Abstract
Establishment of cell polarity is crucial for many biological processes including cell migration and asymmetric cell division. The establishment of cell polarity consists of two sequential processes: an external gradient is first sensed and then the resulting signal is amplified and maintained by intracellular signaling networks usually using positive feedback regulation. Generally, these two processes are intertwined and it is challenging to determine which proteins contribute to the sensing or amplification process, particularly in multicellular organisms. Here, we integrated phenomenological modeling with quantitative single-cell measurements to separate the sensing and amplification components of Wnt ligands and receptors during establishment of polarity of the Caenorhabditis elegans P cells. By systematically exploring how P-cell polarity is altered in Wnt ligand and receptor mutants, we inferred that ligands predominantly affect the sensing process, whereas receptors are needed for both sensing and amplification. This integrated approach is generally applicable to other systems and will facilitate decoupling of the different layers of signal sensing and amplification.
Keywords: Caenorhabditis elegans , cell polarity, phenomenological modeling, Wnt signaling
Introduction
Cell polarity is important for many cellular functions, such as migration, axis formation, and asymmetric cell division (Strutt, 2001; Nelson, 2003; Silhankova and Korswagen, 2007). Conceptually, the establishment of cell polarity in most cells can be thought of as a two-step process: sensing followed by amplification (Weiner, 2002). The sensing machinery is used to detect an external cue. Examples of external cues for unicellular organisms include a pheromone gradient that induces cell polarity in budding yeast and cAMP gradients that directs the chemotactic response of Dictyostelium discoideum (Klein et al, 1988; Arkowitz, 1999). Remarkably, many cells are able to polarize in response to very shallow chemoattractant gradients as small as a 1% change in concentration across the cell diameter (Zigmond, 1977; Baier and Bonhoeffer, 1992). This shallow gradient induces a steep intracellular gradient of signaling molecules and cytoskeleton components allowing the cell to polarize (Parent et al, 1998; Servant et al, 2000). After the sensing machinery detects the external cue, an amplification mechanism sets in to convert this spatial information into a stable polarity axis. These amplification mechanisms are often based on positive feedback regulation (Meinhardt, 1999; Weiner, 2002). For example, during the establishment of cell polarity in budding yeast, activated Cdc42 orients the actin cytoskeleton and directs the delivery of more Cdc42 to membrane sites with high concentrations of the protein (Pruyne and Bretscher, 2000; Wedlich-Soldner et al, 2004). These vesicles are thought to contain Cdc42 therefore reinforcing the polarity axis via positive feedback (Wedlich-Soldner et al, 2003). Amplification through positive feedback has also been reported to be involved in planar cell polarity in multicellular organisms (Tree et al, 2002). In this case, positive feedback serves to polarize a field of cells by amplifying differences between protein levels on adjacent cell surfaces. Amplification ensures robustness and is responsible for the dramatic sensitivity of gradient sensing and the generation of spontaneous cell polarization.
Many mathematical models have been proposed to model the processes of cell polarization (Wedlich-Soldner et al, 2003; Jilkine and Edelstein-Keshet, 2011). Although these models have yielded valuable insights into the underlying molecular mechanisms required for sensing and amplification, they cannot be used to separate and quantify the contributions of the different molecular components to these processes, particularly in the in vivo context of a multicellular organism. Furthermore, construction of such mechanistic models remains challenging and is only possible for a few well-studied system due to the lack of detailed knowledge. On the other hand, phenomenological models that describe the main features of a system through a few important parameters have been used successful in understanding the general features of many systems (Alon, 2007; Mallavarapu et al, 2009). Here, through the integration of quantitative cell polarity measurements with a phenomenological model, we separated the contributions of Wnt ligands and receptors to sensing and amplification during establishment of polarity in the Caenorhabditis elegans’ P cells.
Wnt signaling plays an important role in regulating cell polarity in many organisms and Wnts have been found to be expressed in a gradient (Yoshikawa et al, 2003; Harterink et al, 2011). In C. elegans, loss of Wnt signaling leads to cell migration and cell division defects (Thorpe et al, 1997; Whangbo et al, 2000; Silhankova and Korswagen, 2007). Many of these asymmetric divisions are controlled by the Wnt/β-catenin asymmetry pathway. In this pathway, Wnts first regulate the polarity of the mother cell through asymmetric protein localization (Mizumoto and Sawa, 2007a). After division, the asymmetric protein localization leads to different nuclear levels of POP-1 and SYS-1 in daughter cells (Phillips et al, 2007; Mizumoto and Sawa, 2007b). This mechanism converts the polarity information set up in the mother cell into differential gene expression in the daughters (Mizumoto and Sawa, 2007b). Although major progress has been made in uncovering new Wnt pathways and components, the exact functions of ligands and receptors remain unknown. In many cases, it was found that loss of ligands and receptors leads to different phenotypes (Sternberg and Horvitz, 1988; Herman and Horvitz, 1994). This is unexpected for components acting in the same pathway, suggesting that ligands and receptors may have different functions. But there are also cases where loss of receptors phenotypically mimick the effects of ligand loss. For example, both loss of ligands and receptors cause loss of polarity in the EMS cell (Thorpe et al, 1997). Intriguing, the phenotype is less severe in the ligand/receptor double mutant than the ligand mutant (Bei et al, 2002). This suggests that in the absence of ligands, receptors may be responding to signals that interfere with setting up of the correct polarity.
Here, we found that Wnt signaling also regulates the division of the P cells, which are six pairs of symmetrically placed cells, P1–P12, found at the left and right sides of the worm after hatching. During the first larval stage, P cells migrate to the ventral side and form a single row of cells (Sulston, 1976). After migration, they divide to produce anterior neuronal blast cells, Pn.a, and posterior epithelial daughters, Pn.p, n=1, 2,… 12 (Figure 1A). P cells provide a convenient model system to study cell polarity for the following reasons. First, the establishment of cell polarity leads to a clear observable phenotype of different cell fates (neuronal versus epithelial) in the daughter cells. Second, this is an approximate one-dimensional system with anterior–posterior polarity. Third, the presence of many P cells in the same animal, allow us to study how the establishment of cell polarity varies along the anterior–posterior axis.
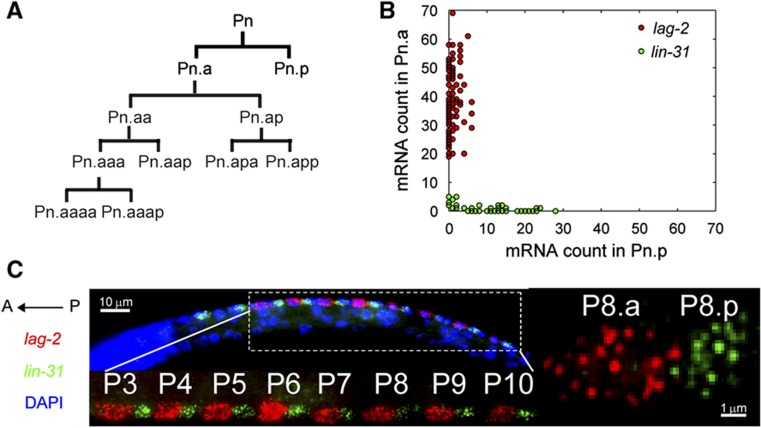
Figure 1.
Identification of Pn.a and Pn.p expression markers. (A) Lineage diagram of P cells. P cells divide to form Pn.a and Pn.p. Pn.a divides to give rise to five different neurons in L1. Pn.p does not divide till the L3 stage. (B) Plot of lag-2 (red) and lin-31 (green) mRNA in Pn.p versus Pn.a. (C) (Top, left) Image of L1 larva after in situ hybridization with lag-2 (red) and lin-31 (green) probes. Dapi is shown in blue. (Top, bottom) Maximum projection image of multiple z-stacks that shows lag-2 (red) and lin-31 (green) expression in Pn.a and Pn.p from the P3–P10 lineages. (Right) Zoom in image of a single stack showing individual lag-2 and lin-31 transcripts in P8.a and P8.p.
Results
lag-2 and lin-31 reliably mark the Pn.a and Pn.p cells, respectively
To identify a marker of P-cell polarity, we quantified the mRNA expression of a panel of 26 genes using single-cell transcript counting (Raj et al, 2008). This panel includes genes that were previously reported to be expressed in P cells and their descendants and also genes from the major signaling pathways (Wnt, Notch, FGF, EGF, and TGF). A set of about 48 single-stranded 20-mer oligonucleotides were designed for visualization of each transcript. These fluorescently labeled oligonucleotides are complementary to the transcript and bind each individual transcript. This becomes visible as a diffraction-limited spot using fluorescence microscopy. Using a custom-written software, we manually segmented the individual cells and computationally determined the transcript number in each cell. The ratio of the expression in the Pn.a versus Pn.p daughters was used to quantify the specificity of the putative markers (Supplementary Table S1). We identified lag-2, a transmembrane protein of the Delta/serrate/lag-2 family, as the most promising Pn.a marker having the largest expression ratio (37.3). lin-31, a gene previously reported as a Pn.p marker (Tan et al, 1998), was found to be the most promising Pn.p marker with lowest expression ratio (0.08). Using these markers, the anterior and posterior daughters of the P3–P10 cells can be scored with high confidence (Figures 1C and 2C; Supplementary Figure 1).
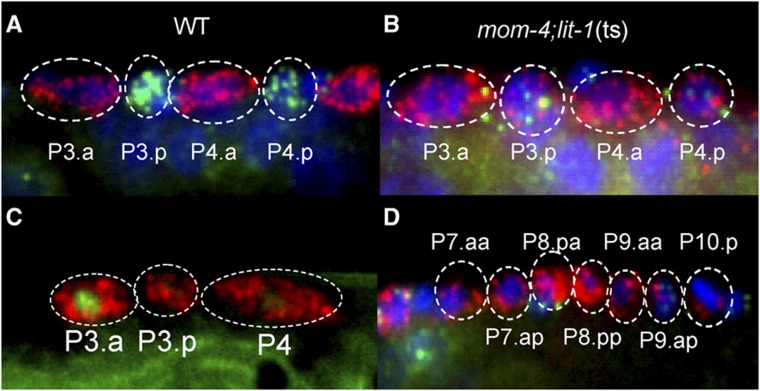
Figure 2.
Inhibition of Wnt/β-catenin asymmetry pathway. (A) In WT animals, lag-2 transcript (red) and lin-12 transcript (green) are found in the Pn.a and Pn.p respectively. (B) In mom-4; lit-1 worms that have been transferred to 25°C for 9 h prior fixation to inhibit the Wnt/β-catenin asymmetry pathway, lag-2 transcript is observed in both the Pn.a and Pn.p. (C) POP-1 localization is high in the Pn.a and low in Pn.p after P-cell division. (D) Both the Pn.a and Pn.p divide in L1, leading to the increase in number of cells in the ventral nerve cord.
Although both daughter cells initially inherit lag-2 mRNA from the P cell, lag-2 is rapidly degraded in the Pn.p cells (Figure 2C). We quantified lag-2 and lin-31 expression by counting individual transcripts in the Pn.p and Pn.a cells excluding cells immediately after division. We observed that in wild-type (WT) animals lag-2 and lin-31 expression is mutually exclusive (Figure 1B). In WT animals, the lag-2 count in Pn.a cells is always much larger than the lag-2 count in Pn.p cells (Figure 1B).
Role of the Wnt/β-catenin asymmetry pathway in regulating P cells’ divisions
Many asymmetric divisions in C. elegans are regulated by the Wnt/β-catenin asymmetry pathway through polarization of the mother cell before division (Mizumoto and Sawa, 2007b). In this pathway, asymmetry localization of many regulatory proteins (including WRM-1 and APR-1) in the mother cell is set up by Wnt ligands and receptors, leading to different nuclear levels of POP-1 and SYS-1 in the daughter cells after division. A high level of POP-1 and low level of SYS-1 in the anterior daughter leads to inhibition of Wnt signaling, whereas a low level of POP-1 and high level of SYS-1 in the posterior daughter leads to transcription of Wnt responsive genes. The amount of nuclear POP-1 in the posterior daughter is regulated by MOM-4 and LIT-1 proteins, which function in exporting POP-1 from the nuclear into the cytoplasm.
To test if the Wnt/β-catenin asymmetry pathway plays a similar role in P cells, we examined the nuclear levels of POP-1 in the daughter cells after division in a POP–GFP translational fusion strain. We found that the anterior daughter has a higher level of nuclear POP-1 than the posterior daughter (Figure 2C). This differential level of POP-1 is also observed in other daughter cells regulated by the Wnt/β-catenin asymmetry pathway (Mizumoto and Sawa, 2007b). Next, we disrupted the POP-1 branch in the asymmetry pathway by a temperature shift in mom-4; lit-1 mutant worms (Takeshita and Sawa, 2005). Similar to WT animals, both Pn.a and Pn.p in mom-4; lit-1 mutant worms inherited lag-2 mRNA from the P cells. But unlike WT animals, Pn.p cells continue to express lag-2 some time after division (Figures 2B and 3B). This lag-2 expression is sustained even after Pn.p cells go on to divide in L1. (Figure 2D). In WT animals, Pn.p cells do not divide until the L3 stage. This suggests that upon inhibition of the Wnt/β-catenin asymmetry pathway, the Pn.p cells have adopted Pn.a-like fates where they expressed lag-2 and divide in L1. This effect is similar to that observed in other cells whereby disruption of the Wnt/β-catenin asymmetry pathway caused the posterior daughter to take on anterior cell fate (Bertrand and Hobert, 2009; Gleason and Eisenmann, 2010). These experiments confirm the role of Wnt/β-catenin asymmetry pathway in polarizing P cells and show that lag-2 expression in the daughter cells after division is a good reflection of the polarity of the mother P cell before division.
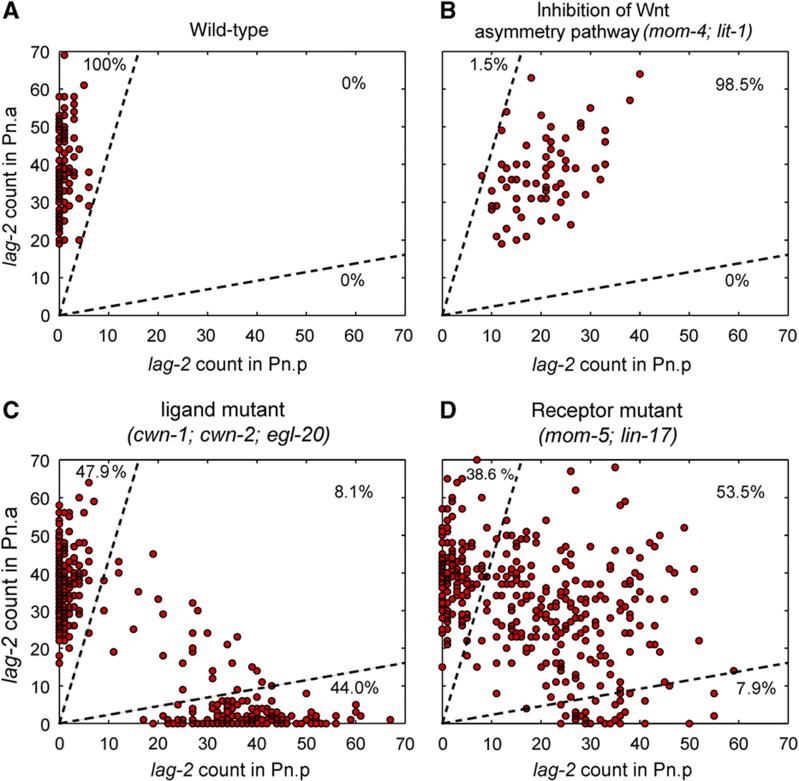
Figure 3.
lag-2 expression in different mutants. Plot of lag-2 mRNA counts in Pn.p versus Pn.a in WT animals. (A) In animals with inhibition of Wnt/β-catenin asymmetry pathway, mom-4; lit-1 (B), multiple-ligand mutant, egl-20; cwn-1; cwn-2, (C) and double-receptor mutant, mom-5; lin-17 (D). Dotted lines divide the plot of lag-2 expression in Pn.p versus Pn.a into regions for correct polarity, loss of polarity and reverse polarity. Fractions of cells that fall into these three regions are indicated on the plots.
Notch signaling is not involved in setting up P-cell polarity
Since Pn.a and Pn.p express high levels of lag-2 and lin-12, respectively (Figure 2A), we tested for the role of Notch signaling in P-cell polarity. To determine if Notch signaling may be required for maintaining different fates in the Pn.a and Pn.p, we inhibited Notch signaling using a lag-1 temperature-sensitive strain (Qiao et al, 1995). However, we did not observe any aberrant development in the Pn.a and Pn.p cells. We also examined animals with lin-12 loss of function mutation, lin-12(n941), and semi-dominant mutation, lin-12(n137), and did not observe any change in cell fates and the expression of lag-2 in Pn.a and Pn.p cells (Greenwald et al, 1983). These results suggest that notch signaling is not involved in setting up P-cell polarity.
Mutations in Wnt ligands induce polarity reversal, whereas mutations in Wnt receptors induce polarity loss
To explore which Wnt ligands and receptors are involved in P-cell polarity, we examined lag-2 and lin-31 expression in Pn.a and Pn.p cells in ligand and receptor mutants. WT polarity was observed in most single- and double-ligand mutants, consistent with previous observations on the redundant role of the Wnt ligands (Zinovyeva et al, 2008) (Supplementary Figure S2). However, in the double-ligand mutant (egl-20; cwn-1), triple-ligand mutant (egl-20; cwn-1; cwn-2), and quintuple-ligand mutant (egl-20; cwn-1; cwn-2; lin-44; mom-2), some Pn.p cells expressed lag-2 and their corresponding Pn.a cells expressed lin-31, suggesting that the P cell had been polarized in the opposite direction (Figure 3C; Supplementary Figure S3). We found that in the triple-ligand mutant, 47.9% correctly polarized, 44.0% exhibited a polarity reversal, and 8.1% showed similar levels of lag-2 in both daughters indicative of a symmetric P-cell division and therefore loss of P-cell polarity. A very different phenotype was observed in the Wnt receptor mutants (Figure 3D). In the mom-5; lin-17 mutant, lag-2 was often observed in both daughter cells (53.5%) and a smaller fraction of cells correctly polarized (38.6%) or reverse polarized (7.9%). The different phenotypes observed in the ligand and receptor mutants suggest that they may play different roles in establishing cell polarity. To determine if the different phenotypes observed is due to the different roles of receptors and ligands in sensing and amplification and to further quantify the phenotypes presented in Figure 3, we constructed a phenomenological model.
Cell–cell signaling has been found to be important in planar cell polarity models (Tree et al, 2002). Since pairs of P cells (P3/P4, P5/P6, P7/P8, and P9/P10) are in contact before division, cell–cell signaling may play a role in their divisions. If cell–cell signaling occurs between pair of P cells, we would expect the polarity of division between these pairs of P cells to be correlated. In WT animals, all the divisions are correct hence WT animals cannot be used for this analysis. Hence, we calculate correlation in the triple-ligand mutant (egl-20; cwn-1; cwn-2) where many of the divisions are polarized in the reversed direction. If there is cell–cell signaling, we would expect pairs of P cells to both divide correctly, leading to both Pn.a daughters having high lag-2 levels, or to both divide with reverse polarity, leading to low levels of lag-2 in both Pn.a daughters. When we examined the lag-2 expression levels in the anterior daughters for the anterior P cell versus the posterior P cell in each pair (Supplementary Figure 4), no significant correlation is found between their expression levels (correlation value=−0.029, P-value=0.70). This suggests that cell–cell signaling does not play a significant role in P–cell division.
A phenomenological model for cell polarity
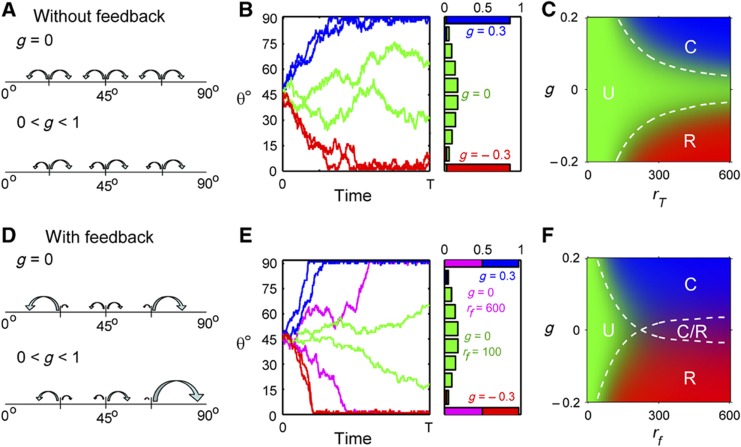
The polarization state of the P cell is denoted by the dynamic variable θ(t), which is an angle that ranges between 0° (reverse polarity) and 90° (WT polarity). We assume that the P cell has initially no polarity (θ(t=0)=45°). For subsequent times, until the P cell divides at time T, we describe the dynamic behavior of θ(t) as a one-dimensional random walk. In the simplest model that only involves the sensing of an external gradient, the probability of taking a right step (increasing θ; towards WT polarity) or left step (decreasing θ; towards reverse polarity) is ((1+g)/2)rTΔt and ((1−g)/2)rTΔt, respectively. Here, g reflects the external gradient that ranges from −1 to 1, rT is the total reaction rate that sets the timescale of the dynamics, Δt is the time step of the numerical simulation. To understand the behavior of the model, we ran stochastic simulations to determine the behavior of individual cells. The chemical master equation is also solved to obtain the distribution for a population of cells at time T. In absence of an external gradient (g=0), θ(t) has equal probability of increasing or decreasing at each step of the simulation (Figure 4A) and performs an unbiased random walk resulting in an approximately Gaussian distribution (symmetric about 45°) at the time of P-cell division (Figure 4B, green). If g>0 or g<0, the probabilities of taking a left and right step are unequal, resulting in a biased random walk towards 90° (blue) or 0° (red), respectively (Figure 4A and B). To summarize the information encoded in the histograms of θ(T) for each set of g and rT, we determine the fraction of the cells that undergoes correct, reverse or loss of polarity based on the following classification rules: θ(T)>75° (correct polarity), θ(T)<15° (reverse polarity), and 15°<θ(T)<75° (loss of polarity). A coordinate on the phase diagram corresponding to the particular set of g and rT, will be colored with different intensities of blue, red, and green depending on the fractions of correct, reverse, and loss of polarity, respectively. (Figure 4C) As expected for g>0 and high rT, values of most cells are correctly polarized. For values of g close to 0 and low values of rT, most of the cells are unpolarized. However, this simple model is unable to explain the coexistence of correct and reverse polarity observed in the triple-ligand mutant (Figure 3C).
Figure 4.
Stochastic modeling. Without feedback (A) Schematics showing the dependence of ra and rp on g. For g=0, ra=rp and for 0<g<1, ra>rp. Values of ra and rp do not depend on θa. (B) (Left) Stochastic simulation of θa from t=0 to t=T (division of P cell) (Right) Histograms of θa(T) for constant rT=0.05 with different values of g=0.3 (blue), g=0 (green), and g=−0.3 (red). (C) Phase diagram showing the parameters space of g and rf giving rise to correct polarity (blue/C), reverse polarity (red/R), and unpolarized divisions (green/U). Dotted lines show the region of parameter space giving rise to 35% correct polarity (top) or 35% reverse polarity (bottom). With feedback (D) Schematics showing the dependence of ra and rp on g and θ. For g=0, ra=rp at θa=45°. For 0<g<1, ra>rp at θa=45°. For both cases, ra increases with θa whereas rp decreases with θa. (E) (Left) Stochastic simulation of θa from t=0 to t=T (division of P cell) (Right) Histograms of θa(T) for g=0.3 and rf=0.06 (blue), for g=−0.3 and rf=0.06 (red), for g=0 and rf=0.06 (magneta) and for g=0 and rf=0.01 (green). (F) Phase diagram showing the parameters space of g and rf giving rise to correct polarity (blue/C), reversed polarity (red/R), unpolarized divisions (green/U), and coexistence of correct and reverse polarity (magneta/C/R). Dotted lines show the region of parameter space giving rise to 35% correct divisions (top) or 35% polarity flips (bottom). The intersection of the two dotted lines shows the coexistence of correct and reverse polarity.
To expand beyond this simple model, we included a mechanism that reinforces a deviation from the non-polarized state. Biologically, this reinforcement could be established by, for example, positive feedback regulation. To introduce amplification, we let the probability of taking a right step increase with θ and the probability of taking a left step decrease with θ. The simplest way to reinforce deviation is to introduce linear dependencies on θ into the probabilities of taking a right or left step. In this expanded model, the probability of taking a right step (increasing θ; towards WT polarity) or left step (decreasing θ; towards reversed polarity) is ((1+g)/2)(θrf+rb)Δt and ((1−g)/2)([90°−θ]rf+rb)Δt, respectively. g performs the same role here as in the simple model and characterized the gradient. On the other hand, rT is replaced by two rates rf and rb. In the absence of a gradient (g=0), θ(t) has equal probability of increasing or decreasing when the cell is unpolarized (θ=45°) similar to the behavior of the simple model. However, if there is any fluctuation that drives the cell away from the unpolarized state, this fluctuation will be reinforced. In other words, the probability that θ will move in the same direction as the fluctuation does, is larger than the probability to move in the opposite direction of the fluctuation (Figure 4D and E, green traces). The parameter rf quantifies this reinforcement. A basal rate independent of θ(t), rb, is included and set to be a constant. The external gradient is superposed on this process and further biases the random walk (Figure 4D and E, blue, red, and magenta traces). Another way to view the polarization process is that initially when the cell is unpolarized (θ=45°), the contributions of rf to the probabilities of taking a right or left step are equal. Hence, the sensing process, represented by g, plays a more important role is setting up polarization. Once sensing has occurred and θ is no longer close to 45°, contributions of amplification represented by rf will become important and act synergistically with the gradient to further set up the polarization. A model assuming additive effects of gradient sensing and amplification is also unable to produce the coexistence of correct and reverse polarity (Supplementary Figure S5).
We repeated the analysis for the expanded model, setting rb as a constant and found a region in parameter space (g versus rf), which allows for the coexistence for correct and reverse polarity (Figure 4F, magenta), which was absent in the simple model (Figure 4C). We then used this model to further quantify the experimental results and explore where the different mutants are located in this parameter space. Note that the present model is a purely phenomenological model (in contrast to a mechanistic molecular model) that allows us to extract parameters, g and rf, from the experimental data.
Loss of Wnt ligands reduces the parameter g, whereas loss of Wnt receptors results in a decreased value of both g and rf
To compare the predictions of the model to the experimental data, we converted the lag-2 transcript count in each pair of daughter P cell to a single angle θlag-2 (Figure 5A). From the earlier experiments, we observed that inhibition of the Wnt/β-catenin asymmetry pathway, which sets up polarity of the P cell, led to expression of lag-2 in both daughter cells. Hence, θlag-2 is a good approximation of the polarization state of the mother P cell at the time of division θ(T) as calculated by the model above.
Figure 5.
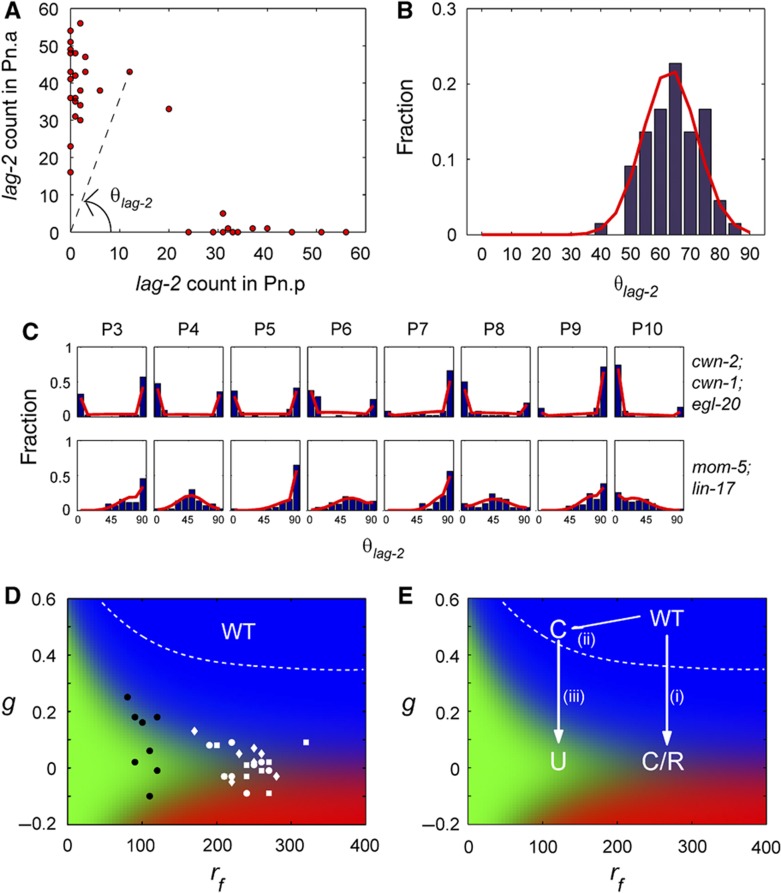
Fitting of ligands and receptors mutants distributions. (A) Plot of lag-2 mRNA counts in P3.p versus P3.a in egl-20; cwn-1; cwn-2 strain and illustration of how θlag-2 is calculated. (B) Histograms for θlag-2 (blue) and maximum likelihood fits (red) for mom-4; lit-1 strain grown at 25°C for 9 h. (C) Histograms for θlag-2 (blue) and maximum likelihood fits (red) for P3–P10 in egl-20; cwn-1; cwn-2 and mom-5; lin-17 strains. (D) Fit parameter, g and rf obtained using maximum likelihood for triple-ligand mutant (egl-20; cwn-1; cwn-2 strain) (white) and receptor mutant (mom-5; lin-17 strain) (black). White dotted line shows the region of parameter space giving rise to above 99.9% correct divisions. Although the parameters of WT P cells cannot be determined exactly as many combinations of parameters can give rise to 100% correct polarity, the parameters of WT P cells must lie above the white dotted line. Hence, values of g obtained for both ligands and receptors mutants are lower than that of WT P cells. (E) Abstraction of parameters’ movement in phase space as a result of mutations. (i) Decreasing ligands in WT P cells leads to reduction of g and converts correct polarity into the coexistence of correct and reverse polarity observed in the triple-ligand mutant. (magneta) (ii) Decreasing receptors slightly will lead to reduction of rf and cells that still polarize correctly if the parameters lie within the region of correct division (white dotted line). (iii) Decreasing ligands in strains with reduced rf would lead to loss of polarity.
To determine the value of rb, we examined the distribution of θlag-2 in the mom-4; lit-1 strain and observed a Gaussian distribution (Figure 5B). Although ligands and receptors are functioning properly in the mom-4; lit-1 strain, Pn.a and Pn.p are unable to execute different cell fates as the MOM-4 and LIT-1 proteins, required for the daughter cells to have different amount of POP-1, lost their function. We observed that the distribution for θlag-2 is centered at 60°. The bias of the θlag-2 distribution is likely to be due to the differential amount of SYS-1 in the two daughter cells. Since information determining through sensing and amplification are not conveyed effectively to the daughter cells as the POP-1 branch is inhibited, we can use this mutant to obtain an estimate for rb=0.006 by setting rf=0. This is likely to be an overestimation as the SYS-1 branch is unaffected. Using this estimation for the value of rb, we determined the values of g and rf that yields maximum likelihood fits for the WT and mutants strains’ distributions.
When we fit the WT distributions, we obtained large uncertainties in the fit parameters. This is expected because the parameter range over which WT polarity is observed is large. Although the WT distribution cannot be fitted uniquely, we know that the parameters lie in the parameter space where 100% correct divisions are observed which is bounded by the dashed line in Figure 5D and E. Next, the experimental distribution of θlag-2 for the different P cells in the ligand and receptor mutants (Figure 5C, blue histograms) were fit to the model (Figure 5C, red lines). The distributions for each of the individual P cells are significantly different; hence, the fit parameters g and rf were determined for each P cell in the ligand mutants and receptor mutants independently. The model is able to reproduce the main features of the experimental distributions. Interestingly, these two parameters segregated out in parameter space (Figure 5D). We found that the gradient parameter g was always lower compared with WT (Figure 5D), suggesting that both Wnt ligands and receptors are important for the sensing of the gradient. However, the feedback parameter rf was significantly smaller when receptors were mutated compared with ligand mutations, suggesting that the receptors might be involved in amplifying the signal.
Decreasing ligand results in symmetric divisions at low rf
From the above analysis, we inferred that Wnt ligands are primarily involved in sensing whereas Wnt receptors function in both sensing and amplification. Mutations in Wnt ligands primarily lead to polarity reversals whereas mutations in Wnt receptors cause loss of polarity. Very similar results have been reported for many tail blast cells in C. elegans, including the T cell, suggesting that our model maybe generally applicable (Sternberg and Horvitz, 1988; Herman and Horvitz, 1994). However, in contrast, it has been reported for the EMS cell in C. elegans that loss of ligands causes loss of polarity apparently inconsistent with our interpretation (Thorpe et al, 1997). However, upon closer inspection of the polarity phase diagram (Figure 5E), we hypothesized that in the EMS cell, the WT parameters are likely to be different. For example, the EMS cell might experience a different gradient or has reduced amplification potential compared with the P cells.
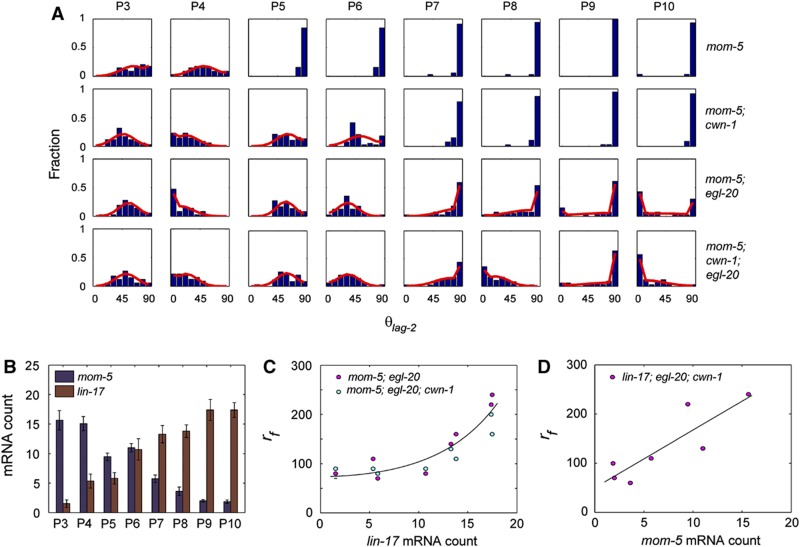
The parameter range over which WT polarity is observed is large and is bounded by the dashed line in Figure 5D and E. If we would reduce rf slightly so that most P cells still correctly polarize (moving from WT to C in Figure 5E) and subsequently remove ligand (moving from C to U in Figure 5E) it should be possible to induce loss of polarity upon loss of Wnt ligand. To test this prediction and access the parameter space of low rf, we looked for mutants for which rf is lower than WT but sufficiently high to yield correct divisions. We chose the mom-5 single receptor mutant as our candidate as there are many P cells with correct divisions in this mutant. In the mom-5 mutant, divisions of the P3 and P4 cells are symmetric whereas the divisions of the P5–P10 cells are correct (Figure 6A). Reducing ligand levels in the mom-5; cwn-1 strain led to symmetric divisions in the P5 and P6 cells (Figure 6A) confirming our prediction. This behavior was also reproduced in the mom-5; egl-20 and mom-5; egl-20; cwn-1 mutants, where divisions in all P3–P10 cells were affected (Figure 6A).
Figure 6.
Effects of reducing ligand in mom-5 background and correlation between receptor level and rf. (A) Histograms of θlag-2 (blue) and maximum likelihood fits (red) for P3–P10 in mom-5 single receptor mutant and mom-5; cwn-1, mom-5; egl-20 and mom-5; cwn-1; egl-20 compound mutants. (B) mom-5 (blue) and lin-17 (red) mRNA counts in P3–P10. (C) Plot of average lin-17 mRNA versus rf for P3–P10 in mom-5; egl-20 and mom-5; cwn-1; egl-20 strains. Exponential fit is shown in black. (D) Plot of average mom-5 mRNA versus rf for P3–P10 in lin-17; cwn-1; egl-20 strains. Linear fit is shown in black.
Positive correlation between receptor levels and rf
In the mom-5; egl-20 and mom-5; egl-20; cwn-1 strains, where divisions in all P3–P10 cells were affected, we observed a high fraction of symmetric divisions in the P3–P6 cells, lower fractions of symmetric divisions in the P7–P8 cells and coexistence of correct and reverse polarity in the P9 and P10 cells (Figure 6A). It is intriguing that all these different phenotypes were observed in the same mutant strain and even in the same mutant animal, suggesting that the values of rf may vary significantly among P cells in the same animal. We hypothesized that the differences in rf are due to the different expression levels of the receptors. We measured transcriptional levels of the receptors genes, mom-5 and lin-17, using single molecule FISH in the P cells and found that anterior P cells expressed higher levels of mom-5 whereas posterior cells expressed higher levels of lin-17 (Figure 6B). We found a positive correlation between the lin-17 count and the parameter rf (Figure 6C) for both the mom-5; egl-20 and mom-5; egl-20; cwn-1 strains. Similarly, a positive correlation was observed between the mom-5 mRNA level and rf in the lin-17; egl-20; cwn-1 strain (Figure 6D). These positive correlations obtained between receptor levels and rf provide quantitative support for our earlier conclusion that receptors may play an important role in amplification. Furthermore, since differences in receptors level could explain most of the differences in rf, it also demonstrates that the ligands’ contribution to amplification is less significant.
Discussion
By combining quantitative single molecule transcript counting with phenomenological modeling, we studied the effects of ligand and receptor loss on P cells’ division. We found that ligand affect primarily the sensing process whereby loss of ligands lead to polarity reversals. Single molecule FISH experiments on Wnt ligands show that ligands are expressed in a gradient. cwn-1 and egl-20 are higher in the posterior region whereas cwn-2 is higher in the anterior region (Harterink et al, 2011). These ligand gradients could potentially serve as the gradients for polarizing the P cells. Indeed, Wnt ligands has been shown to act instructively (Goldstein et al, 2006). But there are also cases where ligands act permissively (Whangbo et al, 2000). To determine if ligands act instructively or permissively, we expressed cwn-2 posteriorly using an egl-20 promoter or uniformly using a myo-3 promoter in a cwn-1; cwn-2; egl-20 triple-ligand mutant (Supplementary Figure 6). We found that ubiquitous expression of cwn-2 using both promoters are able to rescue the polarity reversals, suggesting that it is the ligand level rather than the gradient that is important for the sensing process. However, the recent identification of Wnt inhibitor, sfrp-1, in the anterior region of the worm suggests that ligand profile is not shaped by ligand expression alone (Harterink et al, 2011). Anterior expression of sfrp-1 could potentially convert uniform ligand expression into a ligand gradient. Identification of more Wnt components and quantitative measurement of ligand protein profiles will be necessary to resolve this issue.
Unlike the ligand mutants, polarity loss is observed in the receptor mutants. We reconcile the different phenotypes observed in ligand versus receptor mutants. Ligands affect primarily the sensing process whereas receptors are needed for both sensing and amplification. This suggests that receptors can signal to the downstream proteins needed for amplification regardless of whether they are ligand-bound whereas the function of ligand is to convey information about the directionality of polarization to the cell through the receptors. One possible molecular mechanism is that initially equal amount of receptors bind to both the anterior and posterior sides of a cells. The presence of a ligand gradient would bias the amount of receptors on one side of the cell through mechanisms like reduction in degradation rates for ligand-bound receptors. The higher amount of receptors on one side of the cell may lead to positive feedback to bring even more receptors to the same site. This would eventually cause the cell to become polarized and divide asymmetrically. In the absence of ligand, positive feedback could still lead to a higher amount of receptors on one side of the cell due to initial stochastic differences in the amount of receptors at each side of the cell. However in this case, some fraction of the cells will be polarized in the opposite direction, leading to existence of divisions of both correct and reverse polarity. When receptors are absent, there will not be positive feedback and the cells would divide symmetrically. The observation that receptors cluster asymmetrically before division is consistent with this proposed model (Wu and Herman, 2007). Future work is necessary to elucidate the molecular mechanisms for the differences in roles of ligands and receptors.
In our mom-5;wnt strains (Figure 6A), we observed loss of polarity and polarity reversals for P cells with low and high rf, respectively. This is reminiscent of the different phenotypes observed upon ligand loss in C. elegans. Loss of ligand leads to polarity reversals for many tail blast cells, including the T cell, and loss of polarity in the EMS cell. Our result suggests that these differences are due to low amplification strength in the EMS and high amplification strength in the T cell. It is interesting why some cells have high amplification strength whereas others have low amplification strength. We suggest that low amplification is sufficient for correct division in the EMS as the single MOM-2/ligand source is adjacent to the EMS and likely to provide a steep gradient across the cell. However, for P cells during post-embryonic development, the gradient is shaped by many different ligands and by many ligand-expressing cells all along the worm (Harterink et al, 2011). Therefore, the gradient experienced by the P cells is likely to be shallower and more complicated to interpret. In this case, it will be beneficial for the cell to have higher amplification is to ensure robustness. Hence, we speculate that depending on the gradient and the complexity of the ligand sources, cells may express different receptor levels to balance the tradeoff between robustness of polarity and energy involved in high expression.
Here, we have proposed a general framework for determining the contributions of proteins to sensing and amplification. This method overcomes the need for detailed knowledge of the biochemical reactions and network topology involved in many polarizing system and allows us to extract parameters that quantify the key processes of sensing and amplification.
Materials and methods
C. elegans strains and culturing
General methods for culture, manipulation, and genetics of C. elegans were as described previously (Lewis and Fleming, 1995). Mutations used in this study were: LGI, lin-44(n1792) (Herman and Horvitz, 1994), mom-5(or57) (Thorpe et al, 1997) in mom-5; lin-17 receptor strain and mom-5(gk812) for all other mom-5 strains; LGII, cwn-1(ok546) (Zinovyeva and Forrester, 2005), mom-4(ne1539) (Takeshita and Sawa, 2005); LGIII, lit-1(t1512) (Takeshita and Sawa, 2005); LGIV, cwn-2(ok895) (Zinovyeva and Forrester, 2005), egl-20(n585) (Zinovyeva et al, 2008); LGV, mom-2(ne874ts). Strains were cultured at 20°C except for the mom-4; lit-1 strain, which was cultured at 15°C and transferred to 25°C for 7 or 9 h before fixation and the Wnt quintuple mutant, which was grown at 15°C and shifted to 25°C for 7 h before fixation.
Single molecule mRNA FISH
Probe design and hybridization to perform FISH for single transcript measurement in C. elegans larvae was performed as previously described (Raj et al, 2008). Animals were collected by washing plates with M9 and were fixed in 4% formaldehyde in 1 × PBS for 45 min. Fixed animals were permeabilized in 70% ethanol overnight. All probes for hybridization were coupled to either Cy5 (GE Amersham) or Alexa594 (Invitrogen), depending on the desired gene combinations for image acquisition. Images were taken with a Nikon Ti-E inverted fluorescence microscope equipped with a 100 × oil-immersion objective and a Photometrics Pixis 1024 CCD camera using MetaMorph software (Molecular Devices, Downington, PA, USA). Three-dimensional positions of bright fluorescent spots in each animal were detected with the aid of a custom program written in MATLAB, as described (Raj et al, 2008), which was later manually corrected for further accuracy. Nuclei were visualized with DAPI.
Simulation of histograms of θ
The chemical master equation was solved to obtain the exact solution for each set of parameters. In the simulation, θ takes values between 0 and 90° and increases or decreases in steps of 1°. Next, a transition matrix, M, describing the rates of going from between the different values of θ is constructed. Since there are 91 different values that θ can take, the transition matrix is of size 91 × 91. The rate of going from θ1 to θ2 is represented by M(θ2+1, θ1+1) in the matrix. Furthermore, since θ can only increase or decrease in steps of 1°, only the diagonal terms and values next to the diagonal terms are non-zero in the transition matrix.
For the simple model
 |
For the expanded model
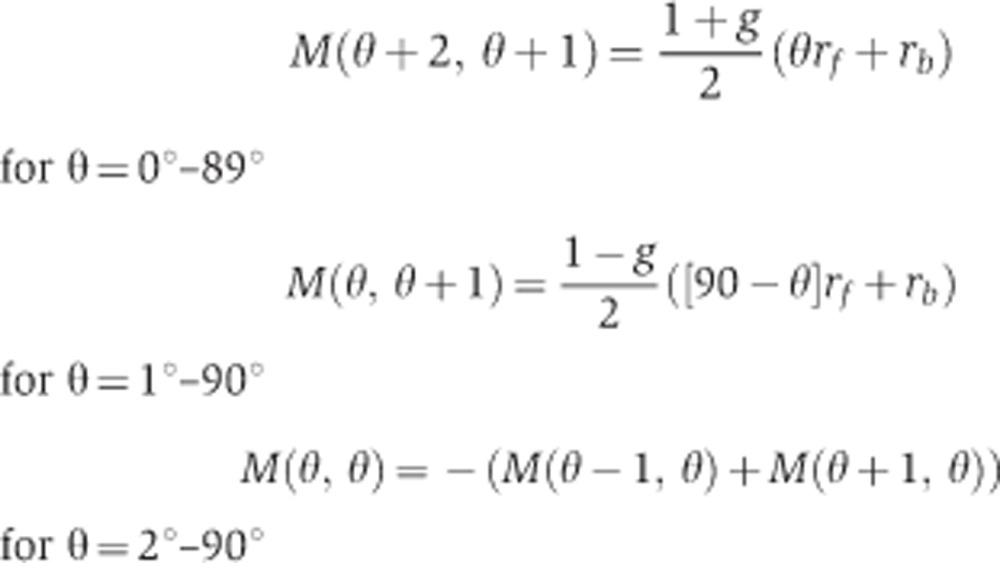 |
Since θ cannot takes values less than 0° and greater than 90°, the boundary conditions at θ=0° and 90° are reflecting.
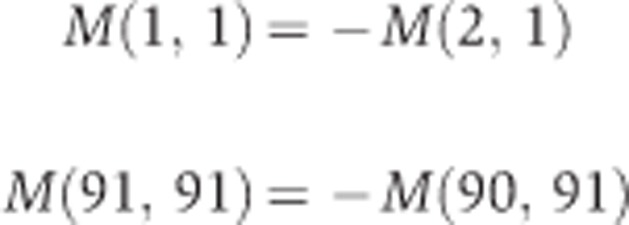 |
The distribution at time t is presented by Pt and is generated by the following equation:
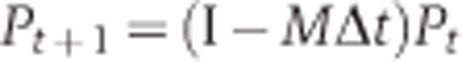 |
where I is the identity matrix. We start the simulation with Pt=0 (θ=45°)=1 as the cells are initially unpolarized. The total simulation time T is underdetermined and is set constant to 1 for all the simulations and different mutants analyzed. Δt is the time step of the numerical simulation and has been chosen to be 10−4 in our simulation. The resulting histograms are independent of the choice of Δt as long as Δt is small enough.
Classification rules for correct polarity, reverse polarity, and loss of polarity
The classification rules are θ(T)>75° (correct polarity), θ(T)<15° (reverse polarity), and 15°≤θ(T)≤75° (loss of polarity). These rules are obtained by finding the angle that best separate the lag-2 expression of Pn.a versus Pn.p between WT animals and the mom-4; lit-1 strain at 25°C (Figure 3A and B).
Stochastic simulation of single θ(t) traces
Monte Carlo simulation were performed to generate many single θ(t) trajectories. The initial condition θ(t=0)=45° was used as the cells are initially unpolarized. For every time t, a random number n is drawn. θ(t+Δt) will be updated based on the following conditions:
Simple model
 |
Expanded model
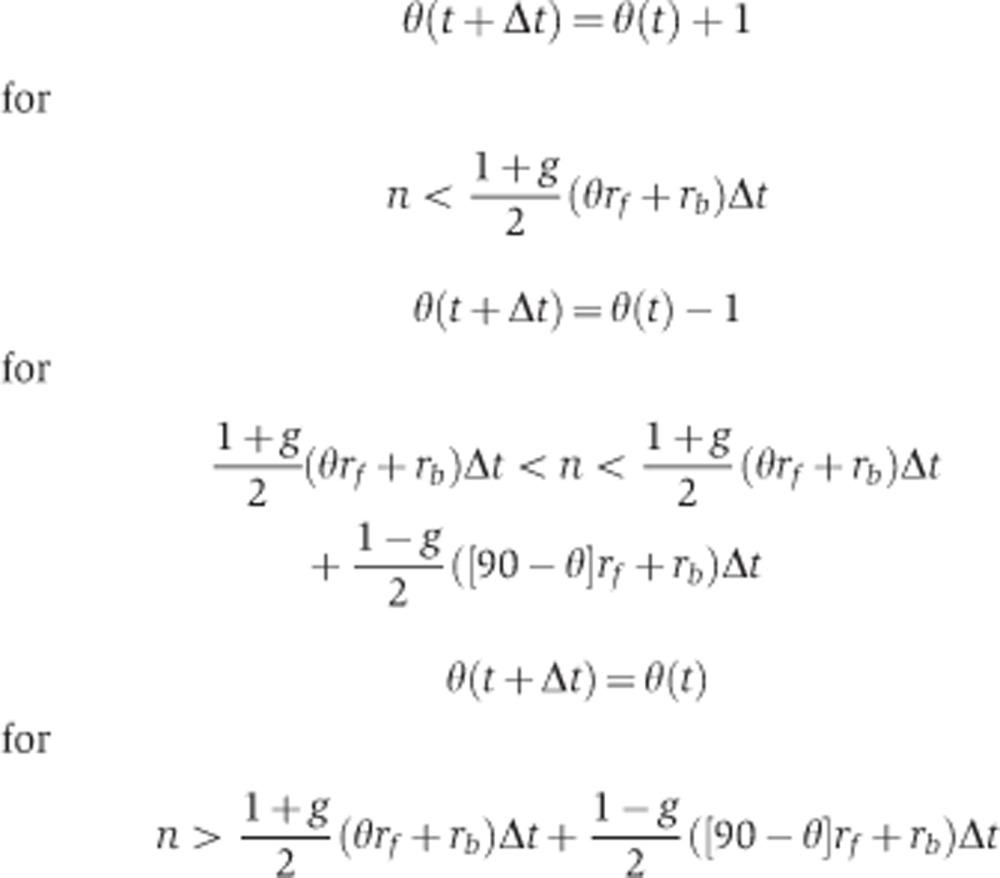 |
The boundary conditions at θ=0° and 90° are reflecting.
Supplementary Material
Acknowledgments
This work was supported by a NIH Pioneer award (1DP1OD003936) and NIH grant R01 GM068957 to AvO and the A*STAR program, Singapore to RZT. Several nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank Jeroen van Zon for discussions on the model. We also thank Stefan Semrau and Jeroen van Zon for comments on the manuscript. We thank Peter Reddien, Erel Levine, Susan Mango, Jeff Gore, Teije Middelkoop, Christoph Engert, Dong Hyun Kim, Apratim Sahay, and Clinton Hansen for general discussions.
Author contributions: RZT and AvO conceived the project. RZT, NJ, and RAM performed the experiments; RZT and AvO constructed the computational model; RZT, NJ, HCK, and AvO wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alon U (2007) Simplicity in biology. Nature 446: 497. [DOI] [PubMed] [Google Scholar]
- Arkowitz RA (1999) Responding to attraction: chemotaxis and chemotropism in Dictyostelium and yeast. Trends Cell Biol 9: 20–27 [DOI] [PubMed] [Google Scholar]
- Baier H, Bonhoeffer F (1992) Axon guidance by gradients of a target-derived component. Science 255: 472–475 [DOI] [PubMed] [Google Scholar]
- Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, Collins J, Mello CC (2002) SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell 3: 113–125 [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hobert O (2009) Linking asymmetric cell division to the terminal differentiation program of postmitotic neurons in C. elegans. Dev Cell 16: 563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason JE, Eisenmann DM (2010) Wnt signaling controls the stem cell-like asymmetric division of the epithelial seam cells during C. elegans larval development. Dev Biol 348: 58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B, Takeshita H, Mizumoto K, Sawa H (2006) Wnt signals can function as positional cues in establishing cell polarity. Dev Cell 10: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR (1983) The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444 [DOI] [PubMed] [Google Scholar]
- Harterink M, Kim DH, Middelkoop TC, Doan TD, van Oudenaarden A, Korswagen HC (2011) Neuroblast migration along the anteroposterior axis of C. elegans is controlled by opposing gradients of Wnts and a secreted Frizzled-related protein. Development 138: 2915–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Horvitz HR (1994) The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120: 1035–1047 [DOI] [PubMed] [Google Scholar]
- Jilkine A, Edelstein-Keshet L (2011) A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput Biol 7: e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein PS, Sun TJ, Saxe CL III, Kimmel AR, Johnson RL, Devreotes PN (1988) A chemoattractant receptor controls development in Dictyostelium discoideum. Science 241: 1467–1472 [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT (1995) Basic culture methods. Methods Cell Biol 48: 3–29 [PubMed] [Google Scholar]
- Mallavarapu A, Thomson M, Ullian B, Gunawardena J (2009) Programming with models: modularity and abstraction provide powerful capabilities for systems biology. J R Soc Interface 6: 257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H (1999) Orientation of chemotactic cells and growth cones: models and mechanisms. J Cell Sci 112: Part 172867–2874 [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H (2007a) Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev Cell 12: 287–299 [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Sawa H (2007b) Two betas or not two betas: regulation of asymmetric division by beta-catenin. Trends Cell Biol 17: 465–473 [DOI] [PubMed] [Google Scholar]
- Nelson WJ (2003) Adaptation of core mechanisms to generate cell polarity. Nature 422: 766–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN (1998) G protein signaling events are activated at the leading edge of chemotactic cells. Cell 95: 81–91 [DOI] [PubMed] [Google Scholar]
- Phillips BT, Kidd AR III, King R, Hardin J, Kimble J (2007) Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci USA 104: 3231–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A (2000) Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci 113: Part 3365–375 [DOI] [PubMed] [Google Scholar]
- Qiao L, Lissemore JL, Shu P, Smardon A, Gelber MB, Maine EM (1995) Enhancers of glp-1, a gene required for cell-signaling in Caenorhabditis elegans, define a set of genes required for germline development. Genetics 141: 551–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S (2008) Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5: 877–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR (2000) Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science 287: 1037–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhankova M, Korswagen HC (2007) Migration of neuronal cells along the anterior-posterior body axis of C. elegans: Wnts are in control. Curr Opin Genet Dev 17: 320–325 [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Horvitz HR (1988) lin-17 mutations of Caenorhabditis elegans disrupt certain asymmetric cell divisions. Dev Biol 130: 67–73 [DOI] [PubMed] [Google Scholar]
- Strutt DI (2001) Asymmetric localization of frizzled and the establishment of cell polarity in the Drosophila wing. Mol Cell 7: 367–375 [DOI] [PubMed] [Google Scholar]
- Sulston JE (1976) Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 275: 287–297 [DOI] [PubMed] [Google Scholar]
- Takeshita H, Sawa H (2005) Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev 19: 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK (1998) MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93: 569–580 [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, Bowerman B (1997) Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90: 695–705 [DOI] [PubMed] [Google Scholar]
- Tree DR, Shulman JM, Rousset R, Scott MP, Gubb D, Axelrod JD (2002) Prickle mediates feedback amplification to generate asymmetric planar cell polarity signaling. Cell 109: 371–381 [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Altschuler S, Wu L, Li R (2003) Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299: 1231–1235 [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R, Wai SC, Schmidt T, Li R (2004) Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol 166: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD (2002) Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr Opin Cell Biol 14: 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo J, Harris J, Kenyon C (2000) Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development 127: 4587–4598 [DOI] [PubMed] [Google Scholar]
- Wu M, Herman MA (2007) Asymmetric localizations of LIN-17/Fz and MIG-5/Dsh are involved in the asymmetric B cell division in C. elegans. Dev Biol 303: 650–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S, McKinnon RD, Kokel M, Thomas JB (2003) Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature 422: 583–588 [DOI] [PubMed] [Google Scholar]
- Zigmond SH (1977) Ability of polymorphonuclear leukocytes to orient in gradients of chemotactic factors. J Cell Biol 75: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyeva AY, Forrester WC (2005) The C. elegans Frizzled CFZ-2 is required for cell migration and interacts with multiple Wnt signaling pathways. Dev Biol 285: 447–461 [DOI] [PubMed] [Google Scholar]
- Zinovyeva AY, Yamamoto Y, Sawa H, Forrester WC (2008) Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans development. Genetics 179: 1357–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.