Allopurinol is the most commonly used drug for the treatment of hyperuricemia and gout. However, allopurinol is also one of the most common causes of severe cutaneous adverse reactions (SCARs), which include drug hypersensitivity syndrome, Stevens–Johnson syndrome, and toxic epidermal necrolysis. A variant allele of the human leukocyte antigen (HLA)-B, HLA-B*58:01, associates strongly with allopurinol-induced SCAR. We have summarized the evidence from the published literature and developed peer-reviewed guidelines for allopurinol use based on HLA-B genotype.
The aim of this article is to provide pharmacogenetic information relevant to the clinical use of HLA-B*58:01 genotyping results in patients with indications for allopurinol use. On the basis of this information, a guideline is recommended for the use of allopurinol when HLA-B*58:01 genotyping results are available. Other areas are beyond the scope of this guideline, including the selection of an alternative to allopurinol or the use of other tests, such as an intradermal skin test and cost effectiveness analyses. The Clinical Pharmacogenetics Implementation Consortium (CPIC) publishes a series of guidelines aimed at incorporating pharmacogenetics into clinical decisions. These guidelines are updated periodically on www.pharmgkb.org based on new developments in the field.
Focused Literature Review
A systematic review was conducted on HLA-B*58:01 and allopurinol (see Supplementary Data online). This guideline was developed based on the literature review by the authors and experts in the field.
Gene: HLA-B
Background
The human major histocompatibility complex (MHC) comprises the genes for human leukocyte antigen (HLA) class I and class II, also known as MHC-I and MHC-II. The MHC gene family is located on chromosome 6 in a region of high linkage disequilibrium. The HLA class I complex consists of three genes, HLA-A, HLA-B, and HLA-C; the HLA class II complex consists of the HLA-DR, HLA-DP, and HLA-DQ genes.1 The HLA genes, particularly HLA-B, are some of the most polymorphic in the human genome with more than 5,000 HLA-I and HLA-II alleles listed in the IMGT/HLA database2 (http://www.ebi.ac.uk/imgt/hla/stats.html).
HLA molecules encoded by the HLA genes play a crucial role in the immune system. They bind endogenous and exogenous peptides that result from the breakdown of self-cells or foreign proteins and present these bound peptide antigens to T cells to initiate their antigen-specific responses. HLA class I molecules present peptides derived from proteins that break down inside cells, whereas HLA class II molecules present antigens encountered outside the cells. The main structural difference between the HLA class I and II molecules lies in the peptide-binding groove, which determines the peptide-binding specificities.3 The diversity of the T-cell repertoire is thus generated by the HLA molecular structures, the peptides bound, and the orientation of peptides within the binding groove. The vast HLA repertoire ensures that an individual is capable of displaying peptides from any invading pathogen and of mounting an effective immune response.4
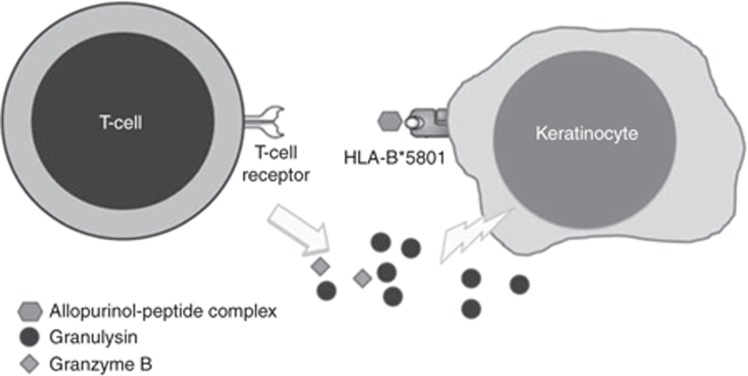
The HLA allele nomenclature has been described in a previous CPIC guideline.5 Although more than 1,500 HLA-B alleles have been identified, this guideline will focus only on the HLA-B*58:01 allele and its association with allopurinol-induced severe cutaneous adverse reaction (SCAR). This strong association suggests that the specific HLA-B molecule is not just a genetic marker, but that it also plays an important role in the pathogenesis of the adverse drug reaction. HLA-B molecules present endogenous or processed exogenous antigens to T cells, thereby eliciting an adaptive immune response. HLA restriction is required for the activation of drug-specific T cells by the culprit drug. The T-cell receptor of the effector T cell is thought to recognize the drug–peptide complex bound by the specific HLA-B molecule on the antigen-presenting cell, resulting in the release of immune mediators and leading to robust adaptive immune reactions in severe adverse drug reactions. For example, CD8+ cytotoxic T lymphocytes are the predominant cells present in the blister fluid of patients with carbamazepine-induced Stevens–Johnson syndrome (SJS)/toxic epidermal necrolysis (TEN). These T cells may induce keratinocyte death by a drug-specific, HLA class I–restricted and granulysin/perforin/granzyme–mediated pathway (Figure 1).6
Figure 1.
Working model for specific human leukocyte antigen (HLA)-B molecule interacting with a drug compound. HLA-B molecules present endogenous or processed exogenous antigens to T cells, thereby eliciting an adaptive immune response. The T-cell receptor of the effector T cell recognizes the drug–peptide complex bound by the specific HLA-B molecule on the antigen-presenting cell, resulting in the release of immune mediators.
Currently, there are three hypotheses to explain how T-cell receptors and HLA interact with drugs and peptides, leading to an immune response. The hapten hypothesis is based on the ability of a drug/metabolite to act as a hapten and thereby covalently bind to proteins or MHC-bound peptides, which are then recognized by T lymphocytes, leading to HLA-restricted T-cell activation. The “p-i concept” (direct pharmacological interaction of drugs with immune receptors) suggests that drugs can bind directly and noncovalently to a matching T-cell receptor, together with MHC molecules.7,8 The latest hypothesis proposes that a drug can interact with a specific HLA allele, for example, abacavir with HLA-B*57:01, which alters the specificity of the HLA allele and thus its peptide-binding repertoire, resulting in T-cell activation.9 The mechanism by which allopurinol interacts with HLA-B*58:01 is unclear.
Genetic test interpretation
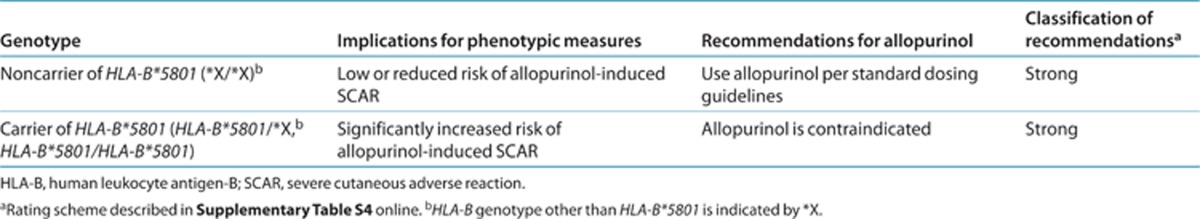
Clinical genotyping tests are available for the interrogation of HLA-B alleles. However, only tests specific for the detection of HLA-B*58:01 are relevant to the risk of developing allopurinol-induced SCAR. The HLA-B alleles do not affect the pharmacokinetics or pharmacodynamics of allopurinol; therefore, further defining the exact HLA-B allele a patient carries is not necessary as it does not provide additional useful clinical information applicable to allopurinol use. The genotyping results will be presented as HLA-B*58:01-positive (at least one copy of HLA-B*58:01 is present) or HLA-B*58:01-negative (no copies of HLA-B*58:01 are detected) with no intermediate genotype. The assignment of the likely phenotype, based on HLA-B*58:01 genotype, is summarized in Table 2. The frequencies of HLA-B*58:01 for major racial/ethnic groups are summarized in Supplementary Tables S1 and S2 online.
Available genetic test options
Testing for HLA-B*58:01 is available from several academic and commercial labs. A diagnostic kit is also available commercially. Refer to the Supplementary Table S3 online and the Pharmacogenetic test section of PharmGKB (http://pharmgkb.org/drug/PA448320#tabview=tab0&subtab=34) for more information on available clinical testing options.
Incidental findings
No other diseases or conditions have been reported to be associated with HLA-B*58:01. However, several other HLA alleles have been associated with other adverse drug reactions. Patients with the HLA-B*15:02 allele are at risk for developing SJS and TEN from carbamazepine in Southeast Asian populations,10 whereas HLA-A*31:01 is associated with carbamazepine-induced hypersensitivity reactions in the Japanese and European populations.11,12 In addition, HLA-B*57:01 is associated with abacavir-induced hypersensitivity reaction.13
Other considerations
A genome-wide association study conducted in the Japanese population identified 21 genetic variants on chromosome 6 that were significantly associated with allopurinol-induced SJS and TEN. Single-nucleotide polymorphisms with the strongest associations were rs2734583, rs3094011, and GA005234.14 In addition, the same study found that rs9263726 was in absolute linkage disequilibrium with HLA-B*58:01 and suggested that this single-nucleotide polymorphism could be used instead of HLA-B*58:01. Six single-nucleotide polymorphisms (rs2844665, rs3815087, rs3130931, rs3130501, rs3094188, and rs9469003) associated with allopurinol-induced SJS/TEN were also identified by a genome-wide association study conducted in the European population.15 Findings of this study have not been replicated in other populations.
Drug: Allopurinol
Background
Allopurinol, an analog of the purine-based hypoxanthine, inhibits the conversion of hypoxanthine and xanthine to uric acid by xanthine oxidase (xanthine oxido-reductase). In 1966, the US Food and Drug Administration approved allopurinol for treating gout, an inflammatory arthritis caused by intra-articular monosodium urate crystals that form in the setting of chronic hyperuricemia, i.e., serum/plasma urate concentration ≥7 mg/dl (0.42 mmol/l).16 The goal of therapy is to maintain serum/plasma urate concentration below 6 mg/dl (0.36 mmol/l) to prevent monosodium urate from crystallizing and to permit the dissolution of tissue monosodium urate deposits (tophi). Allopurinol is also used to treat or prevent uric acid kidney stones and acute uric acid nephropathy during chemotherapy for malignancies (tumor lysis syndrome). However, gout is the main indication, and a significant fraction of affected patients are exposed to allopurinol during the course of their disease.
An increasing worldwide prevalence and incidence of gout has been attributed to epidemic obesity and age-associated comorbidities, such as renal insufficiency and hypertension, which is often treated with diuretics that interfere with renal uric acid excretion.17 Genetic variants of transporters that affect uric acid excretion have been linked to hyperuricemia and gout, but their contribution to the overall burden of disease remains to be established.18 Based on analysis of the 2007–2008 National Health and Nutrition Examination Survey, gout is estimated to affect 8.3 million patients in the United States (6.1 million men and 2.2 million women) or 3.9% of adults.17 Similar or somewhat greater prevalence has been observed in some ethnic groups and other geographic regions.18,19,20 Treatment practices vary worldwide, but an estimated 25–30% of patients with gout in the United States and UK are prescribed allopurinol therapy.21,22 Hyperuricemia, the main risk factor for developing gout, is estimated to occur in about 21% of US adults.17 Not all patients with hyperuricemia develop gout, and allopurinol therapy for “asymptomatic hyperuricemia” is strongly discouraged.23,24
Guidelines for treating gout with allopurinol suggest a starting daily dose of 100 mg, with escalation up to 800 mg as necessary to control hyperuricemia; up to 900 mg is allowed in the UK.24 The most commonly prescribed dose of allopurinol is 300 mg/day, which is often inadequate.23 Moreover, although effective control of gout generally requires indefinite urate-lowering therapy, adherence to allopurinol frequently declines over time.23,25 Patients with renal insufficiency commonly receive doses of 100–200 mg daily, partly because exposure to oxypurinol, the long-circulating active metabolite of allopurinol, increases as renal function declines, and partly because of a concern, not universally shared, that renal insufficiency confers an increased risk of serious adverse reactions to allopurinol.23,26,27,28
The greatest safety concern with allopurinol is an estimated 0.1–0.4% risk of SCAR, also known as allopurinol hypersensitivity syndrome. SCAR is manifested by SJS, TEN, or drug reaction with eosinophilia and systemic symptoms,29,30 and is associated with systemic signs that may include fever, leukocytosis, eosinophilia, hepatitis, and acute renal failure. Allopurinol is a major cause of SCAR, and one of the most serious, with up to 25% mortality. Typically, SCAR develops within weeks or a few months of initiating treatment, but longer delays may occur. Before recent evidence strongly implicated HLA-B*58:01 status (reviewed below), renal insufficiency, diuretic use, concomitant antibiotic treatment, and higher doses of allopurinol were cited as risk factors for SCAR. Because the incidence of SCAR is low, quantifying the magnitude of risk conferred by these factors is difficult. As noted, low (often inadequate) doses of allopurinol are commonly used to treat patients with gout and renal insufficiency to avoid allopurinol hypersensitivity. A recent study showed that starting allopurinol at a lower dose may reduce the risk of allopurinol-induced hypersensitivity syndrome.31
Linking genetic variability to variability in drug-related phenotypes
There is substantial evidence linking HLA-B*58:01 genotype with phenotypic variability. Application of a grading system indicates a high quality of evidence in the majority of cases (Supplementary Table S4 online). The evidence that provides the basis for most of the guideline dosing recommendations is summarized in Table 1.
Table 1. Association of HLA-B*5801 with allopurinol-induced SCAR in different populations.
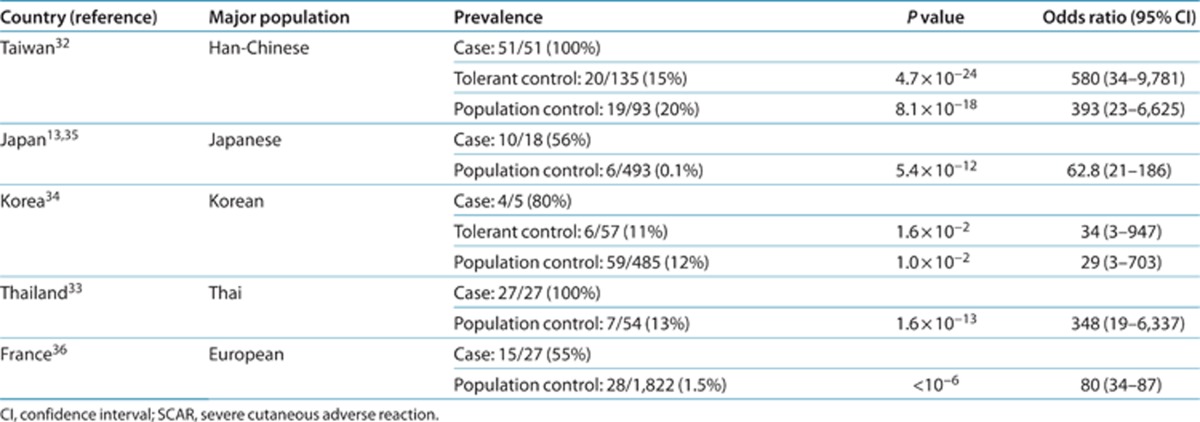
The association of HLA-B*58:01 with allopurinol-induced SCAR was first identified in the Taiwan Han-Chinese population.32 HLA-B*58:01 was present in 100% (51/51) of the patients with allopurinol-induced SCAR, as compared with 15% (20/135) of allopurinol-tolerant controls and 20% (19/93) of the population controls. This strong association was replicated in a Thai population, in which all the patients with allopurinol-induced SCAR (N = 27) carried the allele and only 13% (7/54) of the allopurinol-tolerant controls tested positive for HLA-B*58:01.33 A similar but more modest association was observed in three other populations: 80% (4/5) of Korean cases with SCAR carried the allele vs. 12% (59/485) of healthy Korean controls;34 in a Japanese population, 56% (10/18) of the cases with allopurinol-induced SCAR had HLA-B*58:01 as against 0.61% (6/493) of healthy controls;14,35 a European study identified HLA-B*58:01 in 55% (15/27) of patients with allopurinol-induced SCAR, whereas only 1.5% (18/1,822) of the controls tested positive for the allele (Table 1).36 One recent meta-analysis that consolidated all the published studies gave the odds ratios for allopurinol-induced SCAR in HLA-B*58:01 carriers as 73 and 165 for studies using healthy controls and allopurinol-tolerant controls, respectively.37
Clinical trials testing whether genotyping for the presence of HLA-B*58:01 can reduce the number of allopurinol-induced SCAR cases have not been published; however, one such study is under way in Taiwan. It is a nonrandomized trial design using historical data as control due to ethical considerations. Genotyping of HLA-B*58:01 is performed on patients with indications for allopurinol use. If the participants test positive for HLA-B*58:01, drugs other than allopurinol are used.
Therapeutic recommendations
The Taiwan Department of Health has updated the labeling for allopurinol to include information on HLA-B*58:01 (http://www.doh.gov.tw/EN2006/Newsroom/Press_list.aspx?year=2009&doc_no=72847). The updated label describes the strong association between HLA-B*58:01 and allopurinol-induced SJS/TEN in the Han-Chinese population and recommends testing for the allele before the use of allopurinol. The updated label does not recommend such testing for patients who have had no adverse events after prolonged use of allopurinol. At the time of writing this article, the US Food and Drug Administration had not updated the labeling for allopurinol. However, given the high specificity for allopurinol-induced SCAR, allopurinol should not be prescribed to patients who have tested positive for HLA-B*58:01. Alternative medication should be considered for these patients to avoid the risk of developing SCAR. For patients who have tested negative, allopurinol may be prescribed as usual (Table 2). However, testing negative for HLA-B*58:01 does not totally eliminate the possibility of developing SCAR, especially in the European population.
Table 2. Recommended therapeutic use of allopurinol by HLA-B genotype.
Several clinical factors have been reported to be associated with an increased risk for allopurinol hypersensitivity. Renal dysfunction is the most significant nongenetic factor, and patients with renal insufficiency were four times more likely to develop adverse events than those with normal renal function.32 The risk of allopurinol-induced hypersensitivity is also reported to increase with the concomitant use of ampicillin or amoxicillin.
In addition to SCAR, allopurinol therapy is also associated with a 2–3% incidence of less severe rashes unassociated with systemic symptoms or organ damage. Food and Drug Administration guidelines recommend discontinuing allopurinol if a rash develops. Until recently, only uricosuric drugs such as probenecid and benzbromarone were available for the patients who need to discontinue allopurinol due to skin rash. However, probenecid is often less effective than allopurinol, particularly in patients with renal insufficiency, and benzbromarone is not an approved drug in many countries. This has prompted attempts to induce tolerance to allopurinol by rechallenge with gradual escalation of low doses as tolerated. This unreliable approach has not been widely accepted, and its use may decline because alternative urate-lowering therapies are now available. Febuxostat, which received Food and Drug Administration approval in 2009, is available in 19 countries. It is a nonpurine xanthine oxidase inhibitor that is primarily metabolized in the liver to inactive glucuronide and excreted into the urine and bile. Therefore, mild to moderate renal impairment might have little impact on the pharmacokinetics of febuxostat. It was reported to be tolerated in 12 of 13 patients with a history of severe allopurinol hypersensitivity.38 In 2010, the Food and Drug Administration approved pegloticase, a PEGylated urate oxidase, as an orphan drug for treating patients with refractory chronic gout who had an inadequate response to, or were intolerant of, other urate-lowering drugs.39 Newer urate-lowering drugs are in clinical trials.
Recommendations for incidental findings
Not applicable.
Other considerations
Not applicable.
Potential benefits and risks for the patient
Results from the Taiwan HLA-B*58:01 clinical study are not yet available. At the time of writing this article, more than 1,600 patients had been recruited, but the incidence of allopurinol-induced SCAR had not been reported.40 Given the high negative predictive value of the allele, especially in patients of Asian descent (>99%), HLA-B*58:01 testing could significantly reduce the incidence and risk for allopurinol-associated SCAR. As with any laboratory test, one potential risk could come from genotyping error. A false-negative genotyping result could lead to allopurinol-induced adverse events. Genotypes are lifelong characteristics; such errors could have a broader health implication if other associations with HLA-B*58:01 are identified in the future.
Caveats: appropriate use and/or potential misuse of genetic tests
The positive predictive value for HLA-B*58:01 is ~1.5% and the negative predictive value is 100% (based on the data from the Han-Chinese and Thai populations).33 Therefore, a significant number of patients carrying the allele will not develop SCAR when they receive allopurinol treatment. New genetic factors may be identified in the future to differentiate the HLA-B*58:01 carriers who are or are not likely to develop SCAR. The most severe SCAR (toxic epidermal necrolysis) carries a 30% mortality rate. However, further study is warranted on the development of SCAR in European populations.
HLA-B*58:01 predicts only allopurinol-induced SCAR, not other adverse events (such as mild skin rash) that a patient might experience during allopurinol treatment. The marker also does not predict the efficacy of treatment with allopurinol. Regardless of the genotyping results, physicians should monitor patients closely.
Disclaimer
CPIC guidelines reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written and are intended only to assist clinicians in decision making, in addition to identifying questions for further research. New evidence may have emerged since a guideline was submitted for publication. Guidelines are limited in scope and are not applicable to interventions or diseases not specifically identified. Guidelines do not account for all individual variation among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the health-care provider to determine the best course of treatment for the patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be solely made by the clinician and the patient. CPIC assumes no responsibility for any injury to persons or damage to property related to any use of CPIC's guidelines, or for any errors or omissions.
Acknowledgments
Helpful comments were kindly provided by Nicola Dalbeth, Lisa K. Stamp, and Naoyuki Kamatani. We would also like to acknowledge the critical input of members of the Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network, funded by the National Institutes of Health (NIH). This work was funded by NIH grant GM61374.
M.S.H. is a co-inventor of pegloticase (Krystexxa) and is a consultant to, and receives royalties from, Savient Pharmaceuticals. M.T.M.L. is a paid consultant of YongLin Healthcare Foundation. The other authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Supplementary Material
References
- Horton R., et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- Robinson J., et al. The IMGT/HLA database Nucleic Acids Res. 37D1013–D1017.2009. 18838392 [Google Scholar]
- Hülsmeyer M., et al. Dual, HLA-B27 subtype-dependent conformation of a self-peptide. J. Exp. Med. 2004;199:271–281. doi: 10.1084/jem.20031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P., Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- Martin M.A., Klein T.E., Dong B.J., Pirmohamed M., Haas D.W., Kroetz D.L. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin. Pharmacol. Ther. 2012;91:734–738. doi: 10.1038/clpt.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.H., Hung S.I., Chen Y.T. Human leukocyte antigens and drug hypersensitivity. Curr. Opin. Allergy Clin. Immunol. 2007;7:317–323. doi: 10.1097/ACI.0b013e3282370c5f. [DOI] [PubMed] [Google Scholar]
- Pichler W.J., et al. Pharmacological interaction of drugs with immune receptors: the p-i concept. Allergol. Int. 2006;55:17–25. doi: 10.2332/allergolint.55.17. [DOI] [PubMed] [Google Scholar]
- Wei C.Y., Chung W.H., Huang H.W., Chen Y.T., Hung S.I. Direct interaction between HLA-B and carbamazepine activates T cells in patients with Stevens-Johnson syndrome. J. Allergy Clin. Immunol. 2012;129:1562–9.e5. doi: 10.1016/j.jaci.2011.12.990. [DOI] [PubMed] [Google Scholar]
- Illing P.T., et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- Chung W.H., et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428:486. doi: 10.1038/428486a. [DOI] [PubMed] [Google Scholar]
- Ozeki T., et al. Genome-wide association study identifies HLA-A*3101 allele as a genetic risk factor for carbamazepine-induced cutaneous adverse drug reactions in Japanese population. Hum. Mol. Genet. 2011;20:1034–1041. doi: 10.1093/hmg/ddq537. [DOI] [PubMed] [Google Scholar]
- McCormack M., et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N. Engl. J. Med. 2011;364:1134–1143. doi: 10.1056/NEJMoa1013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallal S., et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- Tohkin M., et al. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. Pharmacogenomics J. e-pub ahead of print 13 September 2011, doi:10.1038/tpj.2011.41. 2011. [DOI] [PubMed]
- Génin E., et al. Genome-wide association study of Stevens-Johnson Syndrome and toxic epidermal necrolysis in Europe. Orphanet J. Rare Dis. 2011;6:52. doi: 10.1186/1750-1172-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortmann R.L. Gout and Hyperuricemia. In: Kelley's Textbook of Rheumatology 8th edn. (eds. Firestein,G.S., Budd, R.C., Harris, E.D. Jr, McInnes, I.B., Ruddy, S.. & Sergent, J.S.) 1481–1503 (Saunders Elsevier, Philadelphia, PA, 2009).; [Google Scholar]
- Zhu Y., Pandya B.J., Choi H.K. Prevalence of gout and hyperuricemia in the US general population: the National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum. 2011;63:3136–3141. doi: 10.1002/art.30520. [DOI] [PubMed] [Google Scholar]
- Merriman T.R., Dalbeth N. The genetic basis of hyperuricaemia and gout. Joint Bone Spine. 2011;78:35–40. doi: 10.1016/j.jbspin.2010.02.027. [DOI] [PubMed] [Google Scholar]
- Chuang S.Y., Lee S.C., Hsieh Y.T., Pan W.H. Trends in hyperuricemia and gout prevalence: Nutrition and Health Survey in Taiwan from 1993–1996 to 2005–2008. Asia Pac. J. Clin. Nutr. 2011;20:301–308. [PubMed] [Google Scholar]
- Winnard D., et al. National prevalence of gout derived from administrative health data in Aotearoa New Zealand. Rheumatology (Oxford). 2012;51:901–909. doi: 10.1093/rheumatology/ker361. [DOI] [PubMed] [Google Scholar]
- Cea Soriano L., Rothenbacher D., Choi H.K., García Rodríguez L.A. Contemporary epidemiology of gout in the UK general population. Arthritis Res. Ther. 2011;13:R39. doi: 10.1186/ar3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primatesta P., Plana E., Rothenbacher D. Gout treatment and comorbidities: a retrospective cohort study in a large US managed care population. BMC Musculoskelet. Disord. 2011;12:103. doi: 10.1186/1471-2474-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J., Terkeltaub R. A critical reappraisal of allopurinol dosing, safety, and efficacy for hyperuricemia in gout. Curr. Rheumatol. Rep. 2009;11:135–140. doi: 10.1007/s11926-009-0019-z. [DOI] [PubMed] [Google Scholar]
- Jordan K.M., et al. British Society for Rheumatology and British Health Professionals in Rheumatology guideline for the management of gout. Rheumatology (Oxford). 2007;46:1372–1374. doi: 10.1093/rheumatology/kem056a. [DOI] [PubMed] [Google Scholar]
- Sarawate C.A., et al. Gout medication treatment patterns and adherence to standards of care from a managed care perspective. Mayo Clin. Proc. 2006;81:925–934. doi: 10.4065/81.7.925. [DOI] [PubMed] [Google Scholar]
- Dalbeth N., Stamp L. Allopurinol dosing in renal impairment: walking the tightrope between adequate urate lowering and adverse events. Semin. Dial. 2007;20:391–395. doi: 10.1111/j.1525-139X.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Stamp L.K., et al. Relationship between serum urate and plasma oxypurinol in the management of gout: determination of minimum plasma oxypurinol concentration to achieve a target serum urate level. Clin. Pharmacol. Ther. 2011;90:392–398. doi: 10.1038/clpt.2011.113. [DOI] [PubMed] [Google Scholar]
- Stamp L.K., Zhu X., Dalbeth N., Jordan S., Edwards N.L., Taylor W. Serum urate as a soluble biomarker in chronic gout-evidence that serum urate fulfills the OMERACT validation criteria for soluble biomarkers. Semin. Arthritis Rheum. 2011;40:483–500. doi: 10.1016/j.semarthrit.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Cacoub P., et al. The DRESS syndrome: a literature review. Am. J. Med. 2011;124:588–597. doi: 10.1016/j.amjmed.2011.01.017. [DOI] [PubMed] [Google Scholar]
- Halevy S., et al. Allopurinol is the most common cause of Stevens-Johnson syndrome and toxic epidermal necrolysis in Europe and Israel. J. Am. Acad. Dermatol. 2008;58:25–32. doi: 10.1016/j.jaad.2007.08.036. [DOI] [PubMed] [Google Scholar]
- Stamp L.K., et al. Starting dose is a risk factor for allopurinol hypersensitivity syndrome: a proposed safe starting dose of allopurinol. Arthritis Rheum. 2012;64:2529–2536. doi: 10.1002/art.34488. [DOI] [PubMed] [Google Scholar]
- Hung S.I., et al. HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc. Natl. Acad. Sci. U.S.A. 2005;102:4134–4139. doi: 10.1073/pnas.0409500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneeyakul W., et al. Strong association between HLA-B*5801 and allopurinol-induced Stevens-Johnson syndrome and toxic epidermal necrolysis in a Thai population. Pharmacogenet. Genomics. 2009;19:704–709. doi: 10.1097/FPC.0b013e328330a3b8. [DOI] [PubMed] [Google Scholar]
- Kang H.R., et al. Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenet. Genomics. 2011;21:303–307. doi: 10.1097/FPC.0b013e32834282b8. [DOI] [PubMed] [Google Scholar]
- Kaniwa N., et al. HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics. 2008;9:1617–1622. doi: 10.2217/14622416.9.11.1617. [DOI] [PubMed] [Google Scholar]
- Lonjou C., et al. A European study of HLA-B in Stevens-Johnson syndrome and toxic epidermal necrolysis related to five high-risk drugs. Pharmacogenet. Genomics. 2008;18:99–107. doi: 10.1097/FPC.0b013e3282f3ef9c. [DOI] [PubMed] [Google Scholar]
- Zineh I., et al. Allopurinol pharmacogenetics: assessment of potential clinical usefulness. Pharmacogenomics. 2011;12:1741–1749. doi: 10.2217/pgs.11.131. [DOI] [PubMed] [Google Scholar]
- Chohan S. Safety and efficacy of febuxostat treatment in subjects with gout and severe allopurinol adverse reactions. J. Rheumatol. 2011;38:1957–1959. doi: 10.3899/jrheum.110092. [DOI] [PubMed] [Google Scholar]
- Sundy J.S., et al. Efficacy and tolerability of pegloticase for the treatment of chronic gout in patients refractory to conventional treatment: two randomized controlled trials. JAMA. 2011;306:711–720. doi: 10.1001/jama.2011.1169. [DOI] [PubMed] [Google Scholar]
- Lee M., Chen Y., Shen C.A.A prospective study of HLA-B*5801 genotyping for the prevention of allopurinol induced severe cutaneous adverse reactionsAmerican Society for Clinical Pharmacology and Therapeutics 113th Annual Meeting, 12–17 March 2012, National Harbor, Maryland, Abstr. S107.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.