative driver gene, whose expression level may determine the extent to which a pre-implantation embryo can develop. (B) An Oct4-Sall4-Nanog regulatory network controlling DNA methylation in the pre-implantation embryo. The FFL, which is one of the most prevalent motifs or basic building blocks of transcriptional networks (Shen-Orr et al, 2002; Milo et al, 2002; Mangan and Alon, 2003; Shalgi et al, 2007; Tsang et al, 2007; Gerstein et al, 2012), is highlighted by a yellow box.
We found that the DNA methyltransferase Dnmt3b may function as one of the driver genes. The expression of Dnmt3b correlates strongly with the extent to which an embryo depleted of Oct4, Sall4, or Nanog can develop. To determine whether it is a driver gene, we separately knocked down Dnmt3b and discovered that over 60% of the knockdown embryos arrested by the 2-cell stage, which is around the time when the embryonic genome becomes activated in the mouse. In addition, a recent study showed that, although the pre-implantation embryonic genome is in a general state of hypomethylation, the oocyte actually contributes a substantial number of CpG island promoters that remain hypermethylated in the pre-specified embryo (Smith et al, 2012). Hence, it is possible that Dnmt3b is required to maintain the methylation status of these regions during and after the maternal-to-embryonic transition.
A FFL regulates the expression of Dnmt3b in pre-implantation embryos and in ESCs, whereby pluripotency factors can control the expression of Dnmt3b directly or they can regulate its expression indirectly through the miR-290-295 cluster of miRNAs and the Rbl2 transcriptional repressor (Figure 8B). Modeling and simulation of the FFL provided insights into the underlying cause of the increase in gene expression noise during knockdown of a pluripotency factor. Compared with a straightforward two-component design where the pluripotency factor simply activates Dnmt3b in the absence of other pathways, the feed-forward motif allows the expression of Dnmt3b to exhibit a more switch-like behavior, so that the system approaches bistability. Importantly, we note that the switch between an ‘off’ state and an ‘on’ state occurs when the expression level of the pluripotency factors is low. For our set of parameter values (Supplementary File S17), simulation showed that when the expression of the pluripotency factors is high, a 20% fluctuation in their levels causes a 3% change in the expression of Dnmt3b. However, when the expression of the pluripotency factors is low, a 20% fluctuation in their levels causes a 60% change in the expression of Dnmt3b (Figure 6C). Hence, in an uninjected embryo where the levels of Oct4, Sall4, and Nanog are high, the transcriptional output of Dnmt3b is robust to intrinsic noise in the expression of its activators. However, in knockdown embryos, the transcriptional output of Dnmt3b becomes much more sensitive to small changes in the concentration of its activators.
While the FFL may serve as an important paradigm for how pre-implantation mouse embryos ensure their relatively synchronous rate of development, we speculate that there are many other undiscovered network features in the embryo that either buffer its development against intrinsic and extrinsic noise or to provide safeguard measures. Like the miR-290-295 cluster, we found that the most highly expressed miRNA in the pre-implantation embryo, miR-709, also depends on Oct4 and Sall4 for its expression. Interestingly, a recent study showed that miR-709 targets and degrades BORIS mRNA, which has been implicated in the genome-wide erasure of DNA methylation in the germline (Tamminga et al, 2008). Hence, the high expression of miR-709 may provide insurance against the accidental erasure of essential methylation marks in the embryo (Figure 8B).
Two closely related de novo DNA methyltransferases, Dnmt3a and Dnmt3b, are encoded in the mouse genome. Here, we have focused on Dnmt3b for a few reasons. First, although the expression of both Dnmt3a and Dnmt3b was assayed in our single-embryo experiments, only Dnmt3b allowed us to distinguish early-arrested embryos from late-arrested embryos. The expression of Dnmt3b was 2.1-fold higher in embryos that progressed to the 4-cell and multicell stages than in embryos that arrested at the 2-cell and 3-cell stages (Figure 4C), while the expression of Dnmt3a was only 1.1-fold higher (P>0.001). Second, Dnmt3a−/− mice developed to term and appeared grossly normal at birth. In contrast, the Dnmt3b-null allele resulted in embryonic lethality, indicating a more prominent role for Dnmt3b during embryogenesis (Okano et al, 1999a). Third, there is only one known promoter for the Dnmt3b gene, but there are two separate promoters for the Dnmt3a gene (Chen et al, 2002), which can complicate analysis. For example, the level of the full-length Dnmt3a protein is similar between Dicer−/− ESCs and wild-type ESCs, while the level of the smaller Dnmt3a protein, which originates from a downstream intronic promoter, is appreciably lower in Dicer-null ESCs (Sinkkonen et al, 2008). Clearly, the regulatory activities at the two promoters can be different and further work is needed to tease apart the regulation occurring at the two distinct Dnmt3a promoters. In the future, chIP assays to map the genome-wide locations of the DNA methyltransferases and gene knockdown experiments followed by bisulfite sequencing will enable us to understand how Dnmt3a and Dnmt3b contribute to shaping the pre-implantation methylome.
In conclusion, we have deconstructed an Oct4-Sall4-Nanog gene regulatory network in pre-implantation mouse embryos that coordinates various critical cellular events to ensure reliable developmental progression despite intrinsic gene expression noise. Not only have we delineated functional sets of genes dependent on the pluripotency factors for their expression, we have also demonstrated how a particular network design, the FFL, may buffer embryos against fluctuations in the concentration of the hub proteins, so that the embryos can develop in a relatively synchronous and stereotypical manner. Our work has laid the foundation for future studies on other gene networks controlling pre-implantation mammalian embryogenesis and also has important implications for assisted reproduction.
Materials and methods
Embryo collection and culture
All procedures involving animals were performed under our Institutional Animal Care and Use Committee (IACUC) protocol #8315 titled ‘Novel roles of pluripotency regulators in the early mouse embryo’, which was approved by the Administrative Panel on Laboratory Animal Care (APLAC) at Stanford University. Three- to five-week-old wild-type F1 (C57BL6 × DBA/2) females (Charles River) were superovulated with intraperitoneal injections of 5 IU pregnant mare’s serum gonadotropin (PMSG; Sigma) followed by 5 IU human chorionic gonadotropin (hCG; Sigma) 48 h later before being mated overnight with wild-type F1 males. One-cell zygotes were released from oviducts 17 h after hCG injection and cumulus cells were removed by hyaluronidase (Sigma) treatment and pipetting. Embryos were cultured in drops of Human Tubal Fluid with 10% serum supplement (SAGE In-Vitro Fertilization Inc.) under mineral oil (Sigma) in 5% CO2 at 37°C. Each 20 μl drop contained 8–10 embryos.
Single-cell isolation
The zona pellucida was first removed by acid tyrode’s solution and the embryos were then incubated in TrypLE solution (Invitrogen) for 5–10 min at 37°C. The dissociation of individual cells from whole embryos was monitored under the microscope. After rinsing twice in drops of PBS containing 3 mg/ml polyvinylpyrrolidone (PBS-PVP), the dissociated blastomeres were carefully transferred into sample collection tubules with the aid of a finely pulled glass pipette.
Microinjection of antisense MOs
25-nt antisense MOs were purchased from Gene Tools, LLC. The sequences for the MOs are as follows:
Oct4-MO1: 5′-AGT CTG AAG CCA GGT GTC CAG CCA T-3′
Oct4-MO2: 5′-CTC CGA TTT GCA TAT CTG GGC AGG G-3′
Sall4-MO1: 5′-TCC GCC ACC AAT TCC TGG AGT TG-3′
Sall4-MO2: 5′-AAG AGC CCT GGT GAC TTG CCC CCT C-3′
Nanog-MO1: 5′-GGA CCA GGA AGA CCC ACA CTC ATG T-3′
Nanog-MO2: 5′-GCA ACA ACC AAA AAA CTC AGT GTC T-3′
Dnmt3b-MO: 5′-GAT GTC TGC TGT CTC CCT TCA TTG T-3′
Control-MO: 5′-TCC AGG TCC CCC GCA TCC CGG ATC C-3′
The control MO does not target any known sequence in the mouse genome or transcriptome. For each control or knockdown condition, 5–10 pl of 0.6 mM MO was injected into the cytoplasm of each zygote on an inverted microscope (Olympus IX70) equipped with a hydraulic micromanipulation system (Narishige IM300 microinjector). In every experiment, at least 8–10 embryos were used for each of the conditions, namely uninjected, injection of control MO, or injection of MO targeting one of the pluripotency factors.
Immunocytochemical staining
Embryos were first fixed in 4% paraformaldehyde for 30 min and then permeabilized in 0.4% Triton X-100 for 30 min. Next, they were blocked in 10% normal goat serum for 1 h before incubation with antibodies. The embryos were incubated in 1:100 diluted primary antibody overnight at 4°C, in 1:5000 diluted secondary antibody for 1 h, and in 3 μM DAPI for 10 min, before being mounted on slides using Fluoromount-G (SouthernBiotech). All antibodies were diluted in blocking solution. The primary antibodies used were mouse monoclonal anti-Oct4 (Santa Cruz Biotechnologies, sc-5279), mouse monoclonal anti-Sall4 (Santa Cruz Biotechnologies, sc-101147), and rabbit polyclonal anti-Nanog (Cosmo Bio, RCAB0001P). Secondary antibodies were purchased from Invitrogen/Molecular Probes (Alexa Fluor 594 donkey anti-mouse IgG (A-21203) and Alexa Fluor 594 donkey anti-rabbit IgG (A-21207)). Embryos were washed in between the various steps with PBS-PVP and were imaged on the Leica SP2 AOBS confocal laser scanning microscope.
Synthesis of mRNAs for rescue experiments
The overall strategy was to clone the coding region from plasmids using primers that contained the T7 promoter. The PCR products then served as templates for in vitro transcription, which was performed using the mMessage mMachine T7 Ultra Kit (Ambion).
For Oct4, the plasmid MMM1013-98478834 (Open Biosystems) was first modified so that the final mRNA cannot be targeted by the corresponding MO1. The third basepairs of the DNA sequence coding for the second to eighth amino acids were changed using the QuikChange Lightning Kit (Agilent) and the following mutagenic primers: 5′-CCC ACC TTC CCC ATG GCA GGT CAT CTC GCA TCT GAT TTC GCC TTC TCA CCC CC-3′ and 5′-GGG GGT GAG AAG GCG AAA TCA GAT GCG AGA TGA CCT GCC ATG GGG AAG GTG GG-3′. The template for the transcription reaction was then amplified from the modified plasmid using the following primers: 5′-CCA AGT AAT ACG ACT CAC TAT AGG GAC AAG AGG TAA TCC ACC CCC GGC TCG-3′ and 5′-CAA AAT GAT GAG TGA CAG ACA GGC CAG GCT-3′ (the T7 promoter is underlined).
For Sall4, the plasmid OMM5896-99848745 (Open Biosystems) was used for cloning. Even though the gene insert does not include the MO target region, it lacks a STOP codon. The STOP codon and a FLAG tag were inserted at the C’-terminus via two rounds of site-directed mutagenesis using the QuikChange Lightning Kit (Agilent). The primers for the first round of mutagenesis were 5′-TAA GAT TGC TGT CAG CGA CGA CAA GTA ATC AGG CCT CAT GGG CC-3′ and 5′-GGC CCA TGA GGC CTG ATT ACT TGT CGT CGC TGA CAG CAA TCT TA-3′, while the primers for the second round of mutagenesis were 5′-AAA TAA GAT TGC TGT CAG CGA CTA CAA GGA CGA CGA CGA CAA GTA ATC AGG CC-3′ and 5′-GGC CTG ATT ACT TGT CGT CGT CGT CCT TGT AGT CGC TGA CAG CAA TCT TAT TT-3′. The template for the transcription reaction was then amplified from the modified plasmid using the following primers: 5′-CCA AGT AAT ACG ACT CAC TAT AGG GCG CGC ACC ATG TCG AGG CGC AAG CAG GC-3′ and 5′-AAA GCT GGG CCC ATG AGG CCT GAT TAC TT-3′.
For Nanog, the plasmid EMM1002-99866141 (Open Biosystems) was modified so that the final mRNA cannot be targeted by the corresponding MO1. The third basepairs of the DNA sequence coding for the second to seventh amino acids were changed using the QuikChange Lightning Kit (Agilent) and the following mutagenic primers: 5′-CCA TCA CAC TGA CAT GAG CGT CGG ACT ACC AGG ACC CCA CAG TTT GCC TAG T-3′ and 5′-ACT AGG CAA ACT GTG GGG TCC TGG TAG TCC GAC GCT CAT GTC AGT GTG ATG G-3′. The template for the transcription reaction was then amplified from the modified plasmid using the following primers: 5′-CCA AGT AAT ACG ACT CAC TAT AGG GTT GCC TAA AAC CTT TTC AGA AAT CCC TTC C-3′ and 5′-CCG ACT GCT CTT CCG AAG GTC AGG AGT TC-3′.
For Dnmt3b, the plasmid MMM1013-99827219 (Open Biosystems) was also modified so that the final mRNA cannot be targeted by the corresponding MO. The third basepairs of the DNA sequence coding for the second to seventh amino acids were changed using the QuikChange Lightning Kit (Agilent) and the following mutagenic primers: 5′-CCA GCC TCA CGA CAG GAA ACA ATG AAA GGC GAT AGT AGG CAC CTG AAT GAA GAA GAG GGT G-3′ and 5′-CAC CCT CTT CTT CAT TCA GGT GCC TAC TAT CGC CTT TCA TTG TTT CCT GTC GTG AGG CTG G-3′. The template for the transcription reaction was then amplified from the modified plasmid using the following primers: 5′-CCA AGT AAT ACG ACT CAC TAT AGG GCG GCC CAA GTA AAC GTA GCG CAG CGA T-3′ and 5′-GGG GAA CGA TGT GCT TCT GGT CCT AGC TTC-3′.
Cell culture and transfections
The Dicer+/+ (wild-type) and Dicer−/− ESCs (Calabrese et al, 2007) were grown in knockout DMEM (Life Technologies) supplemented with 15% FBS (Thermo Scientific Hyclone), Glutamax (Life Technologies), non-essential amino acids (Life Technologies), penicillin/streptomycin (Life Technologies), β-mercaptomethanol (Life Technologies), and LIF (Millipore). Cells were transfected with Lipofectamine 2000 (Life Technologies) according to manufacturer’s instructions. The media was replaced 4–6 h after transfection.
Microarray analysis of pooled embryos
For each experiment, RNA was extracted from 20 embryos using the PicoPure RNA Isolation Kit (Arcturus). The RNA was reverse transcribed and amplified using the WT-Ovation One-Direct Amplification System (NuGEN). The cDNA was then fragmented and labeled using the FL-Ovation cDNA Biotin Module (NuGEN) and hybridized to the Mouse Genome 430 2.0 Array (Affymetrix). All manufacturers’ instructions were followed. The expression data were submitted to the NCBI Gene Expression Omnibus (GEO) ( http://www.ncbi.nlm.nih.gov/geo) under accession number GSE42135.
The raw data of 42 GeneChips were normalized and processed using dChip (Li and Wong, 2001). The estimations of expression level, fold change between two samples, and the FDR were calculated based on the methodology developed by Johnson and Wong (see http://www.stanford.edu/group/wonglab/doc/NewGeneScoreBiostatDraft.pdf). In comparison with the traditional moderated t-statistic, this methodology takes into account both the fold change and the expression magnitude and calculates a statistic based on fold change, logged fold change, and unpaired t-statistic. We defined differential expression by a threshold of 0.1 FDR estimated from 5000 random permutations across the samples. At least one of the following two conditions has to be satisfied for a gene to be considered as being regulated by a pluripotency factor: (1) significant in uninjected sample versus knockdown sample but not significant in uninjected sample versus control injected sample or (2) significant in control injected sample versus knockdown sample. We converted accession IDs of the results to gene names using tools available on the Mouse Genome Informatics (MGI) website.
Analysis of chIP sequencing (chIP-seq) data sets
The genomic regions bound by Oct4 and Nanog were obtained from a data set published by Chen et al (2008b). The genes regulated by Oct4 and Nanog were obtained in two ways. First, we searched for the closest gene to the bound chromosomal position. Second, we obtained a list of regulated genes using the approach developed by Ouyang et al (2009) with a transcription factor-association strength (TFAS) scores threshold of 3. The list of genes directly regulated by Sall4 was pooled from two published data sets (Lim et al, 2008; Yang et al, 2008). The accession IDs from Yang et al were converted to gene names using the tools on the MGI website. We used CisGenome (Ji et al, 2008) to calculate motif enrichment and to perform de novo motif discovery based on mouse genome mm8. The bound loci from Chen et al were given as single nucleotides, so we extended them to 400 bp regions centered at those loci (200 bp upstream and 200 bp downstream) for our motif analysis. Control regions were randomly selected and the number of control regions was five times that of the targets.
GO analysis
The Gene Expression Data Analysis tool within the PANTHER resource ( http://www.pantherdb.org/tools/genexAnalysis.jsp) (Thomas et al, 2003) was used to identify functional categories in our microarray results. The default set of Mus Musculus genes was used as the reference. Since some common categories are ‘noisy’ and will always appear regardless of the input list, we also randomly generated subsets of genes from all the genes that were tiled on the Affymetrix microarray and performed GO analysis on these random gene lists. The categories or biological processes that showed up repeatedly during the GO analysis of these random lists were then discarded.
Expression profiling of miRNAs
For each experiment and each condition, 20 embryos were harvested directly into DNaseI-containing Lysis Solution from the TaqMan MicroRNA Cells-to-Ct Kit (Applied Biosystems) and the lysates were stored at −80°C. We used the Megaplex™ Primer Pools (Rodents; Applied Biosystems) for reverse transcription and pre-amplification. Reverse transcription was performed at 42°C for 30 min using the MultiScribe RT and pre-amplification was performed for either 12 or 16 cycles (95°C for 15 s and 60°C for 4 min) using the TaqMan PreAmp Master Mix (Applied Biosystems). Real-time PCR was performed on the 7900HT System (Applied Biosystems) using the TaqMan Rodent MicroRNA Array Set (also called TaqMan Low Density Arrays; Applied Biosystems).
Our pipeline for converting Ct values to expression levels is as follows. We set a threshold Ct of 32, above which we considered the miRNA to be absent. The data were normalized to the average Ct values for the U6 RNA and the expression level was taken to be 2−Ct. For each biological sample, we repeated the experiment twice, once with 12 cycles of pre-amplification and once with 16 cycles of pre-amplification. If the expression level from 12 cycles of pre-amplification was 0, then we kept the expression level from 16 cycles of pre-amplification unless there was evidence of non-specific amplification (typically an unusually small Ct outlier). Otherwise, we would use the expression level from 12 cycles of pre-amplification, as there would be less amplification bias with fewer cycles. To compare knockdown embryos with uninjected embryos or embryos injected with a control MO, we performed a regularized t-test (Baldi and Long, 2001) to determine the differentially expressed miRNAs (P<0.05).
MiRNA-target predictions
We first downloaded a set of target site predictions from www.microrna.org (August 2010 release), which was based on the miRanda algorithm (Betel et al, 2008), to identify the miRNAs that may potentially regulate the protein-coding genes that were differentially expressed during knockdown of a pluripotency factor. Subsequently, to increase confidence in the predictions, we used the miRWalk database (Dweep et al, 2011) to identify the miRNA-target pairs that were predicted not only by miRanda but also by at least one other program, including DIANA-microT (Maragkakis et al, 2009a), miRDB (Wang and El Naqa, 2008; Wang, 2008), miRWalk (Dweep et al, 2011), PITA (Kertesz et al, 2007), RNA22 (Miranda et al, 2006), and TargetScan (Lewis et al, 2005; Friedman et al, 2009). Default settings were used for the minimum seed length and the P-value.
Microfluidic quantitative real-time PCR
For reverse transcription and pre-amplification, TaqMan gene expression assays (20 × , Applied Biosystems) were pooled, so that each assay was at a final concentration of 0.2 × . Each individual embryo was harvested directly into a RT-PreAmp Master Mix (5.0 μl CellsDirect 2 × Reaction Mix (Invitrogen), 2.5 μl 0.2 × Assay Mix, 0.5 μl SuperScript III RT/Platinum Taq Mix (Invitrogen), 1.4 μl TE Buffer, and 0.1 μl SUPERase-In (Ambion)). The harvested samples were first reverse transcribed at 50°C for 20 min and then pre-amplified for 20 cycles (95°C for 15 s and 60°C for 4 min). The pre-amplified products were diluted four-fold with TE buffer before real-time PCR on the BioMark System (Fluidigm). We used 48.48 Dynamic Arrays (Fluidigm), TaqMan Universal PCR Master Mix (Applied Biosystems), and TaqMan gene expression assays (Applied Biosystems) in these real-time PCR experiments. The arrays were primed, loaded, and run according to manufacturer’s instructions. Ct values were calculated from the BioMark System’s software using default settings.
Single-embryo and single-cell data processing
To test for normality, we applied the Shapiro–Wilk test. If the two data sets under consideration (either original Ct values or relative expression levels) passed the test, then we would use the Student’s t-test to compare them. If not (which means either one or both data sets did not come from a normal distribution), then we would apply the Wilcoxon Rank Sum test. In addition, we applied a two-sample Kolmogorov–Smirnov test for all comparisons. Expression levels were calculated using a ΔΔCt method.
Since individual cells are derived from whole embryos, fluctuations in single-cell gene expression values can be due to embryo-to-embryo variability (inter-embryo variability) or due to differences between cells of the same embryo (intra-embryo variability). To determine the contribution of inter-embryo variability to gene expression fluctuations observed between individual cells, we first computed the total expression level of an embryo from the Ct values of the four cells that constituted that embryo and then calculated the standard deviation between whole embryos. To determine the contribution of intra-embryo variability to single-cell gene expression fluctuations, we first computed for each gene the standard deviation of the set of Ct values from cells of the same embryo and then calculated the mean of these standard deviations for each experimental condition (uninjected embryos, embryos injected with a control MO, or embryos injected with a MO targeting a pluripotency factor).
Supplementary Material
Acknowledgments
We thank Annett Hahn-Windgassen and Nicholas Johnson for technical assistance, Grace Zheng for providing the wild-type and Dicer knockout ESCs and for technical advice, Yul Yang for technical advice, and Yue Wan and Elizabeth Pollina for critical reading of the manuscript. We also thank Aaron Hsueh and Jin Billy Li for use of laboratory resources. This work was supported by National Institutes of Health Grant R01 HD057970 (to MWMY and WHW).
Author contributions: MHT and MWMY conceived the study. MHT and DEL performed the experiments with initial help from KF. KFA and MHT analyzed the data, with help from WHW. MHT wrote the manuscript with inputs from KFA, WHW, and MWMY. WHW and MWMY supervised the research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA (2008) A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol 15: 268–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA (2007) RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc Natl Acad Sci USA 104: 18097–18102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, DeZiel CE, Gross RH (2007) SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res 35: W259–W264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty A, Carlson JM, Khetani RS, Gross RH (2007) A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinformatics 8: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Xie S, Li E (2002) A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J Biol Chem 277: 38746–38754 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008a) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008b) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N (2011) miRWalk–database: prediction of possible miRNA binding sites by ‘walking’ the genes of three genomes. J Biomed Inform 44: 839–847 [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R, Fan G (2008) Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2: 160–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foygel K, Choi B, Jun S, Leong DE, Lee A, Wong CC, Zuo E, Eckart M, Reijo Pera RA, Wong WH, Yao MW (2008) A novel and critical role for Oct4 as a regulator of the maternal-embryonic transition. PLoS ONE 3: e4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, Min R, Alves P, Abyzov A, Addleman N, Bhardwaj N, Boyle AP, Cayting P, Charos A, Chen DZ, Cheng Y et al. (2012) Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P (2010) Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 18: 675–685 [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS (2004) Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6: 117–131 [DOI] [PubMed] [Google Scholar]
- Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, Lufkin T, Ng HH, Tam WL, Lim B (2010) Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 463: 1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH (2008) An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol 10: 731–739 [DOI] [PubMed] [Google Scholar]
- Lim CY, Tam WL, Zhang J, Ang HS, Jia H, Lipovich L, Ng HH, Wei CL, Sung WK, Robson P, Yang H, Lim B (2008) Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 3: 543–554 [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U (2003) Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA 100: 11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Reczko M, Simossis VA, Alexiou P, Papadopoulos GL, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG (2009a) DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res 37: W273–W276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA, Sethupathy P, Vergoulis T, Koziris N, Sellis T, Tsanakas P, Hatzigeorgiou AG (2009b) Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics 10: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U (2002) Network motifs: simple building blocks of complex networks. Science 298: 824–827 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999a) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999b) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Ouyang Z, Zhou Q, Wong WH (2009) ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci USA 106: 21521–21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, Nishinakamura R (2006) The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 133: 3005–3013 [DOI] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput Biol 3: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15: 259–267 [DOI] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A (2012) A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Tamminga J, Kathiria P, Koturbash I, Kovalchuk O (2008) DNA damage-induced upregulation of miR-709 in the germline downregulates BORIS to counteract aberrant DNA hypomethylation. Cell Cycle 7: 3731–3736 [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA (2010) Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 6: 468–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Bao S, Lee C, Wang X, Tuch BB, Heard E, Lao K, Surani MA (2011) Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS ONE 6: e21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N (2008) Autophagy is essential for preimplantation development of mouse embryos. Science 321: 117–120 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E (2006) Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development 133: 1183–1192 [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M (2004) A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell 6: 133–144 [DOI] [PubMed] [Google Scholar]
- Wang X (2008) miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA 14: 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, El Naqa IM (2008) Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics 24: 325–332 [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, Ng HH (2006) Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem 281: 24090–24094 [DOI] [PubMed] [Google Scholar]
- Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S (2010) Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res 20: 804–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Chai L, Fowles TC, Alipio Z, Xu D, Fink LM, Ward DC, Ma Y (2008) Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc Natl Acad Sci USA 105: 19756–19761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wu J (2007) MicroRNAs and regenerative medicine. DNA Cell Biol 26: 257–264 [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM (2004) Transcript profiling during preimplantation mouse development. Dev Biol 272: 483–496 [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M, Morris SA, Bruce AW (2009) Making a firm decision: multifaceted regulation of cell fate in the early mouse embryo. Nat Rev Genet 10: 467–477 [DOI] [PubMed] [Google Scholar]
- Zheng GX, Ravi A, Calabrese JM, Medeiros LA, Kirak O, Dennis LM, Jaenisch R, Burge CB, Sharp PA (2011) A latent pro-survival function for the mir-290–295 cluster in mouse embryonic stem cells. PLoS Genet 7: e1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH (2007) A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci USA 104: 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.