Abstract
Zinc finger nucleases (ZFN) can facilitate targeted gene addition to the genome while minimizing the risks of insertional mutagenesis. Here, we used a previously characterized ZFN pair targeting the chemokine (C-C motif) receptor 5 (CCR5) locus to introduce, as a proof of concept, the enhanced green fluorescent protein (eGFP) or the microdystrophin genes into human myoblasts. Using integrase-defective lentiviral vectors (IDLVs) and chimeric adenoviral vectors to transiently deliver template DNA and ZFN respectively, we achieved up to 40% targeted gene addition in human myoblasts. When the O6-methylguanine-DNA methyltransferaseP140K gene was co-introduced with eGFP, the frequency of cells with targeted integration could be increased to over 90% after drug selection. Importantly, gene-targeted myoblasts retained their mitogenic activity and potential to form myotubes both in vitro and in vivo when injected into the tibialis anterior of immune-deficient mice. Altogether, our results could lead to the development of improved cell therapy transplantation protocols for muscular diseases.
Keywords: dystrophin, gene addition, gene therapy, myoblast, zinc finger nuclease
Introduction
Genetic engineering represents an interesting strategy for the treatment of inherited disorders. Most current gene therapy protocols aiming to achieve long-term expression of a therapeutic transgene rely on integrating vectors such as retroviruses and lentiviruses. However, insertional mutagenesis associated with random integration of the transgene represents a real biosafety concern for which an alternative is needed.1
The use of zinc finger nucleases (ZFN) can greatly facilitate the integration of a transgene at a precise genomic locus.2,3,4,5 Indeed, ZFN can greatly stimulate homologous-directed recombination by creating a double strand break at a desired DNA sequence. In particular, a pair of ZFN was previously shown to recognize and disrupt with high specificity the major HIV-1 co-receptor, the chemokine (C-C motif) receptor 5 (CCR5), in human T cells and CD34+ cells.2,5,6 The CCR5 locus is considered a potential safe harbor for gene addition as humans carrying a homozygous 32-bp deletion within exon 3 of the CCR5 gene exhibit no deleterious phenotype.7,8,9 We previously showed that ZFN-driven targeted integration at the CCR5 locus of the erythropoietin gene can be achieved at high levels in human mesenchymal stem cells.10 Moreover, biologically relevant secreted levels of erythropoietin were observed after modified mesenchymal stem cell were transplanted in immune-deficient mice.10
Duchenne muscular dystrophy (DMD) is the most severe childhood muscular dystrophy. Various mutations into the dystrophin gene have been shown to be responsible for the disease, making DMD patients candidates for gene therapy.11 Dystrophin plays an important role in maintaining myofiber integrity and stability, the absence of a functional dystrophin protein in the myofibers makes them highly vulnerable to the damage during contractions leading to a continuous degeneration-regeneration process until the exhaustion of endogenous muscle stem cells. As a result, muscle tissue is progressively replaced by fat and connective tissue.12,13
Adeno-associated virus-mediated in vivo gene transfer of the microdystrophin gene, a truncated but functional form of the protein, was recently shown to be feasible in dystrophic dogs.14 Although promising, this strategy requires the in vivo administration of virus and immunosuppression of the host, procedures that can lead to multiple adverse effects.14,15,16 An alternative approach to in vivo gene transfer would be to use targeted integration to safely introduce the microdystrophin gene ex vivo into cultured myoblasts or muscle progenitor cells and then to transplant these cells into the patient.17,18 However, whether targeted gene transfer is feasible with high efficiency in human myoblasts is unknown.
Here, we demonstrate as a proof of concept that ZFN-mediated targeted gene addition into human myoblasts of the enhanced green fluorescent protein (eGFP) or microdystrophin genes at the CCR5 locus can be done at high efficiency. Moreover, we show that such efficiency can be easily increased by the in vitro selection of myoblasts following the co-addition of the mutated form of the O6-methylguanine-DNA methyltransferase (MGMTP140K) drug resistance gene. Finally, we also confirm that ZFN-modified myogenic cells preserve their potential to differentiate in vitro and in vivo upon intramuscular transplantation into immune-deficient mice.
Results
Targeted addition of the eGFP or microdystrophin genes into human myoblasts
We selected the CCR5 locus to carry out ZFN-mediated targeted gene addition in human myoblasts. This is justified by the fact that a pair of highly specific ZFN targeting this locus has already been developed and is currently being used in the clinic to disrupt the CCR5 locus in autologous CD4 T cells from HIV patients (clinicaltrials.gov identifier: NCT00842634). We have also previously shown that we can achieve adequate transgene expression from genes inserted into the endogenous CCR5 locus using the CCR5 ZFN.10 These two characteristics greatly increase the translational potential of our approach. Using non-integrating chimeric adenoviral vector (Ad5/F35) for the expression of ZFN and integrase-defective lentiviral vector (IDLV), carrying a phosphoglycerate kinase (PGK)–eGFP template DNA flanked by CCR5 homology arms (Figure 1a), we first determined the best vector ratio necessary to achieve targeted gene addition in human myoblasts (Figure 1b). We found that when myoblasts were simultaneously exposed to Ad5/F35 vectors at an multiplicity of infection of 2,000 and to 100 ng of p24 IDLV.GFP donor, we observed a high frequency (close to 40%) of targeted gene addition in absence of any drug selection (Figure 1c,d). Targeted gene addition was determined by flow cytometry at selected intervals up to 30 days post-transduction, the minimum time required to dilute out the residual eGFP expression from nonintegrated IDLV.
Figure 1.
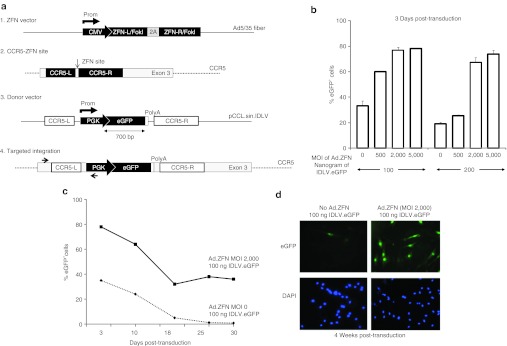
Targeted addition of the eGFP gene in human myoblasts. (a) Schematic representation of (1) Ad5/F35 chimeric adenoviral vector containing the expression cassette for the CCR5-specific ZFN, (2) the location of the ZFN target site in exon 3 of the endogenous CCR5 locus, (3) IDLV donor vector containing the PGK-eGFP expression cassette flanked by CCR5 homology arms, (4) the expected result after targeted gene addition at the CCR5 locus. PCR primers (black arrows) used for targeted integration analysis are indicated. (b) Levels of targeted addition obtained in human myoblasts as determined by flow cytometry on day 3 post-transduction with increasing doses of Ad5/F35 ZFN and IDLV.eGFP donor vector. Shown is the average and SD of three independent experiments. (c) Long-term profile of eGFP gene expression in myoblasts transduced with the optimal dose of donor DNA and Ad.ZFN vectors. Note the dilution over time of the eGFP expression in absence of Ad.ZFN. (d) Representative photographs showing the expression of eGFP in myoblasts 4 weeks following their transduction with the indicated vectors. Nuclei were stained with DAPI. Results are representative of three independent experiments. Original magnification: ×100. CMV, cytomegalovirus; DAPI, 4′,6-diamidino-2-phenylindole; eGFP, enhanced green fluorescent protein; IDLV, integrase-defective lentiviral vector; MOI, multiplicity of infection; PGK, phosphoglycerate kinase; ZFN, zinc finger nuclease.
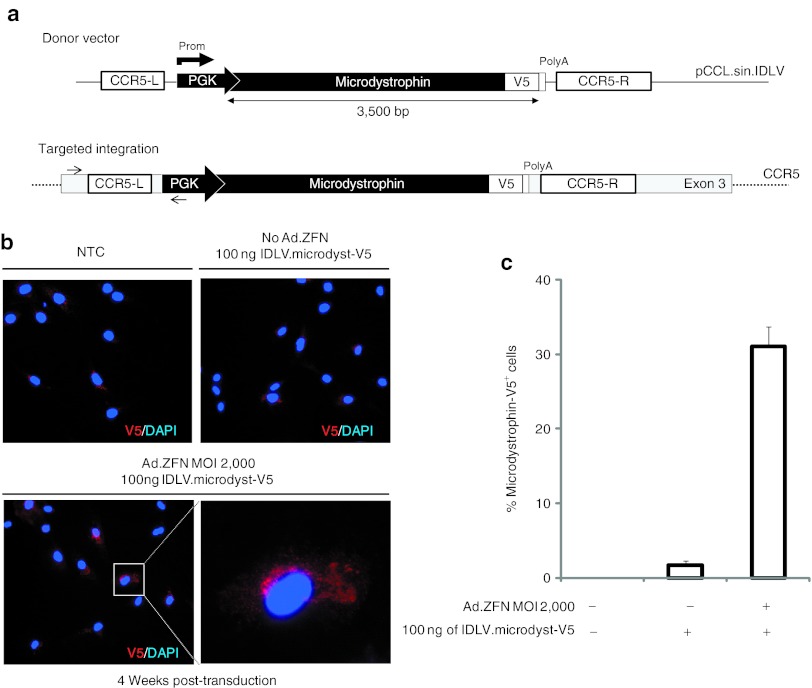
We next wanted to determine whether it was possible to introduce, with a similar efficiency, a DNA template containing the microdystrophin gene that is about five times the size of the eGFP gene (Figure 2a). Using the same optimized transduction protocol described above, we achieved up to 30% targeted addition of the microdystrophin gene into human myoblasts, a level obtained without any drug selection. Targeted integration was determined by immunocytochemistry 4 weeks post-transduction (Figure 2b,c). These results are in agreement with previous work by Moehle et al., who showed that ZFNs can drive targeted integration of a large DNA fragment (up to 8 kb) without a significant loss in recombination efficiency.3 Importantly, we also confirmed that targeted integration of both the eGFP and microdystrophin genes occurred at the CCR5 locus only in cells that were cotransduced with vectors delivering both ZFNs and the template DNA. This was established by PCR using specific primers recognizing the 5′ integration junction (Figure 3a). The specificity of the CCR5-ZFN was also determined by performing the surveyor nuclease Cel-1 assay at the CCR2 and ABLIM2 loci, the two most frequent off-target loci identified for this ZFN pair.19,20 As expected, and in opposition to the CCR5 locus where we could detect over 30% cleavage activity, no cleavage was observed at these off-target loci, confirming the specificity of the CCR5-ZFN in myoblasts (Figure 3b).
Figure 2.
Targeted addition of the microdystrophin gene in human myoblasts. (a) Top: Schematic of IDLV donor DNA vector containing the PGK-microdystrophin-V5 expression cassette flanked by CCR5 homology arms. Bottom: Schematic of the expected result after targeted gene addition at the CCR5 locus of the microdystrophin-V5 donor DNA. PCR primers (black arrows) used for targeted integration analysis are indicated. (b) Myoblasts were transduced with the indicated viral vectors and the expression of the microdystrophin-V5 was detected by immunofluorescence using a V5-specific antibody (in red). Nuclei were counterstained with DAPI. Myoblasts were immunostained 4 weeks post-transduction to allow for the dilution of the IDLV vectors. Original magnification: ×200. (c) Quantitative representation of the levels of microdystrophin-V5 gene-targeted myoblasts 4 weeks post-transduction. The proportion of V5-positive cells was determined by counting manually a total of 300 treated cells in randomly selected fields. Results are representative of three independent experiments. DAPI, 4′,6-diamidino-2-phenylindole; IDLV, integrase-defective lentiviral vector; MOI, multiplicity of infection; NTC, non-transduced cells; PGK, phosphoglycerate kinase; ZFN, zinc finger nuclease.
Figure 3.
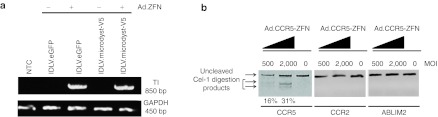
Targeted integration of the eGFP and the microdystrophin genes at the CCR5 locus. (a) Myoblasts were transduced with the indicated vectors and expanded in vitro for 4 weeks before genomic DNA was collected. Targeted integration (TI) was determined by PCR using a set of primers specific for the 5′ integration junction (as shown in Figures 1a and 2a). The bottom gel shows the amplification of the GAPDH gene used as an internal control. (b) Myoblasts were transduced as described above using the indicated vector concentrations and evidence for ZFN-targeted gene disruption at the CCR5, CCR2, and ABLIM2 loci determined by the Cel-1 surveyor assay. eGFP, enhanced green fluorescent protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IDLV, integrase-defective lentiviral vector; MOI, multiplicity of infection; NTC, non-transduced cells; ZFN, zinc finger nuclease.
Targeted integration of the MGMTP140K drug resistance gene allows for the in vitro selective enrichment of modified myoblasts
To maximize the chance of success of cell therapy, it certainly would be advantageous to obtain a population of myoblasts that would be nearly if not fully gene-modified. Hence, despite the remarkably high level of targeted gene addition we achieved in myoblasts in absence of drug selection, we explored the possibility of co-targeting the O6-methylguanine-DNA methyltransferaseP140K (MGMTP140K) gene, which expression can confer a selective growth advantage both in vitro and in vivo.21,22,23 In brief, cells which express the MGMTp140K gene, have a selective growth advantage when exposed to the wild-type MGMT inhibitor O6-benzylguanine (BG) in combination with low dose of 1,3-bis (2-chloroethyl)-N-nitrosourea (BCNU).
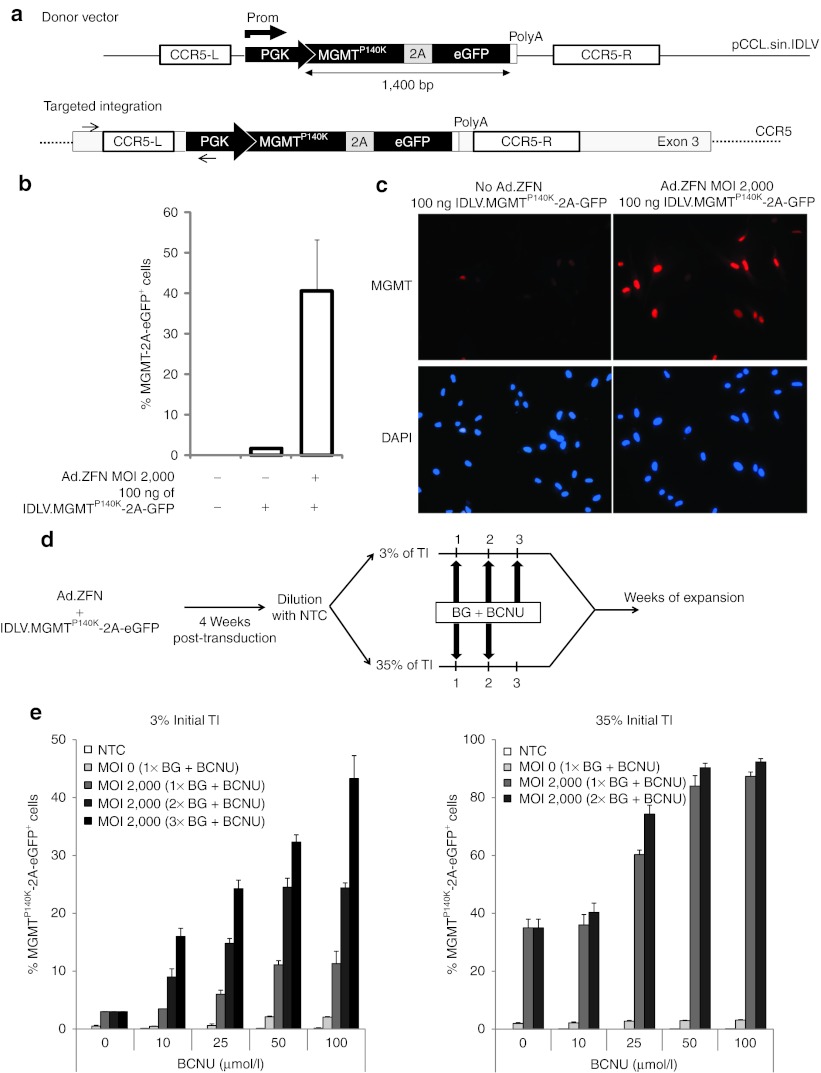
Consequently, we next verified whether ZFN-based expression of the MGMTP140K, along with the eGFP reporter gene to facilitate tracking of transduced cells, was sufficient to confer myoblasts protection against treatments with BG and BCNU (Figure 4a). Using IDLV to deliver a MGMTP140K–eGFP template DNA flanked by CCR5 homology arms, we obtained 40–50% gene addition in myoblasts in absence of drug selection (Figure 4b,c). However, to fully evaluate the selection potential of the BG/BCNU combination, we thought it would be more appropriate to work with a population containing a much lower level of gene addition. As such, we generated two populations containing either 3 or 35% MGMTP140K-2A-GFP gene-targeted myoblasts using non-transduced cells for dilution. Both populations were exposed to 20 µmol/l BG for 1 hour followed by increasing doses of BCNU (10, 25, 50, and 100 µmol/l) for 2 hours (Figure 4d). Myoblasts were then allowed to recover for 1 week after which the proportion of eGFP-positive cells was analyzed by flow cytometry, a procedure defined as one cycle of drug selection. Our results demonstrate that the proportion of MGMTP140K-2A-GFP–expressing cells increased from 3 to 11% following only one cycle using 100 µmol/l BCNU (Figure 4d). After three cycles of selection using the same concentration of BCNU, up to 45% of the myoblasts were positive for eGFP, representing a 15-fold enrichment over the initial population. However, additional cycle of selection did not allow us to further increase the proportion of eGFP-positive cells, suggesting acquired resistance of non-modified cells against BCNU toxicity. In comparison, when starting with a population containing already 35% of gene-targeted myoblasts, over 85% of the cells were expressing eGFP after only one cycle of selection using 50 µmol/l of BCNU (Figure 4d). Altogether these results suggest that the efficiency of ZFN-mediated gene addition at the CCR5 locus, when combined with one round in vitro drug selection, is similar to the one obtained using a vesicular stomatitis virus-G–pseudotyped lentiviral integrating vector (data not shown).
Figure 4.
Selective in vitro enrichment of gene-targeted myoblasts using the MGMTP140K drug resistance gene. (a) Top: Schematic of IDLV donor DNA vector containing the PGK-MGMTP140K-2A-GFP expression cassette flanked by CCR5 homology arms. Bottom: Schematic of the expected result after targeted gene addition at the CCR5 locus of the MGMTP140K-2A-GFP donor DNA. (b) Histogram showing the levels of targeted MGMTP140K-2A-GFP gene expression obtained in human myoblasts transduced with the indicating doses of vectors as determined by flow cytometry 4 weeks post-transduction. Shown are the average and SD of three independent experiments. (c) Representative photographs showing MGMTP140K-2A-GFP expression as detected by immunofluorescence using an anti-mouse MGMT-specific antibody (in red) 4 weeks post-transduction of myoblasts to allow dilution of the IDLV vectors. Nuclei were stained with DAPI. Original magnification: ×200. (d) Schematic of the in vitro drug selection procedure using two populations of myoblasts containing 3 or 35% of MGMTP140K-2A-GFP gene targeted integration (TI). (e) Left and right panels show the enrichment profile starting with respectively 3 or 35% of MGMTP140K-2A-GFP–positive cells following the indicated number of BG and BCNU drug selection cycles. Each value is a mean of three cultures. BCNU, 1,3-bis (2-chloroethyl)-N-nitrosourea; BG, O6-benzylguanine; DAPI, 4′,6-diamidino-2-phenylindole; eGFP, enhanced green fluorescent protein; IDLV, integrase-defective lentiviral vector; MOI, multiplicity of infection; NTC, non-transduced cells; PGK, phosphoglycerate kinase; ZFN, zinc finger nuclease.
ZFN-driven gene addition mediates no toxicity in human myoblasts
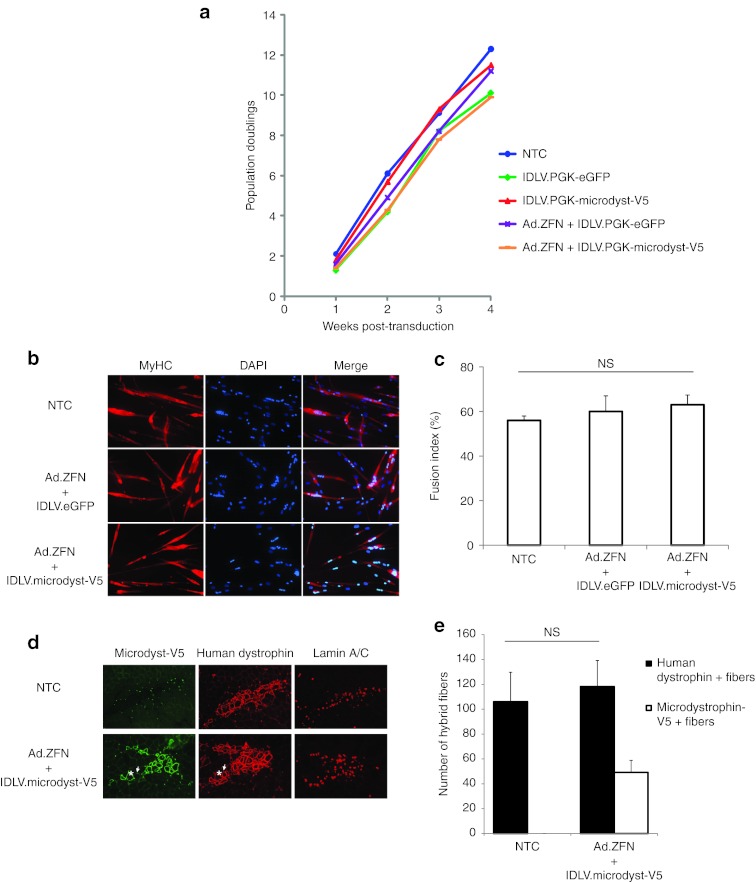
We next wanted to verify the impact that the cotransduction (Ad5/35 and IDLV) and subsequent transient ZFN expression may have on the growth and function of myoblasts. We first found that when passaged every 3 days for up to 4 weeks post-transduction, that gene-targeted myoblasts experience very little cell toxicity based on the kinetics of their population doublings (Figure 5a). Modified myoblasts were also placed for 5 days in medium containing 2% serum and their ability to fuse in vitro evaluated by immunocytochemistry based on the expression of the muscle heavy chain myosin (MyHC) (Figure 5b). As for cell division, we found ZFN-mediated gene addition did not interfere with MyHC expression and fusion potential of myoblasts in vitro (Figure 5c).
Figure 5.
ZFN-mediated gene addition does not compromise the growth and fusion potential of human myoblasts in vitro and in vivo. (a) Shown is the proliferation of myoblasts expressed in terms of population doublings over time following their transduction or not with the indicated vectors. (b) Representative photographs of the fusion potential of myoblasts evaluated in vitro 4 weeks post-transduction. Myotube formation was determined by staining myoblasts for the expression of myosin heavy chain (MyHC) 5 days post-induction of differentiation using 2% serum containing medium. Nuclei were stained with DAPI. Original magnification: ×100. (c) Quantification of the fusion potential of myoblasts' populations shown inb as determined by counting the proportion of nuclei forming MyHC-positive myotubes over the total number of nuclei from randomly selected fields. (d) Tibialis anterior muscles of immune-deficient NSG mice were transplanted wi th either non-transduced or microdystrophin-V5 gene-targeted myoblasts and muscle fibers formation evaluated 4 weeks later. Human fibers were detected by immunofluorescence on successive cryosections using an anti-human dystrophin antibody (in red) and gene-targeted fibers identified using an anti-V5 antibody (in green). Human nuclei were also stained with an anti-Lamin A/C antibody (in red). The asterisks show a fiber positive for both the full-length dystrophin and the microdystrophin gene. The arrows show a fiber positive only for the full-length dystrophin. (e) Quantification of the number of fibers formed in vivo (as shown in d). Fibers were counted from muscle cryosections collected from n = 6 transplanted muscles for each group. eGFP, enhanced green fluorescent protein; IDLV, integrase-defective lentiviral vector; NS, not significant; NTC, non-transduced cells; PGK, phosphoglycerate kinase; ZFN, zinc finger nuclease.
Engraftment of gene-targeted human myoblasts into the muscle of immune-deficient mice
To fully assess the potential for cell therapy of gene-targeted myoblasts, we also measured their capacity to form muscle fibers in vivo. We found that ZFN-modified myoblasts that express the microdystrophin gene were as competent as their non-modified counterparts in forming muscle fibers when transplanted in the tibialis anterior muscle of immune-deficient NSG mice (Figure 5d,e). Myoblasts engraftment was determined by immunohistochemistry performed on muscle cryosections stained with an antibody against the V5 epitope, which was fused at the 3′ end of the microdystrophin gene24 (Figure 2a). Indeed, we found the ratio of V5-positive fibers over the total number of fibers formed (as detected by staining for the full-length human dystrophin) to be similar to the level of targeted gene addition observed before the transplantation (Figure 5e). Altogether, these results suggest that ZFN-mediated gene addition does not interfere with the functionality of myoblasts in vitro and in vivo.
Discussion
We and others have shown that ZFN-driven genetic modification can be achieved in multiple cell types, including mesenchymal, hematopoietic, and embryonic stem cells.2,5,10,25,26 Here, we found that ZFN-mediated targeted integration of the microdystrophin gene can be performed ex vivo in human myoblasts at a sufficiently high frequency to be considered a valid approach for the treatment of myogenic diseases. Indeed, using a CCR5-specific ZFN pair, our results showed that over 40% targeted gene addition can be obtained in human myoblasts in the absence of any apparent cytotoxicity. A major property of myoblasts is their capacity to fuse with each other or with existing fibers to regenerate damaged muscles. As such, our results showed that the fusion potential of ZFN-targeted myoblasts is not altered in vitro or in vivo following transplantation in mice.
Lee and colleagues recently showed that myoblasts can also be selectively enriched in vitro following retrovirus-induced expression of the MGMTP140K gene.21 Interestingly, we could increase the proportion of MGMTP140K-targeted myoblasts to about 90% following in vitro selection using the combination of BCNU and BG drugs. However, our data also showed that sufficiently high level of gene addition needs to be obtained initially in order to reach this plateau. This may be explained by the rapid emergence of resistance to alkylating drugs as a result of increased expression of the endogenous MGMT gene in non-modified myoblasts, as reported in many other cell types.21,27
An important observation from our work is that the constitutive expression of the microdystrophin gene under the control of the ubiquitous PGK promoter was not toxic to human myoblasts, making the use of a muscle-specific promoter unnecessary for the in vitro expansion of modified myoblasts. This is likely explained by the fact that in ZFN-modified cells limited expression of the microdystrophin gene is observed from a single-targeted locus. In contrast, when lentiviral vectors are used to randomly insert the microdystrophin gene, microdystrophin expression is likely higher and toxic to mononuclear cells due to insertion of multiple copies of the gene.28,29
DMD arises from numerous distinct mutations located in various exons of the dystrophin gene. The design of ZFN pairs specific for the dystrophin gene could theoretically allow for the genetic correction of each individual mutation, keeping the expression of the dystrophin under its endogenous promoter. However, this strategy would require the development of several ZFN pairs to account for the diversity of mutations found in the patient population. On the other hand, targeted gene addition at the CCR5 locus could offer a unique solution to DMD patients. This is particularly attractive in the context of previous reports showing that a mutated form of the dystrophin does not act as dominant negative in presence of a functional protein.30,31
The specificity of the CCR5-ZFN pair used in this study was rigorously evaluated in independent studies, which showed it has minimal off-target cleavage activity.5,20,32 As such, we cannot rule out that low level nonspecific integration of the microdystrophin gene occurred within our mixed population of transduced myoblasts. Moreover, we observed low level (about 2%) long-term expression of the eGFP or MGMTP140K-2A-eGFP cassette in myoblasts transduced with IDLV vectors in the absence of ZFN exposure, suggesting random integration of the template DNA is inevitable in a mixed population of cells where episomal DNA is introduced. Nonetheless, we believe the use of ZFN-mediated targeted gene addition to favor specific integration of the transgene represents a great alternative to the random integration obtained with classical retroviral and lentiviral vectors gene delivery systems.
A major limitation of autologous myoblasts transplantation for the treatment of DMD is that dystrophic myoblasts cannot be largely expanded ex vivo as they become rapidly senescent.13 We believe this could be eventually overcome given that we and others recently demonstrated that induced pluripotent stem cells can be efficiently differentiated into functional myogenic progenitors.33,34,35 As such, a strategy where DMD patient fibroblasts would undergo (i) targeted integration of the dystrophin gene, (ii) differentiation into induced pluripotent stem cells clones, (iii) whole genome sequencing, and (iv) reprogramming into myogenic progenitors; could offer great promises for autologous cell therapy of DMD. Moreover, as this strategy would theoretically only require the isolation of a single clone with a validated targeted integration of the dystrophin gene, this would likely make it possible to achieve ZFN-driven targeted integration of the full-length dystrophin gene. Indeed, the size of the dystrophin gene (14 kb) exceeds the encapsidation limit of IDLV vectors used in this study. However, in a scenario where the efficiency of homologous recombination would not be an issue, a large DNA fragment could be delivered to fibroblasts or myoblasts derived from DMD patients using standard transfection procedures as we have shown before.29
In conclusion, our results show that ZFN-driven targeted addition of the microdystrophin gene in human myoblasts can be achieved with high efficiency, and that this manipulation does not affect their regenerative potential in skeletal muscle. Our data provide further evidence of the potential of genome-editing technologies for the treatment of genetic diseases.
Materials and Methods
Mice. NOD/LtSz-scid/IL2rγ−/− (NSG) mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in the animal care facility at the CHU Sainte-Justine Research Centre under pathogen-free conditions in sterile ventilated racks. All in vivo manipulations were previously approved by the institutional committee for good laboratory practices for animal research (GLPAR) (protocol no. S10-32).
Vectors and lentivirus preparation. ZFNs targeting exon 3 of the human CCR5 gene were previously described.2,5 A ZFN-expressing virus was generated from chimeric human adenovirus 5 vectors containing fiber shaft and knob domains of human species B adenovirus (Ad5/F35).5 IDLV carrying GFP, microdystrophin-V524 or MGMTP140K-2A-GFP donor cassettes were generated from the HIV-derived self-inactivating third-generation transfer construct pCCLsin.cPPT.hPGK.X.BGHpA using an integrase-defective packaging plasmid.2,36 IDLV stocks were prepared as described elsewhere.2,36 Lentiviral particles were quantified upon concentration by ultracentrifugation by HIV-1 Gag p24 Antigen ELISA (ZeptoMetrix, Franklin, MA). Yield ranged from 5 to 20 ng p24/µl, depending on the vector type.
Human myoblast culture and transduction. Human myoblasts were obtained from a postmortem muscle biopsy of a normal 13-month-old male and proliferated in MB-1 medium (Hyclone – X) supplemented with 15% fetal bovine serum (Wisent, Saint-Bruno, Québec, Canada), 1% penicillin–streptomycin (Wisent), 10 µg/l of bFGF (R&D systems, Burlington, Ontario, Canada), 0.4 mg/l of dexamethasone (Sigma, Oakville, Ontario, Canada), and 5 mg/l of insulin (Sigma, St Louis, MO). Myoblasts were exposed simultaneously to Ad5/F35-ZFN (multiplicity of infection range from 500 to 2,000) and IDLV template DNA vectors (100 or 200 ng of p24 per 105 cells) for 18 hours, a procedure that was repeated twice. Transduced cells were then expanded in growth conditions and passaged every 4 days until analysis. For differentiation, cells were expanded in Dulbecco's modified Eagle's medium medium supplemented with 2% fetal bovine serum and antibiotics for 5 days, myotubes were first fixed in 4% paraformaldehyde for 15 minutes, permeabilized three times for 15 minutes with 3% triton X-100 in phosphate-buffered saline (PBS) and then immunostained against the mouse anti-MyHC using the MF20 anti-mouse MyHC antibody at a dilution of (1:100) for 2 hours (Developmental Studies Hybridoma Bank, University of Iowa) and subsequently with an anti-mouse ALEXA fluor 594 at a dilution of (1:300) for 1 hour (Invitrogen, Burlington, Ontario, Canada). Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole). The fusion index (defined as the number of DAPI-stained nuclei inside MyHC+ myotubes in a given field divided by the total number of DAPI-stained nuclei in the same field) of each condition was calculated. The assay was done three times.
Cel-1 assay. The ability of the CCR5-ZFN pair to cut the CCR5 locus was verified using the surveyor nuclease (Cel-1; Transgenomic, Omaha, NE), a nucleotide mismatch selective endonuclease able to detect the presence of mutant clones. Briefly the mismatch assay consists of amplifying the target region from ZFN-treated genomic DNA via standard PCR using the following specific primers (CCR5 Cel-1 forward primer: 5′-AAGATGGATTATCAAGTGTCAAGTCC-3′ CCR5 Cel-1 reverse primer: 5′-CAAAGTCCCACTGGGCG-3′ CCR2 Cel-1 forward primer: 5′-CCACATCTCGTTCTCGGTTTATC-3′ CCR2 Cel-1 reverse primer: 5′-CGCCAAAATAACCGATGTG-3′ and ABLIM2 Cel-1 forward primer: 5′-CGATGACTCTGAGGTCTA CTCG-3′ ABLIM2 Cel-1 reverse primer: 5′-CAAGTGAACACAT GGTTTGCAG-3′) as described.2,3,4,5 PCR products are denatured and allowed to re-anneal. The mismatch sensitive enzyme cuts DNA at the sites where heterogeneous mismatches occur. Reactions are resolved by gel electrophoresis. The presence of digested PCR products indicates ZFN-mediated mutagenesis due to ZFN activity. The assay is sensitive enough to detect single-nucleotide changes and has a linear detection range between 0.69 and 44%.5
Analysis of targeted gene addition efficacy. Efficacy of eGFP gene-targeted integration was determined at days 3, 10, 18, 25, and 30 days post-transduction by immunofluorescence using an Olympus BX51 epifluorescent microscope and by flow cytometry (LSRFortessa cell analyzer; BD, Mississauga, Ontario, Canada). Non-viable cells were identified using 7-aminoactinomycin D (7-AAD) and were excluded from the analysis. Microdystrophin-V5 expression was detected by immunofluorescence using an anti-V5 antibody (1:5,000; Invitrogen) 1 month after transduction. Targeted integration at the CCR5 locus was determined by PCR using 100 ng of genomic DNA with a set of primers against the 5′ junction (forward CCR5 primer: 5′-TTGGAGGGGTGAGGTGAGAGG-3′, reverse hPGK primer: 5′-TGAAGAATGTGCGAGACCCAGG-3′) as follows: 94 °C for 10 minutes, then 30 cycles of 94 °C for 1 minute, 60 °C for 30 seconds, and 72 °C for 1 minute, followed by extension at 72 °C for 10 minutes. The expected amplicon length is 815 bp.2
In vitro selection of myoblasts using the BCNU/BG combination. Four weeks post-transduction, myoblasts containing gene addition of the MGMTP140K-2A-GFP cassette were treated with 50 µmol/l O6BG for 2 hours followed by increasing doses (range from 10–100 µmol/l) of BCNU for 1 hour. Cells were then washed with PBS and allowed to proliferate in the presence of 50 µmol/l O6BG for 7 days before their analysis by fluorescence-activated cell sorting. Where indicated, this procedure was repeated up to two more times. Cells were maintained in the presence of O6BG during their post-treatment expansion to inactivate endogenous MGMT gene.
In vivo transplantation of human myoblasts. Twelve weeks old NSG immune-deficient mice were irradiated at a dose of 9 Gy (1 Gy per minute) using a Faxitron model CP160 at the leg level to inhibit the proliferation of the recipient satellite cells, a condition that favors the participation of grafted cells in recipient muscle regeneration. The day of the transplantation, cultured myoblasts were detached from the flasks using 0.25% Trypsin-EDTA solution and washed twice with PBS. A total of 1 × 106 myoblasts resuspended in 20 µl of PBS containing 10 µg/ml cardiotoxin (Sigma), were implanted in each tibialis anterior through about 20 percutaneous microinjections. The grafted tibialis anterior muscles were harvested 4 weeks after transplantation, embedded in OCT and snap-frozen in liquid nitrogen.
Immune detection of hybrid fibers in muscle sections. Muscle frozen sections (12 µm) were first washed with PBS and nonspecific binding was blocked by incubating the sections with PBS containing 10% of fetal bovine serum for 1 hour. Immunofluorescence to detect human nuclei was performed with a mouse anti-human Lamin A/C antibody (1:100 for 2 hours; Vector Laboratories, Burlington, Ontario, Canada). Incubation with the primary antibody was followed by incubation with an anti-mouse ALEXA fluor 549 (1:300 for 1 hour; Invitrogen). Myofibers expressing the microdystrophin-V5 protein were detected using a mouse anti-V5 antibody (1:200 overnight; Invitrogen) and an anti-mouse ALEXA fluor 488 (1:300 for 1 hour; Invitrogen). All sections were mounted using the Vectashield (Vector Laboratories) mounting medium to prevent loss of fluorescence.
Statistical analysis. All data are expressed as means ± SEM and are representative of at least three separate experiments. The statistical significance of the difference between groups was determined by Student's t-test using GraphPad software (La Jolla, CA). A value of P < 0.05 was considered significant.
Acknowledgments
This work was supported by grants from the ThéCell – FRQS network to C.M.B. and J.P.T. B.F.B. has been supported by a post-doctoral fellowship from the FRQS and C.M.B. is currently supported by a scientist award by the FRQS. We also are grateful to Dr Fairbairn (Paterson Institute for Cancer research, UK) for providing the MGMTP140K cDNA. M.C.H. is an employee of Sangamo Biosciences Inc. The other authors declared no conflict of interest.
References
- Biasco L, Baricordi C., and, Aiuti A. Retroviral integrations in gene therapy trials. Mol Ther. 2012;20:709–716. doi: 10.1038/mt.2011.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA.et al. (2007Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery Nat Biotechnol 251298–1306. [DOI] [PubMed] [Google Scholar]
- Moehle EA, Moehle EA, Rock JM, Rock JM, Lee YL, Lee YL.et al. (2007Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases Proc Natl Acad Sci USA 1043055–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S.et al. (2005Highly efficient endogenous human gene correction using designed zinc-finger nucleases Nature 435646–651. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O.et al. (2008Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases Nat Biotechnol 26808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V.et al. (2010Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo Nat Biotechnol 28839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM.et al. (1996Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene Nature 382722–725. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF.et al. (2011Site-specific integration and tailoring of cassette design for sustainable gene transfer Nat Methods 8861–869. [DOI] [PubMed] [Google Scholar]
- Sadelain M, Papapetrou EP., and, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- Benabdallah BF, Allard E, Yao S, Friedman G, Gregory PD, Eliopoulos N.et al. (2010Targeted gene addition to human mesenchymal stromal cells as a cell-based plasma-soluble protein delivery platform Cytotherapy 12394–399. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Brown RH., and, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- Petrof BJ. Molecular pathophysiology of myofiber injury in deficiencies of the dystrophin-glycoprotein complex. Am J Phys Med Rehabil. 2002;81 suppl. 11:S162–S174. doi: 10.1097/00002060-200211001-00017. [DOI] [PubMed] [Google Scholar]
- Blau HM, Webster C., and, Pavlath GK. Defective myoblasts identified in Duchenne muscular dystrophy. Proc Natl Acad Sci USA. 1983;80:4856–4860. doi: 10.1073/pnas.80.15.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Storb R, Halbert CL, Banks GB, Butts TM, Finn EE.et al. (2012Successful regional delivery and long-term expression of a dystrophin gene in canine muscular dystrophy: a preclinical model for human therapies Mol Ther 201501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS.et al. (2007Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression Mol Ther 151160–1166. [DOI] [PubMed] [Google Scholar]
- Yuasa K, Sakamoto M, Miyagoe-Suzuki Y, Tanouchi A, Yamamoto H, Li J.et al. (2002Adeno-associated virus vector-mediated gene transfer into dystrophin-deficient skeletal muscles evokes enhanced immune response against the transgene product Gene Ther 91576–1588. [DOI] [PubMed] [Google Scholar]
- Skuk D, Goulet M, Roy B, Chapdelaine P, Bouchard JP, Roy R.et al. (2006Dystrophin expression in muscles of duchenne muscular dystrophy patients after high-density injections of normal myogenic cells J Neuropathol Exp Neurol 65371–386. [DOI] [PubMed] [Google Scholar]
- Skuk D, Roy B, Goulet M, Chapdelaine P, Bouchard JP, Roy R.et al. (2004Dystrophin expression in myofibers of Duchenne muscular dystrophy patients following intramuscular injections of normal myogenic cells Mol Ther 9475–482. [DOI] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C.et al. (2011An unbiased genome-wide analysis of zinc-finger nuclease specificity Nat Biotechnol 29816–823. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK., and, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kahatapitiya P, Kramer B, Joya JE, Hook J, Liu R.et al. (2009Methylguanine DNA methyltransferase-mediated drug resistance-based selective enrichment and engraftment of transplanted stem cells in skeletal muscle Stem Cells 271098–1108. [DOI] [PubMed] [Google Scholar]
- Jansen M, Sorg UR, Ragg S, Flasshove M, Seeber S, Williams DA.et al. (2002Hematoprotection and enrichment of transduced cells in vivo after gene transfer of MGMT(P140K) into hematopoietic stem cells Cancer Gene Ther 9737–746. [DOI] [PubMed] [Google Scholar]
- Lee K, Gerson SL, Maitra B., and, Koç ON. G156A MGMT-transduced human mesenchymal stem cells can be selectively enriched by O6-benzylguanine and BCNU. J Hematother Stem Cell Res. 2001;10:691–701. doi: 10.1089/152581601753193913. [DOI] [PubMed] [Google Scholar]
- Pichavant C, Chapdelaine P, Cerri DG, Dominique JC, Quenneville SP, Skuk D.et al. (2010Expression of dog microdystrophin in mouse and dog muscles by gene therapy Mol Ther 181002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT.et al. (2011In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases Stem Cells 291717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R.et al. (2011Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations Cell 146318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobola MS, Tseng SH, Blank A, Berger MS., and, Silber JR. Role of O6-methylguanine-DNA methyltransferase in resistance of human brain tumor cell lines to the clinically relevant methylating agents temozolomide and streptozotocin. Clin Cancer Res. 1996;2:735–741. [PubMed] [Google Scholar]
- Quenneville SP, Chapdelaine P, Skuk D, Paradis M, Goulet M, Rousseau J.et al. (2007Autologous transplantation of muscle precursor cells modified with a lentivirus for muscular dystrophy: human cells and primate models Mol Ther 15431–438. [DOI] [PubMed] [Google Scholar]
- Quenneville SP, Chapdelaine P, Rousseau J, Beaulieu J, Caron NJ, Skuk D.et al. (2004Nucleofection of muscle-derived stem cells and myoblasts with phiC31 integrase: stable expression of a full-length-dystrophin fusion gene by human myoblasts Mol Ther 10679–687. [DOI] [PubMed] [Google Scholar]
- Warner LE, DelloRusso C, Crawford RW, Rybakova IN, Patel JR, Ervasti JM.et al. (2002Expression of Dp260 in muscle tethers the actin cytoskeleton to the dystrophin-glycoprotein complex and partially prevents dystrophy Hum Mol Genet 111095–1105. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Crawford RW, DelloRusso C., and, Chamberlain JS. Spectrin-like repeats from dystrophin and alpha-actinin-2 are not functionally interchangeable. Hum Mol Genet. 2002;11:1807–1815. doi: 10.1093/hmg/11.16.1807. [DOI] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C.et al. (2011An unbiased genome-wide analysis of zinc-finger nuclease specificity Nat Biotechnol 29816–823. [DOI] [PubMed] [Google Scholar]
- Salani S, Donadoni C, Rizzo F, Bresolin N, Comi GP., and, Corti S. Generation of skeletal muscle cells from embryonic and induced pluripotent stem cells as an in vitro model and for therapy of muscular dystrophies. J Cell Mol Med. 2012;16:1353–1364. doi: 10.1111/j.1582-4934.2011.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darabi R, Pan W, Bosnakovski D, Baik J, Kyba M., and, Perlingeiro RC. Functional myogenic engraftment from mouse iPS cells. Stem Cell Rev. 2011;7:948–957. doi: 10.1007/s12015-011-9258-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudenege S, Lebel C, Huot NB, Dufour C, Fujii I, Gekas J.et al. (2012Successful transplantation in muscles of myoblasts derived from normal hESCs and hiPSCs obtained from a DMD patient Mol Ther 202153–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ.et al. (2006Effective gene therapy with nonintegrating lentiviral vectors Nat Med 12348–353. [DOI] [PubMed] [Google Scholar]