Abstract
Rate control analysis defines the in vivo control map governing yeast protein synthesis and generates an extensively parameterized digital model of the translation pathway. Among other non-intuitive outcomes, translation demonstrates a high degree of functional modularity and comprises a non-stoichiometric combination of proteins manifesting functional convergence on a shared maximal translation rate. In exponentially growing cells, polypeptide elongation (eEF1A, eEF2, and eEF3) exerts the strongest control. The two other strong control points are recruitment of mRNA and tRNAi to the 40S ribosomal subunit (eIF4F and eIF2) and termination (eRF1; Dbp5). In contrast, factors that are found to promote mRNA scanning efficiency on a longer than-average 5′untranslated region (eIF1, eIF1A, Ded1, eIF2B, eIF3, and eIF5) exceed the levels required for maximal control. This is expected to allow the cell to minimize scanning transition times, particularly for longer 5′UTRs. The analysis reveals these and other collective adaptations of control shared across the factors, as well as features that reflect functional modularity and system robustness. Remarkably, gene duplication is implicated in the fine control of cellular protein synthesis.
Keywords: eukaryotic translation machinery, gene duplication, in vivo rate control, post-transcriptional gene expression, system modularity
Introduction
Protein synthesis is the single most energy-consuming cellular process. In Saccharomyces cerevisiae, it has been estimated to generate up to 13 000 protein molecules per cell per second with high accuracy (von der Haar, 2008). Precise control of the highly complex translation machinery underpins the capacity for the growth and selective competitiveness of living organisms; therefore, the quantitative properties of this machinery and the evolution of its molecular components are tightly coupled. Translation is not only a key point of control for eukaryotic gene expression, but also features in many regulatory responses intrinsic to organism development or triggered by environmental stress (Sonenberg and Hinnebusch, 2009). Moreover, aberrant intracellular translation factor activities correlate with oncogenesis and other disease states (Abbott and Proud, 2004; Cuesta et al, 2009). A great deal is now known about the structure and function of the translation machinery (Kapp and Lorsch, 2004, Sonenberg and Hinnebusch, 2009). However, genetic analysis involves mutations that either modify or abrogate protein functions, while in vitro biochemical experiments do not reproduce the complex environment of the cell. These approaches therefore need to be complemented by analysis of control in vivo.
Initiation (Supplementary Figure 1A) is the phase of eukaryotic translation that is the most complex and that differs most from the counterpart prokaryotic process (McCarthy, 1998). Almost all initiation events in yeast, and most of the initiation events in higher eukaryotes, are of the cap-dependent type, in which recruitment of the small (40S) ribosome subunit to the 5′ end of cellular mRNAs is mediated by a cap-binding complex of initiation factors. Scanning is a poorly understood step in which pre-initiation complexes scan along 5′UTRs that can exceed 1000 nucleotides in length in search of start codons (McCarthy, 1998; Kozak, 2002; Kapp and Lorsch, 2004), and may involve some degree of random-walk-type motion (Berthelot et al, 2004). Once the 40S ribosomal subunit locates a start codon and recruits a large (60S) subunit (accompanied by release of eIFs), elongation of the polypeptide chain proceeds until a stop codon is reached, whereupon the polypeptide chain is hydrolysed off the final tRNA and released (Supplementary Figures 1B and C).
Initiation (Berthelot et al, 2004) and elongation (Arava et al, 2005) are thought to be highly processive. However, there can be many rounds of initiation on an mRNA molecule during the time it takes one ribosome to translate the whole reading frame, and thus the control relationship between these respective steps is not readily predictable. There has been uncertainty whether initiation or elongation is the major site of translational control (Mathews et al, 2000; Wang et al, 2001). Codon sequence in the mRNA open reading frame (ORF) seems to influence translation efficiency largely via its impact on ribosome translocation (McCarthy, 1998; Cannarozzi et al, 2010). This all leaves unresolved whether (and, if so, how) multiple sites of (strong) control across the three phases of translation might co-exist in the pathway.
An intriguing feature of this complex system that we have yet to understand has been highlighted by evidence, indicating that the respective intracellular eIF concentrations in yeast differ markedly from each other (von der Haar and McCarthy, 2002). This has raised the question how the non-stoichiometric intracellular abundance ratios for the respective translation factors relate to the level of control exerted by each protein over the translation initiation rate. Moreover, studies of translation factor levels in transformed mammalian cells have suggested that a single factor can mediate upregulation for the whole system (Cuesta et al, 2009). Analysis of these issues is critical to understand the operational properties, as well as the evolution, of gene expression systems in living cells. Linked to this is another fascinating aspect of the genetics of the translation machinery: six of the translation factors are encoded by duplicated genes (eIF4A, eIF4G, Anb1/Hyp2 (eIF5A), eEF1A, eEF2, and eEF3), whereby three of these gene pairs encode identical proteins (eIF4A (TIF1/TIF2), eEF1A (TEF1/TEF2), and eEF2 (EFT1/EFT2)). The occurrence of multiple (although not always identical) alleles encoding a subset of translation factors is a conserved feature across all the eukaryotes, but the reasons for this are not clear. The characterization of the quantitative control landscape for yeast translation, as described here, now provides the basis for analysing this phenomenon in the context of the system-level control of eukaryotic gene expression.
Results
The distribution of rate control between the respective translation factors in vivo
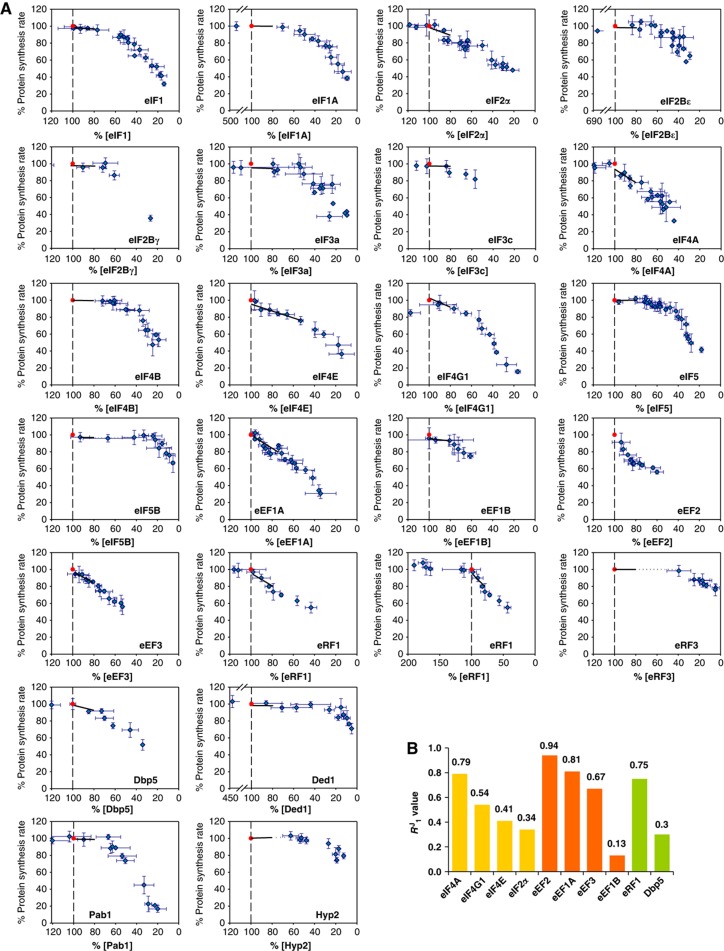
Since control within the translation machinery is shaped by the complex, highly crowded environment of the living cell, quantitative in vivo rate control analysis provides the only route to analyse accurately the control features of this system. We utilized chromosomal tet07 regulatory constructs to determine how steady-state translation factor activity determines protein synthesis rate (Figure 1; Supplementary Figures 2 and 3; Supplementary Tables 1 and 2). In most cases, the tet07 construct was observed to restrict the production of the encoded translation factor to a sub-wild-type level even in the absence of doxycycline because it supported a lower rate of expression than the natural promoter. We ensured that the titrated range of protein could extend back up to the wild-type level by supplementing chromosomal expression using a single-copy plasmid carrying a second copy of the appropriate gene transcribed from one of a selection of constitutive yeast promoters specifically engineered for this purpose (as described in Supplementary Figure 2). This supplementary synthesis of a factor allowed us to cover 80–100% of the physiologically normal abundance for almost all of the factors, as well as a range of >100% for a further subset of factors (Figure 1; Table I; see later). The response coefficient (R1J) was determined from the slope of each plot of global translation rate versus factor concentration in this near-physiological range (100–80% of wild-type factor abundance; Table I). Where R1J was close to zero, we determined fewer points in the near-physiological range since all measurements indicated the maintenance of maximal global protein synthesis for these factors (Figure 1A). Complete fits to the entire data sets (and variance data) are shown (for all factors) in Figure 1 but the transitions to higher RJ values at lower protein synthesis rates are not considered as part of this analysis.
Figure 1.
Response coefficients (R1J values). (A) Plots of protein synthesis against intracellular translation factor abundance, illustrating different types of response profile. Each point in these plots is the average of at least three measurements of percentage change and error bars for these data are shown. The red point in each plot corresponds to the rate of protein synthesis observed in the wild-type strain (100%). In each case, a black line indicates the slope (the R1J value) of the relationship in the range of 100–80%. In the case of eRF1, a second plot shows additional points extending up to almost 200% of the physiological abundance of this factor, confirming the positive influence of the extra factor abundance. (B) A bar chart showing the non-zero R1J values. Source data for this figure is available on the online supplementary information page.
Table 1. Summary of R coefficients for 80–100% (100%+) of intracellular factor concentrations.
| Translation factor (gene) | Strain name | Strain number | R +1 J | R 1 J | R Sp | R Sc |
|---|---|---|---|---|---|---|
| Rps5 (RPS5) | tetO7RPS5 | PTC389 | ND | ND | 0.90 | ND |
| eIF1 (SUI1) | tetO7SUI1 | PTC277 | 0.00 | 0.07 | 0.90 | 0.74 |
| eIF1A (TIF11) | tetO7TIF11 | PTC269 | 0.01 | 0.01 | 0.97 | 0.29 |
| eIF2α (SUI2) | tetO7SUI2 | PTC390 | 0.01 | 0.34 | 1.04 | 0.57 |
| eIF2Bε (GCD6) | tetO7GCD6 | PTC272 | −0.01 | −0.01 | 0.73 | 0.61 |
| eIF2Bγ (GCD1) | tetO7GCD1 | PTC400 | 0.02 | 0.02 | 0.63 | ND |
| eIF3a (RPG1) | tetO7RPG1-60 | PTC391 | −0.05 | 0.00 | 0.95 | 0 |
| eIF3c (NIP1) | tetO7NIP1-60 | PTC401 | 0.00 | 0.02 | 1.04 | ND |
| eIF3j (HCR1) | tetO7HCR1 | PTC270 | 0.00 | 0.00 | ND | ND |
| eIF4A (TIF2) | tetO7TIF2TIF1Δ | PTC393 | −0.02 | 0.79 | 0.89 | 0 |
| eIF4B (TIF3) | tetO7TIF3 | PTC394 | ND | 0.04 | 1.01 | 0 |
| eIF4E (CDC33) | tetO7CDC33 | PTC278 | 0.00 | 0.41 | 0.92 | 0 |
| eIF4G1 (TIF4631) | tetO7TIF4631TIF4632Δ | PTC276 | −0.09 | 0.54 | 1.07 | 0 |
| eIF5 (TIF5) | tetO7TIF5 | PTC268 | −0.01 | −0.01 | 0.95 | 0 |
| eIF5B (FUN12) | tetO7FUN12 | PTC265 | ND | 0.01 | 0.97 | 0 |
| eEF1A (TEF1) | tetO7TEF1TEF2Δ | PTC362 | 0.00 | 0.81 | 0.99 | 0 |
| eEF1B (TEF5) | tetO7TEF5 | PTC363 | ND | 0.13 | 0.63 | 0 |
| eEF2 (EFT1) | tetO7EFT1EFT2Δ | PTC364 | ND | 0.94 | 0.89 | 0 |
| eEF3 (YEF3) | tetO7YEF3 | PTC365 | ND | 0.67 | 0.35 | 0 |
| eRF1 (SUP45) | tetO7SUP45 | PTC366 | 0.08 | 0.75 | 0.45 | 0 |
| eRF3 (SUP35) | tetO7SUP35 | PTC367 | ND | 0.02 | 0.29 | 0 |
| Dbp5 (DBP5) | tetO7DBP5 | PTC398 | −0.00 | 0.30 | 0.64 | 0 |
| Ded1 (DED1) | tetO7DED1-30 | PTC397 | 0.01 | 0.01 | 0.64 | 0.66 |
| Hyp2 (HYP2) | tetO7HYP2 | PTC395 | ND | −0.03 | 0.39 | 0 |
| Pab1 (PAB1) | tetO7PAB1 | PTC271 | 0.02 | 0.02 | 0.19 | 0 |
ND, not determined.
Further experiments demonstrated the specificity and stability of this ‘genetic titration’ strategy (Supplementary Figure 2), and revealed that over the 100–80% translation range there were no disruptive changes in polysome gradient profiles (Supplementary Figure 3A) and steady-state mRNA levels (Supplementary Figure 3B). The polysome gradient profiles were performed using extracts from representative strains that were subject to tet07 regulation via factors in all three phases of translation. Similarly, we examined the intracellular steady-state abundance of a number of mRNA species that are representative of different rates of turnover. We also found that the average cell size was not affected by tet07-mediated suppression (100–80%) of translation factor levels in the constructed strains (Supplementary Figure 4). Since cells whose cell cycle is slowed when the abundance of a translation factor is reduced will manifest a larger average size, measurement of the average cell diameter provides indirect information on potential quantitative effects that might be linked to changes in the cell cycle. We checked whether, in each tet07 strain, the intracellular abundance values of the factors that are not subject to regulation by the chromosomal tet07 construct are affected by the addition of doxycycline (Supplementary Figure 2). Using western blotting and calibrated quantitative mass spectrometry, we observed that, within experimental error, only the abundance of the tet07-regulated factor was reduced over the 100–80% range. These control investigations focused on the factors that have a significantly positive R1J value (Figure 1B), since the zero R1J factors do not affect global protein synthesis in the 100–80% range. The results revealed that, in each case, the change in translation factor abundance was specific to the product of the gene under control of the tet07 regulatory cassette.
The R1J values (Table I) build a map of multiple control foci (Figures 1B and 2A). The protein and gene nomenclatures for the respective translation factors are listed in Table I. The members of the high R1J value (high flux control) group are associated with three major control points: recruitment of mRNA and tRNAi to the 40S ribosomal subunit (promoted by the cap-binding complex factors eIF4A, E, and G and the 5S ternary complex tRNAi (initiator tRNA)-eIF2-GTP), polypeptide elongation (eEFs 1A, 2, and 3), and termination (eRF1, and also Dbp5). The elongation step, which features repeated cycling of elongation factors on and off the ribosome, manifests collectively the highest R1J values (Figure 1). The only apparent exception is Hyp2, which has a low R1J value, but this protein seems to be unusual in that it may be functional in both initiation and elongation (see below). The energy demand of generating large amounts of all these factors will be significant, and it is notable that the cell maintains them all close to the levels required for maximal protein synthesis.
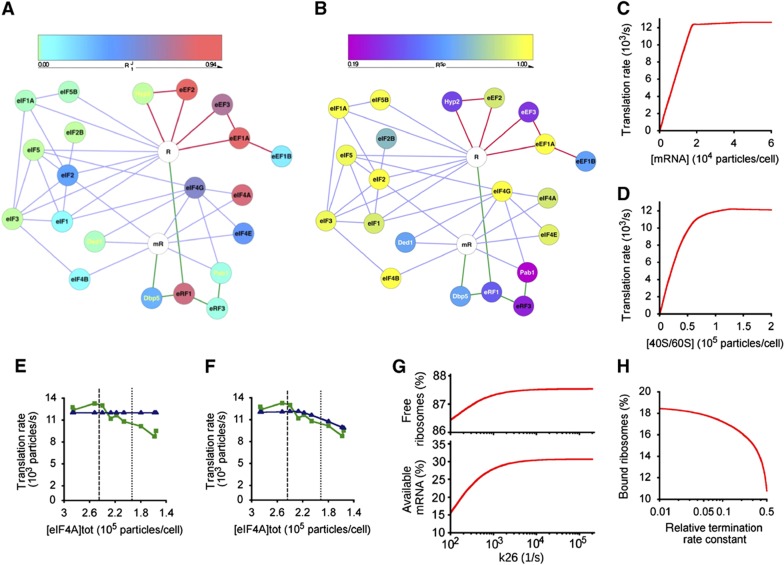
Figure 2.
Control maps and the digital translation model. (A) R1J graph featuring the respective translation factors as nodes (colour-coded according to R1J value) and the confirmed physical/functional interactions as edges (lines colour-coded for initiation (lilac), elongation (red), and termination (green)). The mRNA (mR) and ribosome (R) are represented in white. (B) Equivalent graph for RSp values. (C–H) Predictions of the in silico translation model described in this paper. The model predicts the protein synthesis rate versus intracellular mRNA abundance (C) and versus ribosome content per cell (D) (compare Figure 4B). The digital model calculated the response profile (blue) assuming an obligatory eIF4A:eIF4B intermediate (E), and where eIF4A can function independently of this complex (F). The green line shows the averaged experimental data points (as in Figure 1). Vertical dashed and dotted lines indicate the 100 and 80% points on the eIF4A concentration axis. Changing the elongation rate (magnitude of the rate constant for reaction 26 in Supplementary Figure 1D) affects the intracellular populations of ribosomes and mRNA available for initiation (G). Changing the termination rate (the ratio of the actual termination rate constant versus the standard rate constant used in the model) affects the populations of ribosomal subunits available for initiation (H).
This contrasts with the situation for the low-flux-control group, which includes the scanning-promoting factors (Ded1, eIF1, eIF1A, eIF2B, and eIF5) and the GTPases and guanine nucleotide exchange factors (eIF2, eIF2B, eIF5B, eEF1B, and eRF3). While these factors are of relatively low absolute abundance (and therefore of comparatively low energy cost in terms of their production), in each case the observed intracellular level is in excess of that required to achieve a maximal rate of protein synthesis (i.e., R1J∼0). As a result, these data indicate that the intracellular concentrations of the scanning promoting factors are generally sufficient to ensure that the scanning process occurs as efficiently as possible, and therefore that ribosomes are engaged for the minimal possible time in the scanning phase of initiation, particularly on longer 5′UTRs.
Functional modularity of the translation machinery
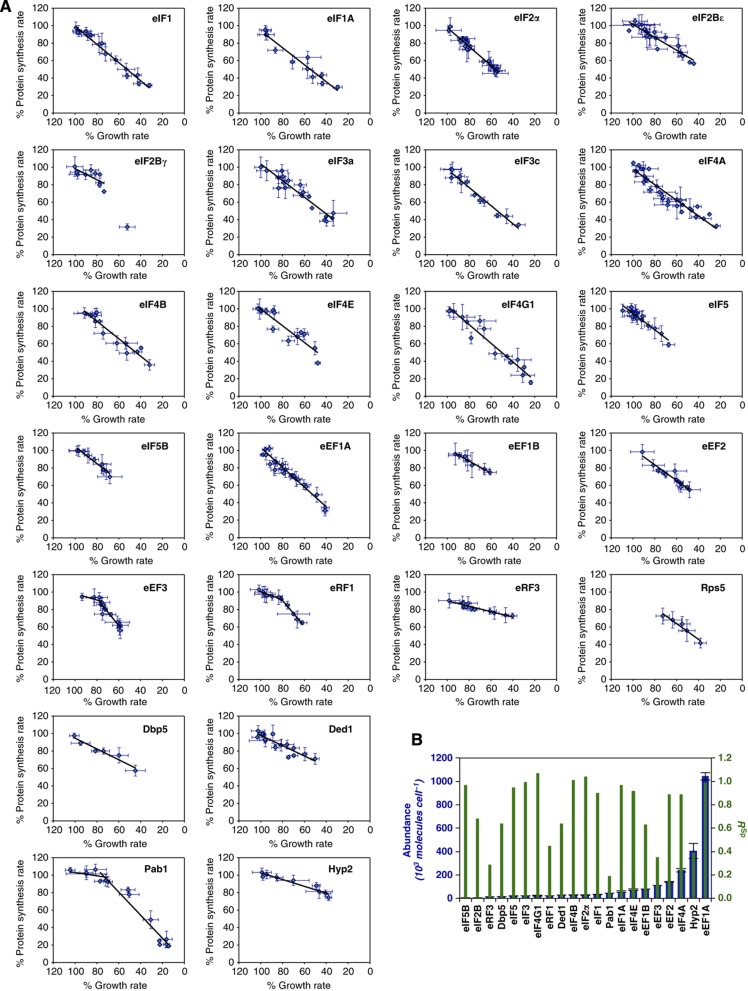
This study focuses on those interactions confirmed by detailed functional and biochemical investigations (Supplementary Table 3), but the potential for multiple physical interactions with other cellular proteins (Krogan et al, 2006) prompted us to assess the functional modularity of the system. We examined how attenuation of the rate of protein synthesis in the cell impacts on growth rate, giving the ‘system specificity ratio’ (RSp; Table I). RSp is calculated by plotting the growth rate for each factor concentration against the equivalent global protein synthesis rate and taking the slope of the resulting relationship (Figure 3). Thus, the value of RSp decreases if a factor has roles in multiple cellular processes because, as its intracellular abundance decreases, the growth rate is more strongly suppressed than is protein synthesis (Figure 3). We observed a linear relationship for almost all of the formally recognized factors with RSp≈1, reflecting a high degree of dedication to the translation system (as observed for Rps5). The RSp plots for Pab1, eEF3, and eRF1 were exceptional in that they were biphasic (Figure 3A). Since this study focuses on the near-physiological range (100–80%) of global protein synthesis, we have utilized the RSp value corresponding to the linear slope of each plot that lies closest to the wild-type factor abundance (Figure 3B; Table I). The network graph for the distribution of RSp values across the translation machinery (Figure 2B) reinforces the view that the translation machinery is largely self-contained in terms of control. Four formally recognized translation factors (eIF2B, eEF1B, eRF1, and eRF3), together with four multifunctional factors (see below), manifest sub-maximal RSp values in the near-physiological range.
Figure 3.
Specificity coefficients (RSp values). (A) Plots of protein synthesis against growth rate. The black lines (determined by regression analysis fitting) give the RSp values. (B) A bar chart comparing RSp and abundance values (Spearman rank correlation coefficient (rS)=−0.1).
Control by additional (multifunctional) translation factors
In this study, we have included a group of additional (essential) proteins that are not formally classified as translation factors in yeast. Dbp5, Ded1, and Pab1 have been shown to be required for normal translation (Sachs and Davis, 1989; Chuang et al, 1997; de la Cruz et al, 1997; Gross et al, 2007). Interestingly, it has been suggested previously that Pab1 be classified as a translation factor in mammalian protein synthesis (Kahvejian et al, 2005). The role of the hypusine-containing protein eIF5A has been uncertain. It was originally categorized as an initiation factor on the basis of its influence on a methionyl-puromycin synthesis assay (Kemper et al, 1976), but more recent work has indicated that it actually promotes translation elongation (Saini et al, 2009), although a role in initiation has not been ruled out (Henderson and Hershey, 2011). eIF5A is encoded by two genes, HYP2 and ANB1, whereby the latter is expressed only under anaerobic conditions.
The quantitative analysis performed here contributes to a more differentiated view of the roles of all of these four factors. Ded1, Pab1, and Hyp2 show R1J values typical of one of the low-flux-control initiation factors while Dbp5 has a mid-range R1J value. The relatively low RSp values of Dbp5, Ded1, Hyp1, and Pab1 suggest that they have functions in addition to a role in protein synthesis (Figures 2B and 3; Table I), and indeed previous work has indicated that these four proteins have a range of non-translational roles in the cell (Amrani et al, 1997; Valentini et al, 2002; Cole and Scarcelli, 2006; Urakov et al, 2006; Halls et al, 2007; Pittman et al, 2009; Merritt et al, 2010). On this basis, we classify Dbp5, Ded1, Pab1, and Hyp2 as multifunctional translation factors.
Translation in silico
An in silico mathematical model of protein synthesis (see the reaction path in Supplementary Figure 1D and the reaction equations in Supplementary information; the model has been deposited in the Biomodels database) was developed as a tool for enhancing analysis of pathway behaviour. The predicted response curve output for each high R1J factor as it nears its physiological abundance (Supplementary Figure 5C) reveals that the sharp convergence on the observed wild-type maximal protein synthesis rate is an intrinsic property of the translational machinery. Global protein synthesis in the cell is predicted to be working very close to maximal capacity in exponentially growing cells (Figures 2C and D). The plot of translation rate versus mRNA abundance shows a linear dependency up to about 17 000 mRNA molecules per cell, and then the rate becomes essentially constant. The plot is close to biphasic, with a relatively sharp transition between linear and constant rates at a critical concentration of mRNA; this is in contrast to normal enzyme kinetics where the dependency is hyperbolic or sigmoidal. Below the critical concentration of mRNA, the rate is linear because there are ribosomes and factors available to attach to mRNA for translation. Beyond the critical point, all ribosomes are attached to mRNA and excess mRNA cannot engage in translation. We also used the model to analyse the role of eIF4A, the eIF with the highest R1J value. Forcing this factor to act via an obligatory eIF4A:eIF4B complex prevents the model from fitting the real data (Figure 2E). Taking into account the measured differences in internal abundance (Figure 4), the modelling output (Figure 2F) indicates that eIF4A has the ability to engage with the translation machinery independently of eIF4B.
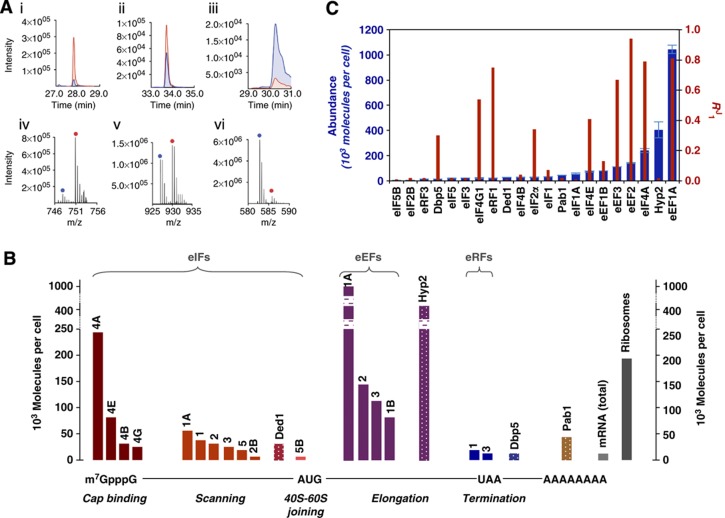
Figure 4.
Abundance versus rate control. (A) Exemplar mass spectrometry-based quantitation of translation factors either using SRM (i–iii) or accurate mass retention time (AMRT) analyses (iv–vi), whereby the two methods of quantitation yielded very similar results (Supplementary Figure 6). Proteins are (i, iv) eIF2Bε; (ii, v) eIF3a; and (iii, vi) eIF4A. The analyte is labelled in blue and the quantotypic reference peptide is labelled in red. The complete set of mass spectrometry data is available in Supplementary Tables 4–6. (B) Bar graph featuring the abundance values determined by mass spectrometry (see Supplementary Table 6) grouped according to function on the protein synthesis pathway. The patterned bars correspond to the subset of additional (multifunctional) factors. Breaks in two of the bars (eEF1A and Hyp2) correspond to non-linear spans in the y axis. (C) Plot of R1J versus factor abundance (Spearman rank correlation coefficient (rS)=0.44). The error data for the abundance values represented in (B) are indicated in (C) and are given numerically in Supplementary Table 6.
Exploring the interdependence of the steps of the pathway, the model shows how efficient peptide elongation maximizes the available capacity for initiation (the ‘free’ ribosome pool; Figure 2G). The current form of the model incorporates 20 codons into the mRNA ORF. This length allows the key properties of translation, including the localized effects of the physical size of the ribosome on the accessibility of the start codon and the potential for ‘queuing’ upstream of the stop codon, to be modelled. At the same time, the chosen ORF size limits calculation times to reasonable levels so that it can readily be used by the majority of users. On the other hand, this version of the model does not precisely reproduce the absolute numbers of ‘free’ ribosomes in real cells because it assumes a short reading frame and ignores ribosomes that are not productively associated with mRNA. Nevertheless, it still serves as a powerful tool for analysing the relationships between different steps of the pathway. Inefficient termination is predicted to cause accumulation of ‘backed-up’ ribosomes, thus reducing the available capacity for initiation (Figure 2H). These results illustrate the fine balance in the cell between elements of control in the respective phases of translation. Indeed, they reveal that control of the respective phases of translation in living cells is more tightly coupled than is apparent in cell-free systems.
Scanning competence, a step exercising strong control?
The stability of the ribosomal pre-initiation complex scanning along the 5′UTR will affect the performance of the translation machinery. The output of our scanning assay (the RSc value; Table I) reflects the overall efficiency of the scanning process as a function of translation factor activity (Supplementary Figure 5B). eIF1, eIF1A, eIF2, eIF2B, and Ded1 were found to promote stable and efficient scanning on the longer 5′UTR utilized here, and we have accordingly classified these proteins as ‘scanning-efficiency-promoting factors’. This represents a sub-category of the larger group of eukaryotic initiation factors. Comparison of Ded1’s RJ, RSp, and RSc values (Table I) reveals that this DEAD-box helicase promotes near-maximal global translation rates over a broad range of abundance (see also the corresponding plot in Figure 1A), yet the translation of mRNAs bearing long 5′UTRs (as in the scanning assay, Supplementary Figure 5B) is differentially sensitive to reductions in Ded1 activity. This is consistent with the previously observed phenotypes of Ded1 mutations (de la Cruz et al, 1997) and with the conclusion drawn from single-molecule force measurements that there is a degree of functional compartmentalization between the RNA helicases Ded1 and eIF4A (Marsden et al, 2006), whereby eIF4A is more involved in 40S recruitment to the 5′ end. Overall, this work has highlighted the key roles of three helicases (Dbp5, Ded1, and eIF4A) in translational control.
Intracellular abundance and system control
Earlier work (von der Haar and McCarthy, 2002; von der Haar, 2008) raised the question how the intracellular factor levels relate to the control exerted by each protein over the translation rate. Moreover, studies of transformed mammalian cells have suggested that overproduction of a single factor can mediate upregulation for the whole system (Cuesta et al, 2009). Global estimations of protein abundance have previously relied on protein tagging (Ghaemmaghami et al, 2003). However, TAP fusions can cause growth inhibition, and tagging in general can affect protein stability (Yen et al, 2008). We therefore determined the absolute intracellular abundance of the (untagged) translation factors in physiologically normal cells using internal multiplexed standards that comprise concatamers of quantotypic tryptic peptides containing [13C6]-arginine and [13C6]-lysine (Beynon et al, 2005; Figure 4A; Supplementary Table 4). Plots of the resulting intracellular protein abundance values reveal large variations in abundance across the respective functional phases of translation (Figure 4B; Supplementary Tables 5 and 6) and very limited correlation between abundance and RSp, or between abundance and R1J, respectively (Figures 3B and 4C; Spearman rank correlation coefficients of −0.1 and 0.44).
There is no obvious generic principle governing the R1J values of subunits of the complexes on this macromolecular assembly pathway. For example, the components of the eIF4F complex, that is, eIF4A, eIF4B, eIF4E, and eIF4G, manifest very different intracellular abundance values (Figure 4B; Supplementary Table 6), yet within this subset of factors there is no apparent pattern relating abundance to flux control (Figure 4C). This may to some degree reflect partial redundancy in the mode of action of eIF4F factors. The discrepancy in component factor levels is less marked for the MFC factors but they are not exactly stoichiometric (eIF1, eIF2, eIF3, and eIF5; Figure 4B). Comparison of the wide range of intracellular levels and R1J values of the eIF4F and MFC factors therefore suggests that progression through the translation pathway is not strictly linked to prior assembly of these key complexes in the initiation phase. This conclusion is supported by in silico experiments of the type shown in Figure 2 and by other experimental studies (Park et al, 2011; Sokabe et al, 2012). In contrast, the consistent R1J values for the subunits of eIF2, eIF2B, and eIF3 indicate that their functional contributions are primarily realized within the respective assembled complexes (i.e., not as separate subunits with independent functions).
Exploring beyond naturally evolved translation factor ratios
Supplementing genomic expression using specially designed plasmids (Supplementary Figure 2D), we observed that translation factors generally do not cause yeast to exceed the physiological maximum protein synthesis rate when their abundance is increased above 100% (i.e., R+1J≈0; Table I). The sharp transitions to this plateau show that the physiologically measured rate of protein synthesis observed at normal translation factor levels represents an absolute ceiling as far as the influence of individual translation factor abundance is concerned. Thus, co-evolution of functional capabilities and abundance in the translation machinery has optimized efficiency. Exceptionally, the overproduction of eRF1 does lead to a measurable, albeit small, increase in both global protein synthesis and growth. The significance of this effect was confirmed by multiple experiments that extended up to almost 200% of the physiological abundance of eRF1 (see R+1J plot (red line) in Figure 1). The negative R+1J value for eIF4G, on the other hand, may reflect marked sensitivity of the pathway to suboptimal redistribution between key eIF4G-containing complexes at super-physiological levels of this factor.
The role of gene duplication in rate control
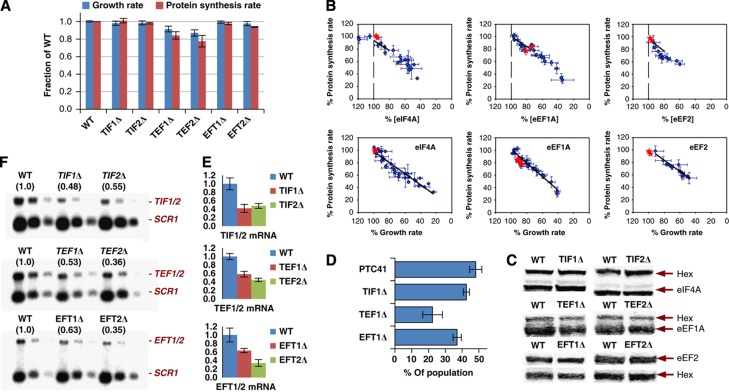
eIF4A (TIF1/TIF2), eEF1A (TEF1/TEF2), and eEF2 (EFT1/EFT2) are each encoded by duplicated genes that encode identical proteins. Multiple alleles encoding a subset of translation factors are frequently observed in eukaryotic genomes. eIF4A, eEF1A, and eEF2 are very abundant factors in yeast (Figure 4) with the highest individual R1J values of the translation machinery (Figures 1 and 2; Table I). Remarkably, deletion of one allele from each pair reduced global protein synthesis and growth rate only minimally (Figure 5A). Mapping these data points (together with protein quantitation data) onto the RJ and RSp plots reveals that the single allele deletion strains perform far better than would be the case if the respective alleles in each pair would contribute equally to maintain maximal protein synthesis (Figure 5B). Western blotting reveals that the total abundance of each encoded factor changed by <10% in the respective single allele deletion strains (eIF4A, TIF1Δ or TIF2Δ; eEF1A, TEF1Δ or TEF2Δ; eEF2, EFT1Δ or EFT2Δ; Figure 5C). The competitive viability advantage conferred by duplication of the respective alleles varies in line with the impact of the respective single allele deletions on protein synthesis and growth (Figure 5D). Analysis of the intracellular mRNA abundance values determined by RT–qPCR (Figure 5E) and northern blotting (Figure 5F) revealed that single allele deletions resulted in reduced intracellular levels of the corresponding mRNA. Comparison of the respective mRNA and protein abundance values indicates that post-transcriptional compensation partially underpins this effect. Overall, rather than simply enhancing the production of these factors to levels unobtainable by single genes, gene duplication seems to function more as a fine control mechanism.
Figure 5.
Identical duplicated genes contribute to fine control. (A) Growth rates and protein synthesis rates of the allele deletion strains compared to those of the wild-type strain. (B) Mapping of the data points from (A) onto the corresponding RJ and RSp plots (from Figures 1 and 3). (C) The relative amounts of the respective translation factors were determined as band intensity ratios in western blots using hexokinase as internal standard. (D) Competitive growth experiments yielded percentage survival rates for each single deletion strain remaining in the total population of cells. (E) RT–qPCR and northern blots (F) to determine transcript levels in the different single deletion strains compared to the wild-type strain. In (F), three different amounts of each sample (800, 400, and 200 ng total RNA) were applied to the gel to ease comparison. The average intensities of the respective northern blot bands relative to the wild-type levels (normalized to SCR1) are given in parentheses—these match closely the normalized values from RT–qPCR (E).
Discussion
We have mapped out the quantitative control landscape that governs the rate of protein synthesis in exponentially growing yeast cells using a minimally disruptive ‘genetic titration’ procedure. This reveals a network of contributions from the components of the translation machinery as well as insight into the regulatory potential of individual factors. Thus, each R1J value featured in Table I is directly relevant to the overall status of control in growing cells and is also indicative of the sensitivity of the pathway to changes in each factor’s activity in the near-physiological domain. It is notable that the high-flux-control factors manifest high RSp values, so that rate control is primarily exercised by dedicated components of the translation machinery. On the other hand, many features of this landscape of control are counterintuitive. For example, elongation is the most dominant control step in the translation pathway, while certain steps in initiation and termination also feature strongly in the control map. Moreover, the factors showing the very highest levels of flux control (in elongation and initiation) are the most abundant components of the translation machinery. The catalytic mode of action of some factors together with the partial redundancy of certain interactions both contribute to the complex nature of the control landscape, and it is notable that scanning is promoted by low-flux-control factors. The observation that the R+1J values are almost all approximately zero also tells us that the yeast cell generally has no means of upregulating protein synthesis (or growth) by enhancing the activity of an individual factor.
eRF1, however, is an exception in this regard. We suspect that the intracellular abundance of eRF1 in the yeast cell reflects a naturally selected balance between positive and negative effects of this protein on termination efficiency, reinitiation efficiency and potentially other poorly understood functions (Merritt et al, 2010). While exceeding the 100% level of this factor can allow small increases in the rate of global protein synthesis under the exponential growth conditions described here, we speculate that the exact composition of the resulting proteome at levels of eRF1 >100% may be modified in ways that are not optimal for growth under all growth conditions. On the other hand, one possible explanation for the deleterious effect of eIF4G overexpression is that this leads to overactivation of eIF4A, with which it is known to form a complex (Schütz et al, 2008). These hypotheses should be subjected to detailed analysis in future work.
Our interpretation of this large-scale in vivo rate control study assumes that the modulation of the abundance of each translation factor between 100 and 80% of its physiological level equates to modulation of its functional activity over the same range. We believe that this is a justifiable assumption in the context of current understanding since previous work in yeast has identified only eIF2 as being subject to regulation (via phosphorylation; Hinnebusch, 2005); modification of other yeast translation factors has generally not been associated with regulatory phenomena (van den Heuvel et al, 1995; Zanchin and McCarthy, 1995). Moreover, we are not aware of any evidence that small degrees of direct modulation of individual translation factor levels in the near-physiological range will influence the modification states or specific activities of those factors in exponentially growing yeast, but further work will be necessary to assess this accurately.
One major consequence of the R1J=0 values of the scanning efficiency-promoting factors is that they are predicted to minimize the time that ribosomal subunits remain associated with mRNA molecules by reducing the likelihood that scanning will be delayed through the lack of availability of essential factors. This maximization of the efficiency of mRNA-bound functions is expected, in turn, to optimize the pool size of free ribosomes available to re-engage with free mRNA 5′ends, thus maximizing overall translation efficiency. Examination of the rate control plots also suggests that the performance of the translation pathway in wild-type cells is likely to be comparatively robust with respect to small natural stochastic fluctuations in the abundance of the low-flux-control factors, since the global protein synthesis rate is relatively insensitive to variations in their intracellular concentrations near the average physiological level. This effect is expected to be effective at the population level because it will operate in all cells.
The reported correlations between overproduction of translation factors in certain mammalian cells and cell transformation (Cuesta et al, 2009) raise questions about the possible differences in control between the respective translation machineries in yeast and mammalian systems. For example, if transformation is induced by enhanced global protein synthesis rates in mammalian cells overproducing a specific factor, then this suggests that all the remaining components of the translation machinery have additional capacity above that required for physiological translation rates. This ‘universal’ additional capacity is not apparent in the yeast translation machinery. An alternative possibility is that the link between overproduction of a translation factor and transformation in mammalian cells may relate to more specific effects of the overproduced factor on the expression of a subset of genes and/or on other cellular processes. These interesting apparent differences between the respective systems merit further investigation.
It is of interest to compare the significance of intracellular subunit concentrations and stoichiometries in other types of molecular machine. For example, the quantities of the respective subunits of the H+-ATPase that are synthesized in the microbial cell are fully utilized in the same stoichiometry in the assembly of the membrane-associated complex (α3β3γ1δ1ε1a1b2c10; McCarthy, 1998). Overall, it is apparent that the functional significance of the relative stoichiometry of the components of a complex multisubunit molecular machine only emerges through detailed quantitative analysis. The evolution of optimal function in the translation machinery has evidently been achieved using highly disparate component stoichiometries.
Another distinctive feature of in vivo translation control that has emerged from our rate control analysis relates to the role of duplicated genes in determining system function. Comparison with the large body of duplicated genes that encode ribosomal proteins in S. cerevisiae is informative. In contrast to the translation factor genes considered here, the majority of ribosomal genes are not only duplicated but also contain introns. The introns seem to be involved in mechanisms of intra-and inter-genic regulation that, while not required for normal growth under laboratory conditions, can impact upon cell competitiveness, especially under stress conditions (Parenteau et al, 2011). In contrast, we have now found that eIF4A, eEF1A, and eEF2 are encoded by duplicated genes that contribute (asymmetrically) in an intron-independent manner to the maintenance of optimal levels of these factors. Given the unexpected characteristics of expression of these translation factor genes, one possible explanation of the observed selective advantage of gene duplication for the cell may involve increased consistency in the respective intracellular factor concentrations sustained over the cell cycle, and we believe that this hypothesis should be tested in future work.
The rate control analysis we describe is not only complementary to the wealth of other biochemical, biophysical, and genetic studies of translation but it will also facilitate the interpretation of these diverse sources of data in terms of system function. For example, the existence of a system control model makes it possible to interpret the impact of mutations in a holistic context. Future work will be able to elucidate precisely the important deterministic relationship between regulatory changes in the activity of specific factors and the resulting modulation of protein synthesis in yeast cells in response to environmental stress. For example, physical and/or functional interactions between the kinase General-control-non-derepressible-2 (Gcn2) and the elongation factors eEF1A and eEF3 seem to mediate modulation of the amino-acid starvation response in S. cerevisiae (Visweswaraiah et al, 2011, 2012), and in vivo rate control analysis could help to quantitate the regulatory impact of these interactions. The current work also represents a starting point for equivalent analyses of the distinct control properties of the protein synthesis pathways in animals and plants and how these react to stress and disease. For example, an analogous investigative strategy could help to test the hypothesis that there is a direct causal relationship between the onset of cancer and enhanced global translation induced by overproduction of eIF2, eIF3 subunits, eIF4A, eIF4E, or eIF4G in transformed cells (Cuesta et al, 2009), and aid characterization of the respective control features of cap-dependent and cap-independent translation in higher eukaryotic cells, although any analysis would need to take account of the effects of post-translational modifications on factor activities (Mahoney et al, 2009).
Materials and methods
R J , R Sp , and R Sc values
Chromosomal integration, analysis of tet07constructs (Bellí et al, 1998; Supplementary Figure 2), and gene disruption (Güldener et al, 1996) were performed as described previously. Growth rate, protein synthesis rate (measured as 35S-L-methionine incorporation), and relative translation factor abundance values were all determined in parallel for the same set of cultures (maximum of eight strains in one set, one of which was the wild-type reference strain PTC41). Cultures were incubated with doxycycline until a stable steady-state level of the tet07-construct-encoded translation factor had been reached (17 h). The in vivo protein synthesis rate was estimated as 35S-L-methionine incorporation into total protein. In order to determine the intracellular concentration of translation factors, cells were harvested by centrifugation from 10 ml cultures grown to OD600=0.4 (∼1.5 × 107 cells/ml) and protein was extracted from cells for western blotting (von der Haar, 2007) and for QconCAT-calibrated quantitation (Beynon et al, 2005). For scanning efficiency measurements, PTC41 and tetO7 strains containing the pDLV-L2/L0 plasmid (Supplementary Figure S6A) were grown in YNBD-Met-His medium, and luciferase activities were measured using a Berthold Technologies Lumat LB 9507 luminometer (McNabb et al, 2005; Supplementary Figure S6B). RNA was isolated by a hot phenol extraction method (Schmitt et al, 1990).
Quantitative mass spectrometry
QconCAT peptides (Supplementary Table 4) were used as internal standards for mass spectrometric absolute protein quantification (Beynon et al, 2005; Brownridge et al, 2011, 2012). For accurate mass (AM) peptide precursor analysis by the Thermo-Fisher LTQ-Orbitrap Velos, tryptic peptides were separated over three 90 min linear liquid chromatography gradients (3–40% acetonitrile plus 0.1% formic acid) per sample, delivering gas-phase fractionation MS acquisitions at m/z; 350–600, 590–800, and 790–2000. The raw data were processed using Proteome Discoverer (Thermo-Fisher, version 1.3) and searched (Mascot, version 2.3) against a S. cerevisiae-specific database with peptide and MSMS tolerances set at 10 p.p.m. and 0.5 Da, respectively. Modifications were set as fixed carbamidomethylation of cysteine and variable methionine oxidation. For selected reaction monitoring (SRM), peptides analysed by the Waters Xevo triple quadrupole mass spectrometer were separated over a 30-min gradient (3–40% acetonitrile plus 0.1% formic acid) and SRM data were acquired using previously determined scheduled transitions (Supplementary Table 5), with optimized collision energies and dwell times, for the unlabelled analyte and labelled QconCAT peptides. SRM data acquired using the transition lists were processed using ApexTrack peak integration, with smoothing enabled (2 smooths over 2 scans) and baseline thresholds of ∼5% with a relative area response threshold of 30. The abundances of the proteins of interest were determined from either the MS peak intensities (AM) or the chromatographic peak areas (SRM) of both the analyte peptide and the corresponding labelled QconCAT peptide (see Figure 4A as an example). As the QconCAT peptides are present in each sample at a known amount, it is possible to calculate the abundance of the analyte peptide from the ratio between the labelled and unlabelled MS/chromatographic peaks (Beynon et al, 2005; Supplementary Table 6). Samples containing QconCAT at 100, 5, 1, and 0.1 fmol per 100 000 cells were analysed to cover the full dynamic range of the proteins in the mixture. As a further check on the accuracy of our methods, we compared the abundance values predicted on the basis of the AM and SRM data (Supplementary Figure 6). For a small number of proteins (see Supplementary Table 6) not included in the QconCAT design, absolute abundance values were obtained by label-free quantification using a Waters Synapt G2 mass spectrometer in MSE acquisition mode (Silva et al, 2006).
Western and northern blotting
Multiple dilutions of protein extracts (von der Haar, 2007) from tetO7 strains and the PTC41 reference strain were used for western blotting (Towbin et al, 1979; Sangthong et al, 2007). Each translation factor was detected by a specific primary antibody (overnight incubation, dilution ranging from 1:500 to 1:10 000). Quantitation of translation factor signal via IRDye-800CW-tagged secondary antibodies was normalized against hexokinase using an Odyssey scanner (LI-COR), thus generating values for each translation factor that were expressed as a percentage of the wild-type physiological abundance (the 100% value, determined in absolute terms as molecules per cell via mass spectrometry). Northern blots were performed essentially as described previously (Sambrook and Russell, 2001) using formaldehyde/formamide gels and Hybond-N+ membranes (GE Healthcare); quantitation of radioactivity was performed using a PharosFX Molecular Imager (Bio-Rad). RT-qPCR was performed according to a protocol adapted from Teste et al (2009) using a Qiagen Rotorgene Q qPCR thermocycler. Primers for qPCRs were design using Clon Manager software, with the exception of UBC6-specific primers. Cell-culture images were analysed using Cellometer M10 software (Supplementary Figure 4).
Computational modelling
A detailed mathematical model (Supplementary Figure 1D), based on ordinary differential equations, was built in the software COPASI (Hoops et al, 2006). The model assumes that 15 codons are occupied by each ribosome, as reflected in reaction R18 in initiation and also reaction R26 (ribosome translocation during elongation). The kinetic functions of these two reactions are based on MacDonald et al (1968) and Heinrich and Rapoport (1980). All other kinetic functions follow mass-action kinetics. The concentrations of transfer RNA species (Met-tRNA, aa-tRNA, and tRNA in the model) are kept constant, while the other species’ concentrations can change in the course of the simulation. The model describes the translation of a short mRNA with 20 codons. Therefore, all reactions in the elongation cycle (R22, R23, R25, R26, R28, and R29) and the corresponding species are replicated accordingly to model the species with ribosomes bound at different positions. The initial concentrations of the various factors were made equal to the protein concentrations determined by the quantitative mass spectrometry method. In total, the model contains 165 different species and 141 reactions. COPASI was used to estimate the value of the 56 parameters (rate constants) involved in the reactions of this model by fitting against the experimental data of modulation of the various translation factors, a total of 212 data points (Figure 1; Supplementary Figure 3). The large number of observations relative to the number of parameters estimated indicates that the problem is identifiable. The minimization of the least-squares objective function was carried out with parallel random search using the Condor-COPASI system (Kent et al, 2012) on a pool of 2500 CPU cores, consuming a total of 17 290 CPU hours.
Supplementary Material
Supplementary figures S1-6, Supplementary tables SI-VI
This Table (parts a and b) is a dataset in Excel.
Acknowledgments
We thank Helen Bryant, Dominik Skiba, Shirley Tait, Kathryn Blount, and Simon Mitchell (technical support); Simon Hubbard, Craig Lawless, and Julian Selley (QconCAT design); the BBSRC for funding. Antibodies were kindly donated by Charles Cole (Dartmouth Medical School, USA), Ernie Hannig (UT Dallas, USA), Alan Hinnebusch (NICHD, Maryland, USA), Terry Kinzy (Rutgers NJ, USA), Patrick Linder (Geneva, Switzerland), Graham Pavitt (Manchester, UK), and Tobias von der Haar/Mick Tuite (Kent, UK). Anne Willis (Leicester, UK) kindly provided Renilla luciferase DNA.
Author contributions: JEGM designed and supervised this study, analysed the data, and wrote the manuscript. HF performed most of the experimental work and analysed the data. SK and JD performed parts of the experimental work (rate control analysis and mRNA analysis). RB helped design the quantitative mass spectrometry experiments and analyze the data. AC performed the quantitative mass spectrometry and completed the data analysis. JP and PM developed the computational model. HW contributed to the design of the study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abbott CM, Proud CG (2004) Translation factors: in sickness and in health. Trends Biochem Sci 29: 25–31 [DOI] [PubMed] [Google Scholar]
- Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F (1997) Yeast Pab1 interacts with Rna15 and participates in the control of the poly(A) tail length in vitro. Mol Cell Biol 17: 3694–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Boas FE, Brown PO, Herschlag D (2005) Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res 33: 2421–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellí G, Garí E, Aldea M, Herrero E (1998) Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast 14: 1127–1138 [DOI] [PubMed] [Google Scholar]
- Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JEG (2004) Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol Microbiol 51: 987–1001 [DOI] [PubMed] [Google Scholar]
- Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ (2005) Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods 2: 587–589 [DOI] [PubMed] [Google Scholar]
- Brownridge P, Holman SW, Gaskell SJ, Grant CM, Harman VM, Hubbard SJ, Lanthaler K, Lawless C, O’Cualain R, Sims P, Watkins R, Beynon RJ (2011) Global absolute quantification of a proteome: Challenges in the deployment of a QconCAT strategy. Proteomics 11: 2957–2970 [DOI] [PubMed] [Google Scholar]
- Brownridge PJ, Harman VM, Simpson DM, Beynon RJ (2012) Absolute multiplexed protein quantification using QconCAT technology. Methods Mol Biol 893: 267–293 [DOI] [PubMed] [Google Scholar]
- Cannarozzi G, Cannarrozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y (2010) A role for codon order in translation dynamics. Cell 141: 355–367 [DOI] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH (1997) Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275: 1468–1471 [DOI] [PubMed] [Google Scholar]
- Cole CN, Scarcelli JJ (2006) Unravelling mRNA export. Nat Cell Biol 8: 645–647 [DOI] [PubMed] [Google Scholar]
- Cuesta R, Gupta M, Schneider RJ (2009) The regulation of protein synthesis in cancer. Prog Mol Biol Transl Sci 90: 255–292 [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P (1997) The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 94: 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H (2007) The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649 [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24: 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM (2007) Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol 365: 835–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich R, Rapoport TA (1980) Mathematical modelling of translation of mRNA in eukaryotes; steady state, time-dependent processes and application to reticulocytes. J Theor Biol 86: 279–313 [DOI] [PubMed] [Google Scholar]
- Henderson A, Hershey JW (2011) Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 108: 6415–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U (2006) COPASI—a COmplex PAthway SImulator. Bioinformatics 22: 3067–3074 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N (2005) Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR (2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657–704 [DOI] [PubMed] [Google Scholar]
- Kemper WM, Berry KW, Merrick WC (1976) Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Balpha and M2Bbeta. J Biol Chem 251: 5551–5557 [PubMed] [Google Scholar]
- Kent E, Hoops S, Mendes P (2012) Condor-COPASI: High-throughput computing for biochemical networks. BMC Syst Biol 6: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene 299: 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrín-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- MacDonald CT, Gibbs JH, Pipkin AC (1968) Kinetics of biopolymerization on nucleic acid templates. Biopolymers 6: 1–5 [DOI] [PubMed] [Google Scholar]
- Mahoney SJ, Dempsey JM, Blenis J (2009) Cell signalling in protein synthesis: ribosome biogenesis and translation initiation and elongation. Prog Mol Biol Transl Sci 90: 53–107 [DOI] [PubMed] [Google Scholar]
- Marsden S, Nardelli M, Linder P, McCarthy JEG (2006) Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J Mol Biol 361: 327–335 [DOI] [PubMed] [Google Scholar]
- Mathews MB, Sonenberg N, Hershey JWB (2000) Origins and principles of translational control. In Translational Control of Gene Expression Sonenberg N, Hershey JWB, Mathews M (eds) pp 1–31Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, [Google Scholar]
- McCarthy JEG (1998) Posttranscriptional control of gene expression in yeast. Microbiol Mol Biol Rev 62: 1492–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb DS, Reed R, Marciniak RA (2005) Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot Cell 4: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt GH, Naemi WR, Mugnier P, Webb HM, Tuite MF, von der Haar T (2010) Decoding accuracy in eRF1 mutants and its correlation with pleiotropic quantitative traits in yeast. Nucleic Acids Res 38: 5479–5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Morin G, Gagnon J, Lucier J-F, Wellinger RJ, Chabot B, Elela SA (2011) Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147: 320–331 [DOI] [PubMed] [Google Scholar]
- Park E-H, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG (2011) Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics 12: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman YR, Kandl K, Lewis M, Valente L, Kinzy TG (2009) Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha. J Biol Chem 284: 4739–4747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW (1989) The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58: 857–867 [DOI] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE (2009) Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, [Google Scholar]
- Sangthong P, Hughes J, McCarthy JEG (2007) Distributed control for recruitment, scanning and subunit joining steps of translation initiation. Nucleic Acids Res 35: 3573–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U (2008) Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA 105: 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JWB (2012) The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res 40: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste M-A, Duquenne M, François JM, Parrou J-L (2009) Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol 10: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakov VN, Valouev IA, Kochneva-Pervukhova NV, Packeiser AN, Vishnevsky AY, Glebov OO, Smirnov VN, Ter-Avanesyan MD (2006) N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination. BMC Mol Biol 7: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE (2002) Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics 160: 393–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel JJ, Lang V, Richter G, Price N, Peacock L, Proud C, McCarthy JEG (1995) The highly acidic C-terminal region of the yeast initiation factor subunit 2α (eIF-2α) contains casein kinase phosphorylation sites and is essential for maintaining normal regulation of GCN4. Biochim Biophys Acta 1261: 337–348 [DOI] [PubMed] [Google Scholar]
- Visweswaraiah J, Lageix S, Castilho BA, Izotova L, Kinzy TG, Hinnebusch AG, Sattlegger E (2011) Evidence that eukaryotic translation elongation factor 1A (eEF1A) binds the Gcn2 protein C terminus and inhibits Gcn2 activity. J Biol Chem 286: 36568–36579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaraiah J, Lee SJ, Hinnebusch AG, Sattlegger E (2012) Overexpression of eukaryotic translation elongation factos 3 (eEF3) impairs Gcn2 activation. J Biol Chem 287: 37757–37768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T (2007) Optimized protein extraction for quantitative proteomics of yeasts. PLoS ONE 2: e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T (2008) A quantitative estimation of the global translational activity in logarithmically growing yeast cells. BMC Syst Biol 2: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, McCarthy JEG (2002) Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol 46: 531–544 [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG (2001) Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 20: 4370–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ (2008) Global protein stability profiling in mammalian cells. Science 322: 918–923 [DOI] [PubMed] [Google Scholar]
- Zanchin N, McCarthy JEG (1995) Characterization of the in vivo phosphorylation sites of the mRNA-cap-binding complex proteins eukaryotic initiation factor-4E and p20 in Saccharomyces cerevisiae. J Biol Chem 270: 26505–26510 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures S1-6, Supplementary tables SI-VI
This Table (parts a and b) is a dataset in Excel.